

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.6
(780 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
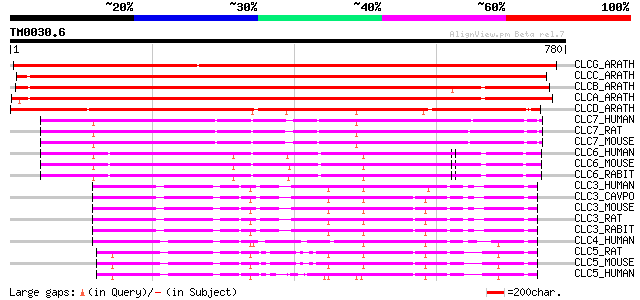
Sequences producing significant alignments: (bits) Value
CLCG_ARATH (P60300) Putative chloride channel-like protein CLC-G 1148 0.0
CLCC_ARATH (Q96282) Chloride channel protein CLC-c (AtCLC-c) 946 0.0
CLCB_ARATH (P92942) Chloride channel protein CLC-b (AtCLC-b) 822 0.0
CLCA_ARATH (P92941) Chloride channel protein CLC-a (AtCLC-a) 815 0.0
CLCD_ARATH (P92943) Chloride channel protein CLC-d (AtCLC-d) 640 0.0
CLC7_HUMAN (P51798) Chloride channel protein 7 (ClC-7) 404 e-112
CLC7_RAT (P51799) Chloride channel protein 7 (ClC-7) 402 e-111
CLC7_MOUSE (O70496) Chloride channel protein 7 (ClC-7) 402 e-111
CLC6_HUMAN (P51797) Chloride channel protein 6 (ClC-6) 328 3e-89
CLC6_MOUSE (O35454) Chloride channel protein 6 (ClC-6) 326 1e-88
CLC6_RABIT (Q9TT16) Chloride channel protein 6 (ClC-6) 320 1e-86
CLC3_HUMAN (P51790) Chloride channel protein 3 (ClC-3) 197 7e-50
CLC3_CAVPO (Q9R279) Chloride channel protein 3 (ClC-3) 197 7e-50
CLC3_MOUSE (P51791) Chloride channel protein 3 (ClC-3) 197 9e-50
CLC3_RAT (P51792) Chloride channel protein 3 (ClC-3) 197 1e-49
CLC3_RABIT (O18894) Chloride channel protein 3 (ClC-3) 197 1e-49
CLC4_HUMAN (P51793) Chloride channel protein 4 (ClC-4) 192 3e-48
CLC5_RAT (P51796) Chloride channel protein 5 (ClC-5) 191 5e-48
CLC5_MOUSE (Q9WVD4) Chloride channel protein 5 (ClC-5) 191 6e-48
CLC5_HUMAN (P51795) Chloride channel protein 5 (ClC-5) 187 7e-47
>CLCG_ARATH (P60300) Putative chloride channel-like protein CLC-G
Length = 763
Score = 1148 bits (2970), Expect = 0.0
Identities = 562/763 (73%), Positives = 654/763 (85%), Gaps = 2/763 (0%)
Query: 6 LSNGDSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGS 65
+ N +E + PLL S R NS+SQVAIVGANVCPIESLDYEI EN+FFKQDWR R
Sbjct: 1 MPNSTTEDSVAVPLLPSLRRATNSTSQVAIVGANVCPIESLDYEIAENDFFKQDWRGRSK 60
Query: 66 LQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFF 125
++IFQYV MKWLLCF IG I+ LIGF NNLAVENLAGVKFVVTSNMM+ RF++ FV F
Sbjct: 61 VEIFQYVFMKWLLCFCIGIIVSLIGFANNLAVENLAGVKFVVTSNMMIAGRFAMGFVVFS 120
Query: 126 ASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSS 185
+NL LTLFA++ITA +APAAAGSGI EVKAYLNGVDAP IF+L TLI+KIIG+I+AVS+
Sbjct: 121 VTNLILTLFASVITAFVAPAAAGSGIPEVKAYLNGVDAPEIFSLRTLIIKIIGNISAVSA 180
Query: 186 SLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAA 245
SL IGKAGPM+HTGACVA++LGQGGSKRY LTW+WLR+FKNDRDRRDL+ CG+AAGIAA+
Sbjct: 181 SLLIGKAGPMVHTGACVASILGQGGSKRYRLTWRWLRFFKNDRDRRDLVTCGAAAGIAAS 240
Query: 246 FRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIM 305
FRAPVGGVLFALEEM+SW +ALLWR FF+TA+VAI LRA+IDVCLSGKCGLFGKGGLIM
Sbjct: 241 FRAPVGGVLFALEEMSSW--SALLWRIFFSTAVVAIVLRALIDVCLSGKCGLFGKGGLIM 298
Query: 306 FDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACV 365
FD YS + YH DV V +LGV+GGILGSLYN+L +KVLR YN I EKG KILLAC
Sbjct: 299 FDVYSENASYHLGDVLPVLLLGVVGGILGSLYNFLLDKVLRAYNYIYEKGVTWKILLACA 358
Query: 366 ISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTND 425
IS+FTSCLLFGLP+LASCQPCP D +E CPTIGRSG +KK+QCPPGHYNDLASLIFNTND
Sbjct: 359 ISIFTSCLLFGLPFLASCQPCPVDALEECPTIGRSGNFKKYQCPPGHYNDLASLIFNTND 418
Query: 426 DAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGM 485
DAI+NLFSKNTD EF Y S+ +FF+T FFLSI S G+V PAG+FVP+IVTGASYGRFVGM
Sbjct: 419 DAIKNLFSKNTDFEFHYFSVLVFFVTCFFLSIFSYGIVAPAGLFVPVIVTGASYGRFVGM 478
Query: 486 LFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVA 545
L G SNLNHGL+AVLGAASFLGG+MR TVS CVI+LELTNNLLLLP++M+VLL+SK+VA
Sbjct: 479 LLGSNSNLNHGLFAVLGAASFLGGTMRMTVSTCVILLELTNNLLLLPMMMVVLLISKTVA 538
Query: 546 DVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTT 605
D FNANIY+LIMK KG PYL +HAEPYMRQL VGDVVTGPLQ+F+G EKV +V VLKTT
Sbjct: 539 DGFNANIYNLIMKLKGFPYLYSHAEPYMRQLLVGDVVTGPLQVFNGIEKVETIVHVLKTT 598
Query: 606 SHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKR 665
+HNGFPV+D PP + APVL G+ILR H+LTLL+K+VFM +P+A + L QF A++FAK+
Sbjct: 599 NHNGFPVVDGPPLAAAPVLHGLILRAHILTLLKKRVFMPSPVACDSNTLSQFKAEEFAKK 658
Query: 666 GSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIP 725
GS R ++IED+ L++EE++M++DLHPF+NASPYTVVE+MSL KALILFRE+G+RHLLVIP
Sbjct: 659 GSGRSDKIEDVELSEEELNMYLDLHPFSNASPYTVVETMSLAKALILFREVGIRHLLVIP 718
Query: 726 KIPSRSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLRFWQPF 768
K +R PVVGILTRHDF EHILG HP + RS+WKRLR PF
Sbjct: 719 KTSNRPPVVGILTRHDFMPEHILGLHPSVSRSKWKRLRIRLPF 761
>CLCC_ARATH (Q96282) Chloride channel protein CLC-c (AtCLC-c)
Length = 779
Score = 946 bits (2446), Expect = 0.0
Identities = 460/746 (61%), Positives = 563/746 (74%), Gaps = 3/746 (0%)
Query: 10 DSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIF 69
D RQPLL+ R N++SQ+AIVGAN CPIESLDYEIFEN+FFKQDWRSR ++I
Sbjct: 32 DGSVGFRQPLLARNRK--NTTSQIAIVGANTCPIESLDYEIFENDFFKQDWRSRKKIEIL 89
Query: 70 QYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFASNL 129
QY +KW L FLIG GL+GF NNL VEN+AG K ++ N+ML +++ AF F NL
Sbjct: 90 QYTFLKWALAFLIGLATGLVGFLNNLGVENIAGFKLLLIGNLMLKEKYFQAFFAFAGCNL 149
Query: 130 ALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHI 189
L A + A IAPAAAGSGI EVKAYLNG+DA I TL VKI GSI V++ +
Sbjct: 150 ILATAAASLCAFIAPAAAGSGIPEVKAYLNGIDAYSILAPSTLFVKIFGSIFGVAAGFVV 209
Query: 190 GKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAP 249
GK GPM+HTGAC+A LLGQGGSK+Y LTWKWLR+FKNDRDRRDLI CG+AAG+AAAFRAP
Sbjct: 210 GKEGPMVHTGACIANLLGQGGSKKYRLTWKWLRFFKNDRDRRDLITCGAAAGVAAAFRAP 269
Query: 250 VGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAY 309
VGGVLFALEE ASWWR ALLWR FFTTA+VA+ LR++I+ C SG+CGLFGKGGLIMFD
Sbjct: 270 VGGVLFALEEAASWWRNALLWRTFFTTAVVAVVLRSLIEFCRSGRCGLFGKGGLIMFDVN 329
Query: 310 SGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMF 369
SG ++Y D+ + LGVIGG+LGSLYNYL +KVLR Y+ INEKG KI+L +S+
Sbjct: 330 SGPVLYSTPDLLAIVFLGVIGGVLGSLYNYLVDKVLRTYSIINEKGPRFKIMLVMAVSIL 389
Query: 370 TSCLLFGLPWLASCQPCPADPVE-PCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAI 428
+SC FGLPWL+ C PCP E CP++GRS IYK FQCPP HYNDL+SL+ NTNDDAI
Sbjct: 390 SSCCAFGLPWLSQCTPCPIGIEEGKCPSVGRSSIYKSFQCPPNHYNDLSSLLLNTNDDAI 449
Query: 429 RNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFG 488
RNLF+ +++EF S++ IFF+ + L I++ G+ +P+G+F+P+I+ GASYGR VG L G
Sbjct: 450 RNLFTSRSENEFHISTLAIFFVAVYCLGIITYGIAIPSGLFIPVILAGASYGRLVGRLLG 509
Query: 489 KKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF 548
S L+ GL+++LGAASFLGG+MR TVSLCVI+LELTNNLL+LPL+M+VLL+SK+VAD F
Sbjct: 510 PVSQLDVGLFSLLGAASFLGGTMRMTVSLCVILLELTNNLLMLPLVMLVLLISKTVADCF 569
Query: 549 NANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHN 608
N +YD I+ KGLPY+E HAEPYMR L DVV+G L F EKV + LK T HN
Sbjct: 570 NRGVYDQIVTMKGLPYMEDHAEPYMRNLVAKDVVSGALISFSRVEKVGVIWQALKMTRHN 629
Query: 609 GFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSD 668
GFPVIDEPPF+EA L GI LR HLL LL+ K F G +LR A DF K G
Sbjct: 630 GFPVIDEPPFTEASELCGIALRSHLLVLLQGKKFSKQRTTFGSQILRSCKARDFGKAGLG 689
Query: 669 RGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIP 728
+G +IED+ L++EEM+M++DLHP TN SPYTV+E++SL KA ILFR+LGLRHL V+PK P
Sbjct: 690 KGLKIEDLDLSEEEMEMYVDLHPITNTSPYTVLETLSLAKAAILFRQLGLRHLCVVPKTP 749
Query: 729 SRSPVVGILTRHDFTAEHILGTHPYL 754
R P+VGILTRHDF EH+LG +P++
Sbjct: 750 GRPPIVGILTRHDFMPEHVLGLYPHI 775
>CLCB_ARATH (P92942) Chloride channel protein CLC-b (AtCLC-b)
Length = 780
Score = 822 bits (2122), Expect = 0.0
Identities = 409/762 (53%), Positives = 546/762 (70%), Gaps = 18/762 (2%)
Query: 9 GDSE-HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQ 67
GD E + L QPL+ + R++ SS+ +A+VGA V IESLDYEI EN+ FK DWR R Q
Sbjct: 20 GDPESNTLNQPLVKANRTL--SSTPLALVGAKVSHIESLDYEINENDLFKHDWRKRSKAQ 77
Query: 68 IFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFAS 127
+ QYV +KW L L+G GLI NLAVEN+AG K + + + +R+ + +
Sbjct: 78 VLQYVFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGHFLTQERYVTGLMVLVGA 137
Query: 128 NLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSL 187
NL LTL A+++ AP AAG GI E+KAYLNGVD P +F T+IVKI+GSI AV++ L
Sbjct: 138 NLGLTLVASVLCVCFAPTAAGPGIPEIKAYLNGVDTPNMFGATTMIVKIVGSIGAVAAGL 197
Query: 188 HIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFR 247
+GK GP++H G+C+A+LLGQGG+ + + W+WLRYF NDRDRRDLI CGSAAG+ AAFR
Sbjct: 198 DLGKEGPLVHIGSCIASLLGQGGTDNHRIKWRWLRYFNNDRDRRDLITCGSAAGVCAAFR 257
Query: 248 APVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFD 307
+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LR I++C SGKCGLFGKGGLIMFD
Sbjct: 258 SPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLREFIEICNSGKCGLFGKGGLIMFD 317
Query: 308 AYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVIS 367
+ YH D+ V ++GVIGGILGSLYN+L +KVLR+YN INEKG I K+LL+ +S
Sbjct: 318 VSHVTYTYHVTDIIPVMLIGVIGGILGSLYNHLLHKVLRLYNLINEKGKIHKVLLSLTVS 377
Query: 368 MFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDA 427
+FTS L+GLP+LA C+PC E CPT GRSG +K+F CP G+YNDLA+L+ TNDDA
Sbjct: 378 LFTSVCLYGLPFLAKCKPCDPSIDEICPTNGRSGNFKQFHCPKGYYNDLATLLLTTNDDA 437
Query: 428 IRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLF 487
+RNLFS NT +EF S++IFF+ L + + G+ P+G+F+PII+ GA+YGR +G
Sbjct: 438 VRNLFSSNTPNEFGMGSLWIFFVLYCILGLFTFGIATPSGLFLPIILMGAAYGRMLGAAM 497
Query: 488 GKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADV 547
G ++++ GLYAVLGAA+ + GSMR TVSLCVI LELTNNLLLLP+ M+VLL++K+V D
Sbjct: 498 GSYTSIDQGLYAVLGAAALMAGSMRMTVSLCVIFLELTNNLLLLPITMIVLLIAKTVGDS 557
Query: 548 FNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEKVRNVVFVLKTT 605
FN +IYD+I+ KGLP+LE + EP+MR LTVG++ P+ G EKV N+V VLK T
Sbjct: 558 FNPSIYDIILHLKGLPFLEANPEPWMRNLTVGELGDAKPPVVTLQGVEKVSNIVDVLKNT 617
Query: 606 SHNGFPVIDEPPFSE------APVLFGIILRHHLLTLLRKKVFMSAP-MAMGGDVLRQFS 658
+HN FPV+DE + A L G+ILR HL+ +L+K+ F++ +V +F
Sbjct: 618 THNAFPVLDEAEVPQVGLATGATELHGLILRAHLVKVLKKRWFLTEKRRTEEWEVREKFP 677
Query: 659 ADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGL 718
D+ A +R + +D+ +T EM+M++DLHP TN +PYTV+E+MS+ KAL+LFR++GL
Sbjct: 678 WDELA----EREDNFDDVAITSAEMEMYVDLHPLTNTTPYTVMENMSVAKALVLFRQVGL 733
Query: 719 RHLLVIPKIPSRS--PVVGILTRHDFTAEHILGTHPYLVRSR 758
RHLL++PKI + PVVGILTR D A +IL P L +S+
Sbjct: 734 RHLLIVPKIQASGMCPVVGILTRQDLRAYNILQAFPLLEKSK 775
>CLCA_ARATH (P92941) Chloride channel protein CLC-a (AtCLC-a)
Length = 775
Score = 815 bits (2106), Expect = 0.0
Identities = 407/772 (52%), Positives = 547/772 (70%), Gaps = 17/772 (2%)
Query: 3 TNHLSNGDSE------HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFF 56
+N NG+ E + L QPLL R++ SS+ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKR 116
K DWRSR Q+FQY+ +KW L L+G GLI NLAVEN+AG K + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
F + F +NL LTL AT++ AP AAG GI E+KAYLNG+D P +F T++VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
+GSI AV++ L +GK GP++H G+C+A+LLGQGG + + W+WLRYF NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LRA I++C SGKCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGT 356
LFG GGLIMFD + YHA D+ V ++GV GGILGSLYN+L +KVLR+YN IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDL 416
I K+LL+ +S+FTS LFGLP+LA C+PC E CPT GRSG +K+F CP G+YNDL
Sbjct: 368 IHKVLLSLGVSLFTSVCLFGLPFLAECKPCDPSIDEICPTNGRSGNFKQFNCPNGYYNDL 427
Query: 417 ASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTG 476
++L+ TNDDA+RN+FS NT +EF S++IFF L +++ G+ P+G+F+PII+ G
Sbjct: 428 STLLLTTNDDAVRNIFSSNTPNEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 487
Query: 477 ASYGRFVGMLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMM 536
++YGR +G G +N++ GLYAVLGAAS + GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 488 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 547
Query: 537 VLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEK 594
VLL++K+V D FN +IY++I+ KGLP+LE + EP+MR LTVG++ P+ +G EK
Sbjct: 548 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 607
Query: 595 VRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAP-MAMGGDV 653
V N+V VL+ T+HN FPV+D + L G+ILR HL+ +L+K+ F++ +V
Sbjct: 608 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 667
Query: 654 LRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILF 713
+F+ + A +R + +D+ +T EM +++DLHP TN +PYTVV+SMS+ KAL+LF
Sbjct: 668 REKFTPVELA----EREDNFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 723
Query: 714 RELGLRHLLVIPKIPS--RSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLR 763
R +GLRHLLV+PKI + SPV+GILTR D A +IL P+L + + + R
Sbjct: 724 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 775
>CLCD_ARATH (P92943) Chloride channel protein CLC-d (AtCLC-d)
Length = 792
Score = 640 bits (1650), Expect = 0.0
Identities = 362/768 (47%), Positives = 482/768 (62%), Gaps = 38/768 (4%)
Query: 1 MSTNHLSNG-DSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQD 59
M +NHL NG +S++ L + S + + + + + SLDYE+ EN ++++
Sbjct: 1 MLSNHLQNGIESDNLLWSRVPESDDTSTDDITLLNSHRDGDGGVNSLDYEVIENYAYREE 60
Query: 60 WRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSL 119
RG L + YV +KW LIG GL NL+VEN AG KF +T ++ K +
Sbjct: 61 QAHRGKLYVGYYVAVKWFFSLLIGIGTGLAAVFINLSVENFAGWKFALTF-AIIQKSYFA 119
Query: 120 AFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGS 179
F+ + NL L + I APAAAGSGI E+K YLNG+D PG TLI KI GS
Sbjct: 120 GFIVYLLINLVLVFSSAYIITQFAPAAAGSGIPEIKGYLNGIDIPGTLLFRTLIGKIFGS 179
Query: 180 ITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSA 239
I +V L +GK GP++HTGAC+A+LLGQGGS +Y L +W + FK+DRDRRDL+ CG A
Sbjct: 180 IGSVGGGLALGKEGPLVHTGACIASLLGQGGSTKYHLNSRWPQLFKSDRDRRDLVTCGCA 239
Query: 240 AGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFG 299
AG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+AIVA+ +R + C SG CG FG
Sbjct: 240 AGVAAAFRAPVGGVLFALEEVTSWWRSQLMWRVFFTSAIVAVVVRTAMGWCKSGICGHFG 299
Query: 300 KGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYL------WNKVLRIYNAINE 353
GG I++D G Y+ K++ + ++GVIGG+LG+L+N L W + N++++
Sbjct: 300 GGGFIIWDVSDGQDDYYFKELLPMAVIGVIGGLLGALFNQLTLYMTSWRR-----NSLHK 354
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCP---ADPVEPCP-TIGRSGIYKKFQC- 408
KG KI+ AC+IS TS + FGLP L C PCP D CP G G Y F C
Sbjct: 355 KGNRVKIIEACIISCITSAISFGLPLLRKCSPCPESVPDSGIECPRPPGMYGNYVNFFCK 414
Query: 409 PPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGV 468
YNDLA++ FNT DDAIRNLFS T EF S+ F + L++++ G VPAG
Sbjct: 415 TDNEYNDLATIFFNTQDDAIRNLFSAKTMREFSAQSLLTFLAMFYTLAVVTFGTAVPAGQ 474
Query: 469 FVPIIVTGASYGRFVGML---FGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELT 525
FVP I+ G++YGR VGM F KK N+ G YA+LGAASFLGGSMR TVSLCVI++E+T
Sbjct: 475 FVPGIMIGSTYGRLVGMFVVRFYKKLNIEEGTYALLGAASFLGGSMRMTVSLCVIMVEIT 534
Query: 526 NNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGD----- 580
NNL LLPLIM+VLL+SK+V D FN +Y++ + KG+P LE+ + +MRQ+ +
Sbjct: 535 NNLKLLPLIMLVLLISKAVGDAFNEGLYEVQARLKGIPLLESRPKYHMRQMIAKEACQSQ 594
Query: 581 -VVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRK 639
V++ P I +V +V +L + HNGFPVID E V+ G++LR HLL LL+
Sbjct: 595 KVISLPRVI-----RVADVASILGSNKHNGFPVIDHTRSGETLVI-GLVLRSHLLVLLQS 648
Query: 640 KV-FMSAPMAMGGDVLR-QFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASP 697
KV F +P+ + S +FAK S +G IEDIHLT ++++M+IDL PF N SP
Sbjct: 649 KVDFQHSPLPCDPSARNIRHSFSEFAKPVSSKGLCIEDIHLTSDDLEMYIDLAPFLNPSP 708
Query: 698 YTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAE 745
Y V E MSL K LFR+LGLRHL V+P+ PSR V+G++TR D E
Sbjct: 709 YVVPEDMSLTKVYNLFRQLGLRHLFVVPR-PSR--VIGLITRKDLLIE 753
>CLC7_HUMAN (P51798) Chloride channel protein 7 (ClC-7)
Length = 805
Score = 404 bits (1038), Expect = e-112
Identities = 255/718 (35%), Positives = 400/718 (55%), Gaps = 31/718 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ EN+ F ++ R + +W++C LIG + GL+ ++ VENLAG+
Sbjct: 95 ESLDYDNSENQLFLEEERRINHTAFRTVEIKRWVICALIGILTGLVACFIDIVVENLAGL 154
Query: 104 KF-VVTSNMMLHKR---FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
K+ V+ N+ S + + + N A L ++I A I P AAGSGI ++K +LN
Sbjct: 155 KYRVIKGNIDKFTEKGGLSFSLLLWATLNAAFVLVGSVIVAFIEPVAAGSGIPQIKCFLN 214
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV P + L TL++K+ G I +V L +GK GPM+H+G+ +AA + QG S +K
Sbjct: 215 GVKIPHVVRLKTLVIKVSGVILSVVGGLAVGKEGPMIHSGSVIAAGISQGRSTSLKRDFK 274
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF + I
Sbjct: 275 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 334
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAY-SGSLMYHAKDVPLVFILGVIGGILGSLYN 338
L ++ + G GLI F + S + Y ++P+ +GV+GG+LG+++N
Sbjct: 335 TFTLNFVLSI-YHGNMWDLSSPGLINFGRFDSEKMAYTIHEIPVFIAMGVVGGVLGAVFN 393
Query: 339 YLWNKVLRIYNAINEKGTICKILLACVISMFTSCLLFGLPWLA-SCQPCPADPVEPCPTI 397
L N L ++ +++ A +++ T+ + F L + + CQP
Sbjct: 394 AL-NYWLTMFRIRYIHRPCLQVIEAVLVAAVTATVAFVLIYSSRDCQPLQG--------- 443
Query: 398 GRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSI 457
G + C G YN +A+ FNT + ++ +LF + + ++ +F + FFL+
Sbjct: 444 GSMSYPLQLFCADGEYNSMAAAFFNTPEKSVVSLF-HDPPGSYNPLTLGLFTLVYFFLAC 502
Query: 458 LSCGVVVPAGVFVPIIVTGASYGRFVGM----LFGKKSNLNHGLYAVLGAASFLGGSMRT 513
+ G+ V AGVF+P ++ GA++GR G+ L G + G YA++GAA+ LGG +R
Sbjct: 503 WTYGLTVSAGVFIPSLLIGAAWGRLFGISLSYLTGAAIWADPGKYALMGAAAQLGGIVRM 562
Query: 514 TVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYM 573
T+SL VI++E T+N+ IM+VL+ +K V DVF +YD+ ++ + +P+L A
Sbjct: 563 TLSLTVIMMEATSNVTYGFPIMLVLMTAKIVGDVFIEGLYDMHIQLQSVPFLHWEAPVTS 622
Query: 574 RQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTT--SHNGFPVIDEPPFSEAPVLFGIILRH 631
LT +V++ P+ EKV +V VL T +HNGFPV++ ++ L G+ILR
Sbjct: 623 HSLTAREVMSTPVTCLRRREKVGVIVDVLSDTASNHNGFPVVEHADDTQPARLQGLILRS 682
Query: 632 HLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHP 691
L+ LL+ KVF+ G V R+ DF + R I+ IH++Q+E + +DL
Sbjct: 683 QLIVLLKHKVFVERSNL--GLVQRRLRLKDF-RDAYPRFPPIQSIHVSQDERECTMDLSE 739
Query: 692 FTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAEHILG 749
F N SPYTV + SL + LFR LGLRHL+V + +R+ VVG++TR D A + LG
Sbjct: 740 FMNPSPYTVPQEASLPRVFKLFRALGLRHLVV---VDNRNQVVGLVTRKDL-ARYRLG 793
>CLC7_RAT (P51799) Chloride channel protein 7 (ClC-7)
Length = 803
Score = 402 bits (1033), Expect = e-111
Identities = 257/719 (35%), Positives = 403/719 (55%), Gaps = 33/719 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ EN+ F ++ R + +W++C LIG + GL+ ++ VENLAG+
Sbjct: 93 ESLDYDNSENQLFLEEERRINHTAFRTVEIKRWVICALIGILTGLVACFIDIVVENLAGL 152
Query: 104 KF-VVTSNMMLHKR---FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
K+ V+ N+ S + + + N A L ++I A I P AAGSGI ++K +LN
Sbjct: 153 KYRVIKDNIDKFTEKGGLSFSLLLWATLNSAFVLVGSVIVAFIEPVAAGSGIPQIKCFLN 212
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV P + L TL++K+ G I +V L +GK GPM+H+G+ +AA + QG S +K
Sbjct: 213 GVKIPHVVRLKTLVIKVSGVILSVVGGLAVGKEGPMIHSGSVIAAGISQGRSTSLKRDFK 272
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF + I
Sbjct: 273 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 332
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAY-SGSLMYHAKDVPLVFILGVIGGILGSLYN 338
L ++ + G GLI F + S + Y ++P+ +GV+GGILG+++N
Sbjct: 333 TFTLNFVLSI-YHGNMWDLSSPGLINFGRFDSEKMAYTIHEIPVFIAMGVVGGILGAVFN 391
Query: 339 YLWNKVLRIYNAINEKGTICKILLACVISMFTSCLLFGLPWLA-SCQPCPADPVEPCPTI 397
L N L ++ +++ A +++ T+ + F L + + CQP
Sbjct: 392 AL-NYWLTMFRIRYIHRPCLQVIEAMLVAAVTATVAFVLIYSSRDCQPLQ---------- 440
Query: 398 GRSGIYK-KFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLS 456
G S Y + C G YN +A+ FNT + ++ +LF + + ++ +F + FFL+
Sbjct: 441 GSSMSYPLQLFCADGEYNSMAAAFFNTPEKSVVSLF-HDPPGSYNPMTLGLFTLVYFFLA 499
Query: 457 ILSCGVVVPAGVFVPIIVTGASYGRFVGM----LFGKKSNLNHGLYAVLGAASFLGGSMR 512
+ G+ V AGVF+P ++ GA++GR G+ L G + G YA++GAA+ LGG +R
Sbjct: 500 CWTYGLTVSAGVFIPSLLIGAAWGRLFGISMSYLTGAAIWADPGKYALMGAAAQLGGIVR 559
Query: 513 TTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPY 572
T+SL VI++E T+N+ IM+VL+ +K V DVF +YD+ ++ + +P+L A
Sbjct: 560 MTLSLTVIMMEATSNVTYGFPIMLVLMTAKIVGDVFIEGLYDMHIQLQSVPFLHWEAPVT 619
Query: 573 MRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTT--SHNGFPVIDEPPFSEAPVLFGIILR 630
LT +V++ P+ EKV +V VL T +HNGFPV+++ ++ L G+ILR
Sbjct: 620 SHSLTAREVMSTPVTCLRRREKVGIIVDVLSDTASNHNGFPVVEDVGDTQPARLQGLILR 679
Query: 631 HHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLH 690
L+ LL+ KVF+ + G V R+ DF + R I+ IH++Q+E + +DL
Sbjct: 680 SQLIVLLKHKVFVE--RSNMGLVQRRLRLKDF-RDAYPRFPPIQSIHVSQDERECTMDLS 736
Query: 691 PFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAEHILG 749
F N SPYTV + SL + LFR LGLRHL+V + + + VVG++TR D A + LG
Sbjct: 737 EFMNPSPYTVPQEASLPRVFKLFRALGLRHLVV---VDNHNQVVGLVTRKDL-ARYRLG 791
>CLC7_MOUSE (O70496) Chloride channel protein 7 (ClC-7)
Length = 803
Score = 402 bits (1033), Expect = e-111
Identities = 257/719 (35%), Positives = 403/719 (55%), Gaps = 33/719 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ EN+ F ++ R + +W++C LIG + GL+ ++ VENLAG+
Sbjct: 93 ESLDYDNSENQLFLEEERRINHTAFRTVEIKRWVICALIGILTGLVACFIDIVVENLAGL 152
Query: 104 KF-VVTSNMMLHKR---FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
K+ V+ N+ S + + + N A L ++I A I P AAGSGI ++K +LN
Sbjct: 153 KYRVIKDNIDKFTEKGGLSFSLLLWATLNSAFVLVGSVIVAFIEPVAAGSGIPQIKCFLN 212
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV P + L TL++K+ G I +V L +GK GPM+H+G+ +AA + QG S +K
Sbjct: 213 GVKIPHVVRLKTLVIKVSGVILSVVGGLAVGKEGPMIHSGSVIAAGISQGRSTSLKRDFK 272
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF + I
Sbjct: 273 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 332
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAY-SGSLMYHAKDVPLVFILGVIGGILGSLYN 338
L ++ + G GLI F + S + Y ++P+ +GV+GGILG+++N
Sbjct: 333 TFTLNFVLSI-YHGNMWDLSSPGLINFGRFDSEKMAYTIHEIPVFIAMGVVGGILGAVFN 391
Query: 339 YLWNKVLRIYNAINEKGTICKILLACVISMFTSCLLFGLPWLA-SCQPCPADPVEPCPTI 397
L N L ++ +++ A +++ T+ + F L + + CQP
Sbjct: 392 AL-NYWLTMFRIRYIHRPCLQVIEAMLVAAVTATVAFVLIYSSRDCQPLQ---------- 440
Query: 398 GRSGIYK-KFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLS 456
G S Y + C G YN +A+ FNT + ++ +LF + + ++ +F + FFL+
Sbjct: 441 GSSMSYPLQLFCADGEYNSMAAAFFNTPEKSVVSLF-HDPPGSYNPMTLGLFTLVYFFLA 499
Query: 457 ILSCGVVVPAGVFVPIIVTGASYGRFVGM----LFGKKSNLNHGLYAVLGAASFLGGSMR 512
+ G+ V AGVF+P ++ GA++GR G+ L G + G YA++GAA+ LGG +R
Sbjct: 500 CWTYGLTVSAGVFIPSLLIGAAWGRLFGISLSYLTGAAIWADPGKYALMGAAAQLGGIVR 559
Query: 513 TTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPY 572
T+SL VI++E T+N+ IM+VL+ +K V DVF +YD+ ++ + +P+L A
Sbjct: 560 MTLSLTVIMMEATSNVTYGFPIMLVLMTAKIVGDVFIEGLYDMHIQLQSVPFLHWEAPVT 619
Query: 573 MRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTT--SHNGFPVIDEPPFSEAPVLFGIILR 630
LT +V++ P+ EKV +V VL T +HNGFPV+++ ++ L G+ILR
Sbjct: 620 SHSLTAREVMSTPVTCLRRREKVGIIVDVLSDTASNHNGFPVVEDVGDTQPARLQGLILR 679
Query: 631 HHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLH 690
L+ LL+ KVF+ + G V R+ DF + R I+ IH++Q+E + +DL
Sbjct: 680 SQLIVLLKHKVFVE--RSNMGLVQRRLRLKDF-RDAYPRFPPIQSIHVSQDERECTMDLS 736
Query: 691 PFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAEHILG 749
F N SPYTV + SL + LFR LGLRHL+V + + + VVG++TR D A + LG
Sbjct: 737 EFMNPSPYTVPQEASLPRVFKLFRALGLRHLVV---VDNHNQVVGLVTRKDL-ARYRLG 791
>CLC6_HUMAN (P51797) Chloride channel protein 6 (ClC-6)
Length = 869
Score = 328 bits (842), Expect = 3e-89
Identities = 206/605 (34%), Positives = 326/605 (53%), Gaps = 26/605 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ N+ + + + + + +Y +KW++ F IG GL+G + V +
Sbjct: 49 ESLDYDRCINDPYLEVLETMDNKKGRRYEAVKWMVVFAIGVCTGLVGLFVDFFVRLFTQL 108
Query: 104 KFVVTSNMMLHKR----FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
KF V + +L+ + NL A+++ +I P AAGSGI EVK YLN
Sbjct: 109 KFGVVQTSVEECSQKGCLALSLLELLGFNLTFVFLASLLV-LIEPVAAGSGIPEVKCYLN 167
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV PGI L TL+ K++G + +V+ L +GK GPM+H+G+ V A L Q S
Sbjct: 168 GVKVPGIVRLRTLLCKVLGVLFSVAGGLFVGKEGPMIHSGSVVGAGLPQFQSISLRKIQF 227
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF++DRD+RD + G+AAG+AAAF AP+GG LF+LEE +S+W L W+ F +
Sbjct: 228 NFPYFRSDRDKRDFVSAGAAAGVAAAFGAPIGGTLFSLEEGSSFWNQGLTWKVLFCSMSA 287
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGS------LMYHAKDVPLVFILGVIGGIL 333
L G G F GL+ F + S ++ A D+ ++GVIGG+L
Sbjct: 288 TFTLNFFRSGIQFGSWGSFQLPGLLNFGEFKCSDSDKKCHLWTAMDLGFFVVMGVIGGLL 347
Query: 334 GSLYNYLWNKVLRIYNA--INEKGTICKILLACVISMFTSCLLFGLPW-LASCQPCPA-- 388
G+ +N L NK L Y ++ K + ++L + ++S+ T+ ++F L C+ +
Sbjct: 348 GATFNCL-NKRLAKYRMRNVHPKPKLVRVLESLLVSLVTTVVVFVASMVLGECRQMSSSS 406
Query: 389 ----DPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSS 444
D + T + K F CP YND+A+L FN + AI LF + D F +
Sbjct: 407 QIGNDSFQLQVTEDVNSSIKTFFCPNDTYNDMATLFFNPQESAILQLFHQ--DGTFSPVT 464
Query: 445 MFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSNLNH---GLYAVL 501
+ +FF+ F L+ + G+ VP+G+FVP ++ GA++GR V + L H G +A++
Sbjct: 465 LALFFVLYFLLACWTYGISVPSGLFVPSLLCGAAFGRLVANVLKSYIGLGHIYSGTFALI 524
Query: 502 GAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKG 561
GAA+FLGG +R T+SL VI++E TN + IM+ L+V+K D FN IYD+ + +G
Sbjct: 525 GAAAFLGGVVRMTISLTVILIESTNEITYGLPIMVTLMVAKWTGDFFNKGIYDIHVGLRG 584
Query: 562 LPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEA 621
+P LE E M +L D++ L + + +++++V +L+TT H+ FPV+ E +E
Sbjct: 585 VPLLEWETEVEMDKLRASDIMEPNLTYVYPHTRIQSLVSILRTTVHHAFPVVTENRGNEK 644
Query: 622 PVLFG 626
+ G
Sbjct: 645 EFMKG 649
Score = 64.7 bits (156), Expect = 9e-10
Identities = 44/129 (34%), Positives = 68/129 (52%), Gaps = 11/129 (8%)
Query: 622 PVLF-GIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFS--ADDFAKRGSDRGERIEDIHL 678
P+ F G+ILR L+TLL + V S + ++ A+D+ R I D+ L
Sbjct: 738 PLTFHGLILRSQLVTLLVRGVCYSESQSSASQPRLSYAEMAEDYP-----RYPDIHDLDL 792
Query: 679 TQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILT 738
T M +D+ P+ N SP+TV + + + LFR +GLRHL P + + +VGI+T
Sbjct: 793 TLLNPRMIVDVTPYMNPSPFTVSPNTHVSQVFNLFRTMGLRHL---PVVNAVGEIVGIIT 849
Query: 739 RHDFTAEHI 747
RH+ T E +
Sbjct: 850 RHNLTYEFL 858
>CLC6_MOUSE (O35454) Chloride channel protein 6 (ClC-6)
Length = 870
Score = 326 bits (836), Expect = 1e-88
Identities = 205/606 (33%), Positives = 325/606 (52%), Gaps = 27/606 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ N+ + + + + + +Y +KW++ F IG GL+G + +V +
Sbjct: 49 ESLDYDRCINDPYLEVLETMDNKKGRRYEAVKWMVVFAIGVCTGLVGLFVDFSVRLFTQL 108
Query: 104 KFVVTSNMMLHKR----FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
KF V + +L+ + NL A+++ +I P AAGSGI E+K YLN
Sbjct: 109 KFGVVQTSVEECSQKGCLALSLLELLGFNLTFVFLASLLV-LIEPVAAGSGIPEIKCYLN 167
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV PGI L TL+ K+ G + +VS L +GK GPM+H+GA V A L Q S
Sbjct: 168 GVKVPGIVRLRTLLCKVFGVLFSVSGGLFVGKEGPMIHSGAVVGAGLPQFQSISLRKIQF 227
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF++DRD+RD + G+AAG+AAAF AP+GG LF+LEE +S+W L W+ F +
Sbjct: 228 NFPYFRSDRDKRDFVSAGAAAGVAAAFGAPIGGTLFSLEEGSSFWNQGLTWKVLFCSMSA 287
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGS------LMYHAKDVPLVFILGVIGGIL 333
L G G F GL+ F + S ++ A D+ ++GVIGG+L
Sbjct: 288 TFTLNFFRSGIQFGSWGSFQLPGLLNFGEFKCSDSDKKCHLWTAMDLGFFVVMGVIGGLL 347
Query: 334 GSLYNYLWNKVLRIYNA--INEKGTICKILLACVISMFTSCLLFGLPW-LASCQPCPADP 390
G+ +N L NK L Y ++ K + ++L + ++S+ T+ ++F L C+ +
Sbjct: 348 GATFNCL-NKRLAKYRMRNVHPKPKLVRVLESLLVSLVTTVVVFVASMVLGECRQMSSTS 406
Query: 391 VE-------PCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYS 443
+ + K F CP YND+A+L FN+ + AI LF + D F
Sbjct: 407 QTGNGSFQLQVTSEDVNSTIKAFFCPNDTYNDMATLFFNSQESAILQLFHQ--DGTFSPV 464
Query: 444 SMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSNLNH---GLYAV 500
++ +FFI F L+ + G VP+G+FVP ++ GA++GR V + L H G +A+
Sbjct: 465 TLALFFILYFLLACWTFGTSVPSGLFVPSLLCGAAFGRLVANVLKSYIGLGHLYSGTFAL 524
Query: 501 LGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSK 560
+GAA+FLGG +R T+SL VI++E TN + IM+ L+V+K D+FN IYD+ + +
Sbjct: 525 IGAAAFLGGVVRMTISLTVILIESTNEITYGLPIMVTLMVAKWTGDLFNKGIYDVHIGLR 584
Query: 561 GLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSE 620
G+P LE + M +L D++ L + + +++++V +L+TT H+ FPV+ E +E
Sbjct: 585 GVPLLEWETDVEMDKLRASDIMEPNLTYVYPHTRIQSLVSILRTTVHHAFPVVTENRGNE 644
Query: 621 APVLFG 626
+ G
Sbjct: 645 KEFMKG 650
Score = 64.3 bits (155), Expect = 1e-09
Identities = 43/129 (33%), Positives = 68/129 (52%), Gaps = 11/129 (8%)
Query: 622 PVLF-GIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFS--ADDFAKRGSDRGERIEDIHL 678
P+ F G++LR L+TLL + V S + ++ A+D+ R I D+ L
Sbjct: 739 PLTFHGLVLRSQLVTLLVRGVCYSESQSSASQPRLSYAEMAEDYP-----RYPDIHDLDL 793
Query: 679 TQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILT 738
T M +D+ P+ N SP+TV + + + LFR +GLRHL P + + +VGI+T
Sbjct: 794 TLLNPRMIVDVTPYMNPSPFTVSPNTHVSQVFNLFRTMGLRHL---PVVNAVGEIVGIIT 850
Query: 739 RHDFTAEHI 747
RH+ T E +
Sbjct: 851 RHNLTNEFL 859
>CLC6_RABIT (Q9TT16) Chloride channel protein 6 (ClC-6)
Length = 869
Score = 320 bits (819), Expect = 1e-86
Identities = 203/605 (33%), Positives = 323/605 (52%), Gaps = 26/605 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ N+ + + + + Y ++KW + F IG GL+G + V+ +
Sbjct: 49 ESLDYDRCINDPYLEVLETMDHKKGRWYEVVKWTVVFAIGVCTGLVGLFVDFFVQLFTQL 108
Query: 104 KFVVTSNMMLHKR----FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
KF V + +L+ + NL A+++ +I P AAGSGI E+K YLN
Sbjct: 109 KFGVVEASVEECSQKGCLALSLLELLGFNLTFVFLASLLV-LIEPVAAGSGIPEIKCYLN 167
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV PGI L TL+ K+ G + +V+ L +GK GPM+H+GA V A L Q S
Sbjct: 168 GVKVPGIVRLRTLLCKVFGVLFSVAGGLFVGKEGPMIHSGAVVGAGLPQFQSISLRKIQF 227
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF++DRD+RD + G+AAGIAAAF AP+G LF+LEE +S+W L W+ F +
Sbjct: 228 NFPYFRSDRDKRDFVSAGAAAGIAAAFGAPIGATLFSLEEGSSFWNQGLTWKVLFCSMSA 287
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGS------LMYHAKDVPLVFILGVIGGIL 333
L G G F GL+ F + S ++ A D+ ++GVIGG+L
Sbjct: 288 TFTLNFFRSGIQFGSWGSFQLPGLLNFGEFKCSDSDKKCHLWTAMDMGFFVVMGVIGGLL 347
Query: 334 GSLYNYLWNKVLRIYNA--INEKGTICKILLACVISMFTSCLLFGLPW-LASCQPCPADP 390
G+ +N L NK L Y ++ K + ++L + ++S+ T+ ++F L C+ +
Sbjct: 348 GATFNCL-NKRLAKYRMRNVHPKPKLVRVLESLLVSLVTTLVVFVASMVLGECRQMSSSS 406
Query: 391 ------VEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSS 444
++ T + K F CP YND+A+L FN + AI LF + D F +
Sbjct: 407 QISNGSLKLQVTSDVNSSIKAFFCPNDTYNDMATLFFNPQESAILQLFHQ--DGTFSPIT 464
Query: 445 MFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSNLNH---GLYAVL 501
+ +FF+ F L+ + G+ VP+G+FVP ++ GA++GR V + L+H G ++++
Sbjct: 465 LALFFVLYFLLACWTYGISVPSGLFVPSLLCGAAFGRLVANVLKSYIGLSHIYSGTFSLI 524
Query: 502 GAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKG 561
GAA+ LGG +R T+SL VI++E TN + IM+ L+V+K D FN IYD+ + +G
Sbjct: 525 GAAALLGGVVRMTISLTVILIESTNEITYGLPIMITLMVAKWTGDFFNKGIYDIHVGLRG 584
Query: 562 LPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEA 621
+P LE E M +L D++ L + + +++++V +L+TT H+ FPV+ E +E
Sbjct: 585 VPLLEWETEVEMDKLRASDIMEPNLTYVYPHTRIQSLVSILRTTVHHAFPVVTENRGNEK 644
Query: 622 PVLFG 626
+ G
Sbjct: 645 EFMKG 649
Score = 64.3 bits (155), Expect = 1e-09
Identities = 44/129 (34%), Positives = 68/129 (52%), Gaps = 11/129 (8%)
Query: 622 PVLF-GIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFS--ADDFAKRGSDRGERIEDIHL 678
P+ F G+ILR L+TLL + V S + ++ A+D+ R I D+ L
Sbjct: 738 PLTFHGLILRSQLVTLLVRGVCYSESQSSASQPRLSYAEMAEDYP-----RFPDIHDLDL 792
Query: 679 TQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILT 738
T M +D+ P+ N SP+TV + + + LFR +GLRHL P + + +VGI+T
Sbjct: 793 TLLNPRMIVDVTPYMNPSPFTVSPNTHVSQVFNLFRTMGLRHL---PVVNAVGEIVGIVT 849
Query: 739 RHDFTAEHI 747
RH+ T E +
Sbjct: 850 RHNLTYEFL 858
>CLC3_HUMAN (P51790) Chloride channel protein 3 (ClC-3)
Length = 762
Score = 197 bits (502), Expect = 7e-50
Identities = 173/659 (26%), Positives = 290/659 (43%), Gaps = 98/659 (14%)
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
+ + ++ + L+ A + + AP A GSGI E+K L+G G TL++K
Sbjct: 149 YIMNYIMYIFWALSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKT 208
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
I + AV+S L +GK GP++H C + ++ + +Y N+ +R+++
Sbjct: 209 ITLVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSA 258
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
SAAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++
Sbjct: 259 ASAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------N 309
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINE 353
FG L++F + Y + P + +LGV GG+ G+ + N W + + +
Sbjct: 310 PFGNSRLVLFYVEYHTPWYLFELFPFI-LLGVFGGLWGAFFIRANIAWCRRRK-----ST 363
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKF--QCPPG 411
K +L +++ T+ + F P+ + S + K+ C P
Sbjct: 364 KFGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPL 408
Query: 412 HYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMF---IFFITGFFLSILSCGVVVPAGV 468
+ L + N I + YS+++ + I +++ + G+ VP+G+
Sbjct: 409 ESSSLCDYRNDMNASKIVDDIPDRPAGIGVYSAIWQLCLALIFKIIMTVFTFGIKVPSGL 468
Query: 469 FVPIIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGS 510
F+P + GA GR VG+ + + +H GLYA++GAA+ LGG
Sbjct: 469 FIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLGGV 528
Query: 511 MRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHA 569
R TVSL VI+ ELT L + +M ++ SK V D F IY+ ++ G P+L+
Sbjct: 529 TRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDAKE 588
Query: 570 EPYMRQLTVGDVVTGPLQ-------IFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAP 622
E T+ V P + + V ++ ++ TS+NGFPVI E+
Sbjct: 589 EFEFTHTTLAADVMRPRRNDPPLAVLTQDNMTVDDIENMINETSYNGFPVIMS---KESQ 645
Query: 623 VLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEE 682
L G LR L + SA G V GS R + E
Sbjct: 646 RLVGFALRRDLTIAIE-----SARKKQEGIV------------GSSRVCFAQHTPSLPAE 688
Query: 683 MDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHD 741
+ L + SP+TV + + + +FR+LGLR LV ++GI+T+ D
Sbjct: 689 SPRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLV----THNGRLLGIITKKD 743
>CLC3_CAVPO (Q9R279) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 197 bits (502), Expect = 7e-50
Identities = 174/658 (26%), Positives = 291/658 (43%), Gaps = 98/658 (14%)
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
+ + ++ + L+ A + + AP A GSGI E+K L+G G TL++K
Sbjct: 149 YIMNYIMYIFWALSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKT 208
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
I + AV+S L +GK GP++H C + ++ + +Y N+ +R+++
Sbjct: 209 ITLVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSA 258
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
SAAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++
Sbjct: 259 ASAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------N 309
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINE 353
FG L++F + Y + P + +LGV GG+ G+ + N W + + +
Sbjct: 310 PFGNSRLVLFYVEYHTPWYLFELFPFI-LLGVFGGLWGAFFIRANIAWCRRRK-----ST 363
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKF--QCPPG 411
K +L +++ T+ + F P+ + S + K+ C P
Sbjct: 364 KFGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPL 408
Query: 412 HYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMF---IFFITGFFLSILSCGVVVPAGV 468
+ L + N I + YS+++ + I +++ + G+ VP+G+
Sbjct: 409 ESSSLCDYRNDMNASKIVDDIPDRPAGVGVYSAIWQLCLALIFKIIMTVFTFGIKVPSGL 468
Query: 469 FVPIIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGS 510
F+P + GA GR VG+ + + +H GLYA++GAA+ LGG
Sbjct: 469 FIPSMAIGAIAGRIVGIAVEQLAYFHHDWFIFKEWCEVGADCITPGLYAMVGAAACLGGV 528
Query: 511 MRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHA 569
R TVSL VI+ ELT L + +M ++ SK V D F IY+ ++ G P+L+
Sbjct: 529 TRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA-K 587
Query: 570 EPYMRQLTVGDVV-----TGPLQIF-HGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPV 623
E + DV+ PL + V ++ ++ TS+NGFPVI E+
Sbjct: 588 EEFTHTTLAADVMRPRRNDPPLAVLTQDNMTVDDIENMINETSYNGFPVIMS---KESQR 644
Query: 624 LFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEM 683
L G LR L + SA G V GS R + E
Sbjct: 645 LVGFALRRDLTIAIE-----SARKKQEGIV------------GSSRVCFAQHTPSLPAES 687
Query: 684 DMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHD 741
+ L + SP+TV + + + +FR+LGLR LV ++GI+T+ D
Sbjct: 688 PRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLV----THNGRLLGIITKKD 741
>CLC3_MOUSE (P51791) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 197 bits (501), Expect = 9e-50
Identities = 174/658 (26%), Positives = 291/658 (43%), Gaps = 98/658 (14%)
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
+ + ++ + L+ A + + AP A GSGI E+K L+G G TL++K
Sbjct: 149 YIMNYIMYIFWALSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKT 208
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
I + AV+S L +GK GP++H C + ++ + +Y N+ +R+++
Sbjct: 209 ITLVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSA 258
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
SAAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++
Sbjct: 259 ASAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------N 309
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINE 353
FG L++F + Y + P + +LGV GG+ G+ + N W + + +
Sbjct: 310 PFGNSRLVLFYVEYHTPWYLFELFPFI-LLGVFGGLWGAFFIRANIAWCRRRK-----ST 363
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKF--QCPPG 411
K +L +++ T+ + F P+ + S + K+ C P
Sbjct: 364 KFGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPL 408
Query: 412 HYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMF---IFFITGFFLSILSCGVVVPAGV 468
+ L + N I + YS+++ + I +++ + G+ VP+G+
Sbjct: 409 ESSSLCDYRNDMNASKIVDDIPDRPAGVGVYSAIWQLCLALIFKIIMTVFTFGIKVPSGL 468
Query: 469 FVPIIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGS 510
F+P + GA GR VG+ + + +H GLYA++GAA+ LGG
Sbjct: 469 FIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLGGV 528
Query: 511 MRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHA 569
R TVSL VI+ ELT L + +M ++ SK V D F IY+ ++ G P+L+
Sbjct: 529 TRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA-K 587
Query: 570 EPYMRQLTVGDVV-----TGPLQIF-HGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPV 623
E + DV+ PL + V ++ ++ TS+NGFPVI E+
Sbjct: 588 EEFTHTTLAADVMRPRRSDPPLAVLTQDNMTVDDIENMINETSYNGFPVIMS---KESQR 644
Query: 624 LFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEM 683
L G LR L + SA G V GS R + E
Sbjct: 645 LVGFALRRDLTIAIE-----SARKKQEGIV------------GSSRVCFAQHTPSLPAES 687
Query: 684 DMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHD 741
+ L + SP+TV + + + +FR+LGLR LV ++GI+T+ D
Sbjct: 688 PRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLV----THNGRLLGIITKKD 741
>CLC3_RAT (P51792) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 197 bits (500), Expect = 1e-49
Identities = 174/658 (26%), Positives = 291/658 (43%), Gaps = 98/658 (14%)
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
+ + ++ + L+ A + + AP A GSGI E+K L+G G TL++K
Sbjct: 149 YIMNYIMYIFWALSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKT 208
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
I + AV+S L +GK GP++H C + ++ + +Y N+ +R+++
Sbjct: 209 ITLVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSA 258
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
SAAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++
Sbjct: 259 ASAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------N 309
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINE 353
FG L++F + Y + P + +LGV GG+ G+ + N W + + +
Sbjct: 310 PFGNSRLVLFYVEYHTPWYLFELFPFI-LLGVFGGLWGAFFIRANIAWCRRRK-----ST 363
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKF--QCPPG 411
K +L +++ T+ + F P+ + S + K+ C P
Sbjct: 364 KFGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPL 408
Query: 412 HYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMF---IFFITGFFLSILSCGVVVPAGV 468
+ L + N I + YS+++ + I +++ + G+ VP+G+
Sbjct: 409 ESSSLCDYRNDMNASKIVDDIPDRPAGVGVYSAIWQLCLALIFKIIMTVFTFGIKVPSGL 468
Query: 469 FVPIIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGS 510
F+P + GA GR VG+ + + +H GLYA++GAA+ LGG
Sbjct: 469 FIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLGGV 528
Query: 511 MRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHA 569
R TVSL VI+ ELT L + +M ++ SK V D F IY+ ++ G P+L+
Sbjct: 529 TRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA-K 587
Query: 570 EPYMRQLTVGDVV-----TGPLQIF-HGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPV 623
E + DV+ PL + V ++ ++ TS+NGFPVI E+
Sbjct: 588 EEFTHTTLAADVMRPRRSDPPLAVLTQDNMTVDDIENMINETSYNGFPVIMS---KESQR 644
Query: 624 LFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEM 683
L G LR L + SA G V GS R + E
Sbjct: 645 LVGFALRRDLTIAIE-----SARKKQEGVV------------GSSRVCFAQHTPSLPAES 687
Query: 684 DMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHD 741
+ L + SP+TV + + + +FR+LGLR LV ++GI+T+ D
Sbjct: 688 PRPLKLRSILDMSPFTVTDHTPMEIVVDVFRKLGLRQCLV----THNGRLLGIITKKD 741
>CLC3_RABIT (O18894) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 197 bits (500), Expect = 1e-49
Identities = 174/658 (26%), Positives = 291/658 (43%), Gaps = 98/658 (14%)
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
+ + ++ + L+ A + + AP A GSGI E+K L+G G TL++K
Sbjct: 149 YIMNYIMYIFWALSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKT 208
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
I + AV+S L +GK GP++H C + ++ + +Y N+ +R+++
Sbjct: 209 ITLVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSA 258
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
SAAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++
Sbjct: 259 ASAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------N 309
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINE 353
FG L++F + Y + P + +LGV GG+ G+ + N W + + +
Sbjct: 310 PFGNSRLVLFYVEYHTPWYLFELFPFI-LLGVFGGLWGAFFIRANIAWCRRRK-----ST 363
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKF--QCPPG 411
K +L +++ T+ + F P+ + S + K+ C P
Sbjct: 364 KFGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPL 408
Query: 412 HYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMF---IFFITGFFLSILSCGVVVPAGV 468
+ L + N I + YS+++ + I +++ + G+ VP+G+
Sbjct: 409 ESSSLCDYRNDMNASKIVDDIPDRPAGIGVYSAIWQLCLALIFKIIMTVFTFGIKVPSGL 468
Query: 469 FVPIIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGS 510
F+P + GA GR VG+ + + +H GLYA++GAA+ LGG
Sbjct: 469 FIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLGGV 528
Query: 511 MRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHA 569
R TVSL VI+ ELT L + +M ++ SK V D F IY+ ++ G P+L+
Sbjct: 529 TRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA-K 587
Query: 570 EPYMRQLTVGDVV-----TGPLQIF-HGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPV 623
E + DV+ PL + V ++ ++ TS+NGFPVI E+
Sbjct: 588 EEFTHTTLAADVMRPRRNDPPLAVLTQDNMTVDDIENMINETSYNGFPVIMS---KESQR 644
Query: 624 LFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEM 683
L G LR L + SA G V GS R + E
Sbjct: 645 LVGFALRRDLTIAIE-----SARKKQEGIV------------GSSRVCFAQHTPSLPAES 687
Query: 684 DMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHD 741
+ L + SP+TV + + + +FR+LGLR LV ++GI+T+ D
Sbjct: 688 PRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLV----THNGRLLGIITKKD 741
>CLC4_HUMAN (P51793) Chloride channel protein 4 (ClC-4)
Length = 760
Score = 192 bits (488), Expect = 3e-48
Identities = 179/663 (26%), Positives = 291/663 (42%), Gaps = 108/663 (16%)
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
+ L ++ + L A + + AP A GSGI E+K L+G G TL++K
Sbjct: 149 YILNYLMYILWALLFAFLAVSLVRVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLLIKT 208
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
+ + VSS L +GK GP++H C SK Y KN+ RR+++
Sbjct: 209 VTLVLVVSSGLSLGKEGPLVHVACCCGNFFSSLFSK----------YSKNEGKRREVLSA 258
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
+AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++
Sbjct: 259 AAAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFTLRSI---------N 309
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLW---NKVLRIYNA 350
FG L++F + Y A+ P + +LGV GG+ G+L+ N W K R+
Sbjct: 310 PFGNSRLVLFYVEYHTPWYMAELFPFI-LLGVFGGLWGTLFIRCNIAWCRRRKTTRLGK- 367
Query: 351 INEKGTICKILLACVISMFTSCLLFGLPWL-ASCQPCPADPVEPCPTIGRSGIYKKFQCP 409
+L V++ T+ + + P+ S ++ C + S + P
Sbjct: 368 -------YPVLEVIVVTAITAIIAYPNPYTRQSTSELISELFNDCGALESSQLCDYINDP 420
Query: 410 PGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVF 469
++ +D R + +Q + IF I ++I + G+ +P+G+F
Sbjct: 421 --------NMTRPVDDIPDRPAGVGVYTAMWQLALALIFKIV---VTIFTFGMKIPSGLF 469
Query: 470 VPIIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGSM 511
+P + GA GR VG+ + + +H GLYA++GAA+ LGG
Sbjct: 470 IPSMAVGAIAGRMVGIGVEQLAYHHHDWIIFRNWCRPGADCVTPGLYAMVGAAACLGGVT 529
Query: 512 RTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHAE 570
R TVSL VI+ ELT L + +M + SK VAD F IY+ + G P+L+ E
Sbjct: 530 RMTVSLVVIMFELTGGLEYIVPLMAAAVTSKWVADAFGKEGIYEAHIHLNGYPFLDVKDE 589
Query: 571 PYMRQLTVGDVV-----TGPLQIF-HGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVL 624
R L DV+ PL + V +V ++K T +NGFPV+ ++ L
Sbjct: 590 FTHRTLAT-DVMRPRRGEPPLSVLTQDSMTVEDVETLIKETDYNGFPVVVS---RDSERL 645
Query: 625 FGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMD 684
G R L+ ++ +A G V ++ T+E +
Sbjct: 646 IGFAQRRELILAIK-----NARQRQEGIVSNSI------------------MYFTEEPPE 682
Query: 685 M------FIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILT 738
+ + L N SP+TV + + + +FR+LGLR LV ++GI+T
Sbjct: 683 LPANSPHPLKLRRILNLSPFTVTDHTPMETVVDIFRKLGLRQCLV----TRSGRLLGIIT 738
Query: 739 RHD 741
+ D
Sbjct: 739 KKD 741
>CLC5_RAT (P51796) Chloride channel protein 5 (ClC-5)
Length = 746
Score = 191 bits (486), Expect = 5e-48
Identities = 179/661 (27%), Positives = 297/661 (44%), Gaps = 112/661 (16%)
Query: 122 VTFFASNLALTLFATIITAMI---APAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIG 178
V +F L LFA + +++ AP A GSGI E+K L+G G TL++K I
Sbjct: 138 VNYFMYVLWALLFAFLAVSLVKAFAPYACGSGIPEIKTILSGFIIRGYLGKWTLVIKTIT 197
Query: 179 SITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGS 238
+ AVSS L +GK GP++H C +L +K Y KN+ RR+++ +
Sbjct: 198 LVLAVSSGLSLGKEGPLVHVACCCGNILCHCFNK----------YRKNEAKRREVLSAAA 247
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++ F
Sbjct: 248 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFTLRSI---------NPF 298
Query: 299 GKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINEKG 355
G L++F + + + VP + +LG+ GG+ G+L+ N W + + K
Sbjct: 299 GNSRLVLFYVEFHTPWHLFELVPFI-VLGIFGGLWGALFIRTNIAWCRKRKTTQL--GKY 355
Query: 356 TICKILLACVISMFTSCLLFGLPWL-ASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYN 414
+ ++L +++ T+ L F + S ++ C + S + C ++
Sbjct: 356 PVVEVL---IVTAITAILAFPNEYTRMSTSELISELFNDCGLLDSSKL-----CDYENH- 406
Query: 415 DLASLIFNTNDDAIRNLFSKNTDSEFQYSSMF---IFFITGFFLSILSCGVVVPAGVFVP 471
FNT+ YS+M+ + I ++I + G+ +P+G+F+P
Sbjct: 407 ------FNTSKG---GELPDRPAGVGVYSAMWQLALTLILKIVITIFTFGMKIPSGLFIP 457
Query: 472 IIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGSMRT 513
+ GA GR +G+ + + +H GLYA++GAA+ LGG R
Sbjct: 458 SMAVGAIAGRLLGVGMEQLAYYHHDWGIFNSWCSQGADCITPGLYAMVGAAACLGGVTRM 517
Query: 514 TVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHAEPY 572
TVSL VI+ ELT L + +M + SK VAD IYD ++ G P+LE E +
Sbjct: 518 TVSLVVIMFELTGGLEYIVPLMAAAMTSKWVADALGREGIYDAHIRLNGYPFLEA-KEEF 576
Query: 573 MRQLTVGDVV----TGPL--QIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFG 626
+ DV+ PL + V +V ++ T+++GFPV+ E+ L G
Sbjct: 577 AHKTLAMDVMKPRRNDPLLTVLTQDSMTVEDVETIISETTYSGFPVVVS---RESQRLVG 633
Query: 627 IILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDM- 685
+LR L+ + A++ D I+ T+ M
Sbjct: 634 FVLRRDLIISIEN-----------------------ARKKQDGVVSTSIIYFTEHSPPMP 670
Query: 686 -----FIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRH 740
+ L + SP+TV + + + +FR+LGLR LV ++GI+T+
Sbjct: 671 PYTPPTLKLRNILDLSPFTVTDLTPMEIVVDIFRKLGLRQCLV----THNGRLLGIITKK 726
Query: 741 D 741
D
Sbjct: 727 D 727
>CLC5_MOUSE (Q9WVD4) Chloride channel protein 5 (ClC-5)
Length = 746
Score = 191 bits (485), Expect = 6e-48
Identities = 179/661 (27%), Positives = 297/661 (44%), Gaps = 112/661 (16%)
Query: 122 VTFFASNLALTLFATIITAMI---APAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIG 178
V +F L LFA + +++ AP A GSGI E+K L+G G TL++K I
Sbjct: 138 VNYFMYVLWALLFAFLAVSLVKAFAPYACGSGIPEIKTILSGFIIRGYLGKWTLVIKTIT 197
Query: 179 SITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGS 238
+ AVSS L +GK GP++H C +L +K Y KN+ RR+++ +
Sbjct: 198 LVLAVSSGLSLGKEGPLVHVACCCGNILCHCFNK----------YRKNEAKRREVLSAAA 247
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++ F
Sbjct: 248 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFTLRSI---------NPF 298
Query: 299 GKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINEKG 355
G L++F + + + VP + +LG+ GG+ G+L+ N W + + K
Sbjct: 299 GNSRLVLFYVEFHTPWHLFELVPFI-VLGIFGGLWGALFIRTNIAWCRKRKTTQL--GKY 355
Query: 356 TICKILLACVISMFTSCLLFGLPWL-ASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYN 414
+ ++L +++ T+ L F + S ++ C + S + C ++
Sbjct: 356 PVVEVL---IVTAITAILAFPNEYTRMSTSELISELFNDCGLLDSSKL-----CDYENH- 406
Query: 415 DLASLIFNTNDDAIRNLFSKNTDSEFQYSSMF---IFFITGFFLSILSCGVVVPAGVFVP 471
FNT+ YS+M+ + I ++I + G+ +P+G+F+P
Sbjct: 407 ------FNTSKG---GELPDRPAGVGIYSAMWQLALTLILKIVITIFTFGMKIPSGLFIP 457
Query: 472 IIVTGASYGRFVGMLFGKKSNLNH------------------GLYAVLGAASFLGGSMRT 513
+ GA GR +G+ + + +H GLYA++GAA+ LGG R
Sbjct: 458 SMAVGAIAGRLLGVGMEQLAYYHHDWGIFNSWCSQGADCITPGLYAMVGAAACLGGVTRM 517
Query: 514 TVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYLETHAEPY 572
TVSL VI+ ELT L + +M + SK VAD IYD ++ G P+LE E +
Sbjct: 518 TVSLVVIMFELTGGLEYIVPLMAAAMTSKWVADALGREGIYDAHIRLNGYPFLEA-KEEF 576
Query: 573 MRQLTVGDVV----TGPL--QIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFG 626
+ DV+ PL + V +V ++ T+++GFPV+ E+ L G
Sbjct: 577 AHKTLAMDVMKPRRNDPLLTVLTQDSMTVEDVETIISETTYSGFPVVVS---RESQRLVG 633
Query: 627 IILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDM- 685
+LR L+ + A++ D I+ T+ M
Sbjct: 634 FVLRRDLIISIEN-----------------------ARKKQDGVVSTSIIYFTEHSPPMP 670
Query: 686 -----FIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRH 740
+ L + SP+TV + + + +FR+LGLR LV ++GI+T+
Sbjct: 671 PYTPPTLKLRNILDLSPFTVTDLTPMEIVVDIFRKLGLRQCLV----THNGRLLGIITKK 726
Query: 741 D 741
D
Sbjct: 727 D 727
>CLC5_HUMAN (P51795) Chloride channel protein 5 (ClC-5)
Length = 746
Score = 187 bits (476), Expect = 7e-47
Identities = 181/668 (27%), Positives = 300/668 (44%), Gaps = 126/668 (18%)
Query: 122 VTFFASNLALTLFATIITAMI---APAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIG 178
V +F L LFA + +++ AP A GSGI E+K L+G G TL++K I
Sbjct: 138 VNYFMYVLWALLFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLVIKTIT 197
Query: 179 SITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGS 238
+ AVSS L +GK GP++H C +L +K Y KN+ RR+++ +
Sbjct: 198 LVLAVSSGLSLGKEGPLVHVACCCGNILCHCFNK----------YRKNEAKRREVLSAAA 247
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF + A LR++ F
Sbjct: 248 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFTLRSI---------NPF 298
Query: 299 GKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLY---NYLWNKVLRIYNAINEKG 355
G L++F + + + VP + +LG+ GG+ G+L+ N W + + K
Sbjct: 299 GNSRLVLFYVEFHTPWHLFELVPFI-LLGIFGGLWGALFIRTNIAWCRKRKTTQL--GKY 355
Query: 356 TICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYND 415
+ ++L V++ T+ L F P E + S + + +ND
Sbjct: 356 PVIEVL---VVTAITAILAF--------------PNE-YTRMSTSELISEL------FND 391
Query: 416 LASLIFNTNDDAIRNLFSKNTDSEFQ--------YSSMF---IFFITGFFLSILSCGVVV 464
L+ ++ N F+ + E YS+M+ + I ++I + G+ +
Sbjct: 392 -CGLLDSSKLCDYENRFNTSKGGELPDRPAGVGVYSAMWQLALTLILKIVITIFTFGMKI 450
Query: 465 PAGVFVPIIVTGASYGRFVGM-------------LFGKKSN-----LNHGLYAVLGAASF 506
P+G+F+P + GA GR +G+ +F + + GLYA++GAA+
Sbjct: 451 PSGLFIPSMAVGAIAGRLLGVGMEQLAYYHQEWTVFNSWCSQGADCITPGLYAMVGAAAC 510
Query: 507 LGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF-NANIYDLIMKSKGLPYL 565
LGG R TVSL VI+ ELT L + +M + SK VAD IYD ++ G P+L
Sbjct: 511 LGGVTRMTVSLVVIMFELTGGLEYIVPLMAAAMTSKWVADALGREGIYDAHIRLNGYPFL 570
Query: 566 ETHAEPYMRQLTVGDVV----TGPL--QIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFS 619
E E + + DV+ PL + V +V ++ T+++GFPV+
Sbjct: 571 EA-KEEFAHKTLAMDVMKPRRNDPLLTVLTQDSMTVEDVETIISETTYSGFPVVVS---R 626
Query: 620 EAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLT 679
E+ L G +LR L+ + A++ D I+ T
Sbjct: 627 ESQRLVGFVLRRDLIISIEN-----------------------ARKKQDGVVSTSIIYFT 663
Query: 680 QEEMDM------FIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPV 733
+ + + L + SP+TV + + + +FR+LGLR LV +
Sbjct: 664 EHSPPLPPYTPPTLKLRNILDLSPFTVTDLTPMEIVVDIFRKLGLRQCLV----THNGRL 719
Query: 734 VGILTRHD 741
+GI+T+ D
Sbjct: 720 LGIITKKD 727
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.142 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 92,207,670
Number of Sequences: 164201
Number of extensions: 4046209
Number of successful extensions: 12299
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 11854
Number of HSP's gapped (non-prelim): 202
length of query: 780
length of database: 59,974,054
effective HSP length: 118
effective length of query: 662
effective length of database: 40,598,336
effective search space: 26876098432
effective search space used: 26876098432
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0030.6


