

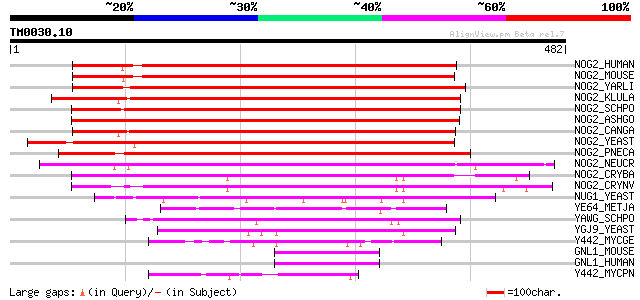
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.10
(482 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NOG2_HUMAN (Q13823) Nucleolar GTP-binding protein 2 (Autoantigen... 408 e-113
NOG2_MOUSE (Q99LH1) Nucleolar GTP-binding protein 2 407 e-113
NOG2_YARLI (Q6C036) Nucleolar GTP-binding protein 2 371 e-102
NOG2_KLULA (Q6CSP9) Nucleolar GTP-binding protein 2 367 e-101
NOG2_SCHPO (O14236) Nucleolar GTP-binding protein 2 365 e-100
NOG2_ASHGO (Q75DA4) Nucleolar GTP-binding protein 2 364 e-100
NOG2_CANGA (Q6FWS1) Nucleolar GTP-binding protein 2 363 e-100
NOG2_YEAST (P53742) Nucleolar GTP-binding protein 2 363 e-100
NOG2_PNECA (Q9C3Z4) Nucleolar GTP-binding protein 2 (Binding-ind... 353 5e-97
NOG2_NEUCR (Q7SHR8) Nucleolar GTP-binding protein 2 344 3e-94
NOG2_CRYBA (Q6TGJ8) Nucleolar GTP-binding protein 2 343 7e-94
NOG2_CRYNV (Q8J109) Nucleolar GTP-binding protein 2 333 4e-91
NUG1_YEAST (P40010) Nuclear GTP-binding protein NUG1 (Nuclear GT... 139 1e-32
YE64_METJA (Q58859) Hypothetical GTP-binding protein MJ1464 130 8e-30
YAWG_SCHPO (Q10190) Hypothetical GTP-binding protein C3F10.16c i... 122 2e-27
YGJ9_YEAST (P53145) Hypothetical GTP-binding protein in SEH1-PRP... 119 2e-26
Y442_MYCGE (Q49435) Hypothetical protein MG442 67 1e-10
GNL1_MOUSE (P36916) Guanine nucleotide-binding protein-like 1 (G... 65 4e-10
GNL1_HUMAN (P36915) Guanine nucleotide-binding protein-like 1 (G... 65 5e-10
Y442_MYCPN (P75135) Hypothetical protein MG442 homolog (K05_orf271) 64 9e-10
>NOG2_HUMAN (Q13823) Nucleolar GTP-binding protein 2 (Autoantigen
NGP-1)
Length = 731
Score = 408 bits (1049), Expect = e-113
Identities = 203/336 (60%), Positives = 248/336 (73%), Gaps = 9/336 (2%)
Query: 55 FTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAG--ASGTAEANEADGFIDL 112
F FGPK++RKRP L A D QSL + A ++ E++++ T NEA
Sbjct: 140 FETTFGPKSQRKRPNLFASDMQSLIENAEMSTESYDQGKDRDLVTEDTGVRNEAQ----- 194
Query: 113 LGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMML 172
+++KGQ+K I GELYKVIDSSDVVVQVLDARDP GTR H+E +LK+ +KH++
Sbjct: 195 --EEIYKKGQSKRIWGELYKVIDSSDVVVQVLDARDPMGTRSPHIETYLKKEKPWKHLIF 252
Query: 173 LLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAIS 232
+LNKCDL+P WATK W+ VLS++YPTLAFHAS+ FGKG+ + +LRQF +L +DK+ IS
Sbjct: 253 VLNKCDLVPTWATKRWVAVLSQDYPTLAFHASLTNPFGKGAFIQLLRQFGKLHTDKKQIS 312
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNKDS 292
VGFIGYPNVGKSSVINTLR+K VC VAPI GETKVWQYITL +RIFLIDCPGVVY ++DS
Sbjct: 313 VGFIGYPNVGKSSVINTLRSKKVCNVAPIAGETKVWQYITLMRRIFLIDCPGVVYPSEDS 372
Query: 293 EADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKL 352
E D+VLKGVV+V +K D IG VL+R E++ + YKI WEN DFL +L TGKL
Sbjct: 373 ETDIVLKGVVQVEKIKSPEDHIGAVLERAKPEYISKTYKIDSWENAEDFLEKLAFRTGKL 432
Query: 353 LKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDL 388
LKGGEPDL T KMVL+DWQRG+IPFFV PP E L
Sbjct: 433 LKGGEPDLQTVGKMVLNDWQRGRIPFFVKPPNAEPL 468
>NOG2_MOUSE (Q99LH1) Nucleolar GTP-binding protein 2
Length = 728
Score = 407 bits (1047), Expect = e-113
Identities = 201/334 (60%), Positives = 250/334 (74%), Gaps = 9/334 (2%)
Query: 55 FTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGA--SGTAEANEADGFIDL 112
F FGPK++RKRP L A D QSL + A ++ E++++ T NEA
Sbjct: 140 FESTFGPKSQRKRPNLFASDMQSLLENAEMSTESYDQGKDRDLVMEDTGVRNEAQ----- 194
Query: 113 LGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMML 172
+++KGQ+K I GELYKVIDSSDVVVQVLDARDP GTR H+E +LK+ +KH++
Sbjct: 195 --EEIYKKGQSKRIWGELYKVIDSSDVVVQVLDARDPMGTRSPHIEAYLKKEKPWKHLIF 252
Query: 173 LLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAIS 232
+LNKCDL+P WATK W+ VLS++YPTLAFHAS+ FGKG+ + +LRQF +L +DK+ IS
Sbjct: 253 VLNKCDLVPTWATKRWVAVLSQDYPTLAFHASLTNPFGKGAFIQLLRQFGKLHTDKKQIS 312
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNKDS 292
VGFIGYPNVGKSSVINTLR+K VC VAPI GETKVWQYITL +RIFLIDCPGVVY ++DS
Sbjct: 313 VGFIGYPNVGKSSVINTLRSKKVCNVAPIAGETKVWQYITLMRRIFLIDCPGVVYPSEDS 372
Query: 293 EADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKL 352
E D+VLKGVV+V +K D IG VL+R E++ + YKI+ WEN DFL +L TGKL
Sbjct: 373 ETDIVLKGVVQVEKIKAPQDHIGAVLERAKPEYISKTYKIESWENAEDFLEKLALRTGKL 432
Query: 353 LKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQE 386
LKGGEPD++T +KMVL+DWQRG+IPFFV PP E
Sbjct: 433 LKGGEPDMLTVSKMVLNDWQRGRIPFFVKPPNAE 466
>NOG2_YARLI (Q6C036) Nucleolar GTP-binding protein 2
Length = 509
Score = 371 bits (952), Expect = e-102
Identities = 183/343 (53%), Positives = 236/343 (68%), Gaps = 6/343 (1%)
Query: 55 FTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGASGTAEANEADGFIDLLG 114
+ FGPK +RK+P L D+ LA A +Q+ FE K S + DG+
Sbjct: 137 YANTFGPKAQRKKPQLAVGDFAELASAADESQQDFEAKKEEDNSW-----KVDGWSQEAK 191
Query: 115 HTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLL 174
+F KGQ+K I ELYKVIDSSDVV+ VLDARDP GTRC +E+++K+ +KH++ +L
Sbjct: 192 EAIFHKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCTSVEQYIKKEAPHKHLIFVL 251
Query: 175 NKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAISVG 234
NKCDL+P W W++ LS++YPTLAFHASI SFGKGSL+ +LRQ++ L D+Q ISVG
Sbjct: 252 NKCDLVPTWVAAAWVKHLSQDYPTLAFHASITNSFGKGSLIQLLRQYSALHPDRQQISVG 311
Query: 235 FIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVV-YQNKDSE 293
FIGYPN GKSS+INTLR K VCK APIPGETKVWQYITL KRIFLIDCPG+V KDSE
Sbjct: 312 FIGYPNTGKSSIINTLRKKKVCKTAPIPGETKVWQYITLMKRIFLIDCPGIVPPSQKDSE 371
Query: 294 ADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKLL 353
D++ +GVVRV ++ I +L+R +HL R Y++ W N +FL ++ + G+LL
Sbjct: 372 TDILFRGVVRVEHVSYPEQYIPALLERCETKHLERTYEVSGWSNATEFLEKIARKHGRLL 431
Query: 354 KGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSEEPIVNG 396
KGGEPD AK++L+D+ RGKIP+FVPPPQ ED+ G
Sbjct: 432 KGGEPDESGIAKLILNDFNRGKIPWFVPPPQAEDMENVKFAAG 474
>NOG2_KLULA (Q6CSP9) Nucleolar GTP-binding protein 2
Length = 513
Score = 367 bits (943), Expect = e-101
Identities = 183/360 (50%), Positives = 239/360 (65%), Gaps = 7/360 (1%)
Query: 37 NGNNDAKMEQSNSRVQRPFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKY--- 93
N + A+ + PF FGPK +RK+P + A + L + FEEK
Sbjct: 119 NEKDSAESPTAKILETEPFEQTFGPKAQRKKPRIAASSLEELISSTSTDNKTFEEKQELD 178
Query: 94 -AAGASGTAEANEADGFIDLLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGT 152
G G E E DG+ +F KGQ+K I ELYKVIDSSDVV+ VLDARDP GT
Sbjct: 179 STLGLMGKQE--EEDGWTQAAKEAIFHKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGT 236
Query: 153 RCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKG 212
RC + ++ +KH++ +LNKCDL+P W W++ LSKE PTLAFHASI SFGKG
Sbjct: 237 RCKSVTDYMTNETPHKHLIYVLNKCDLVPTWVAAAWVKHLSKERPTLAFHASITNSFGKG 296
Query: 213 SLLSVLRQFARLKSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYIT 272
SL+ +LRQF++L D+ ISVGFIGYPN GKSS+INTLR K VC+VAPIPGETKVWQYIT
Sbjct: 297 SLIQLLRQFSQLHKDRHQISVGFIGYPNTGKSSIINTLRKKKVCQVAPIPGETKVWQYIT 356
Query: 273 LTKRIFLIDCPGVV-YQNKDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYK 331
L KRIFLIDCPG+V +KDSE D++ +GVVRV ++ I +LKR ++HL R Y+
Sbjct: 357 LMKRIFLIDCPGIVPPSSKDSEEDILFRGVVRVEHVSHPEQYIPGILKRCKRQHLERTYE 416
Query: 332 IKEWENENDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSEE 391
I W++ DF+ + + G+LLKGGEPD +K +L+D+ RGKIP+FVPPP+++ + E+
Sbjct: 417 ISGWKDSVDFIEMIARKQGRLLKGGEPDESGVSKQILNDFNRGKIPWFVPPPEKDKIEEK 476
>NOG2_SCHPO (O14236) Nucleolar GTP-binding protein 2
Length = 537
Score = 365 bits (937), Expect = e-100
Identities = 185/339 (54%), Positives = 238/339 (69%), Gaps = 3/339 (0%)
Query: 54 PFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGASGTAEANEADGFIDLL 113
PF FGPK++RKRP + LAK++ Q A+EEK A +E+D +
Sbjct: 136 PFENTFGPKSQRKRPKISFDSVAELAKESDEKQNAYEEKIEERI--LANPDESDDVMLAA 193
Query: 114 GHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLL 173
+F KGQ+K I ELYKVIDSSDV++QVLDARDP GTRC +E++L+ +KHM+L+
Sbjct: 194 RDAIFSKGQSKRIWNELYKVIDSSDVLIQVLDARDPVGTRCGTVERYLRNEASHKHMILV 253
Query: 174 LNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAISV 233
LNK DL+P W+++L+KEYPT+AFHASIN SFGKGSL+ +LRQFA L SDK+ ISV
Sbjct: 254 LNKVDLVPTSVAAAWVKILAKEYPTIAFHASINNSFGKGSLIQILRQFASLHSDKKQISV 313
Query: 234 GFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVV-YQNKDS 292
G IG+PN GKSS+INTLR K VC VAPIPGETKVWQY+ L KRIFLIDCPG+V + DS
Sbjct: 314 GLIGFPNAGKSSIINTLRKKKVCNVAPIPGETKVWQYVALMKRIFLIDCPGIVPPSSNDS 373
Query: 293 EADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKL 352
+A+++LKGVVRV N+ + I VL R +HL R Y+I W + +FL +L K G+L
Sbjct: 374 DAELLLKGVVRVENVSNPEAYIPTVLSRCKVKHLERTYEISGWNDSTEFLAKLAKKGGRL 433
Query: 353 LKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSEE 391
LKGGEPD + AKMVL+D+ RGKIP+F+ P ++E
Sbjct: 434 LKGGEPDEASVAKMVLNDFMRGKIPWFIGPKGLSSSNDE 472
>NOG2_ASHGO (Q75DA4) Nucleolar GTP-binding protein 2
Length = 502
Score = 364 bits (934), Expect = e-100
Identities = 182/340 (53%), Positives = 237/340 (69%), Gaps = 3/340 (0%)
Query: 54 PFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGAS-GTAEANEA-DGFID 111
P+ FGPK +RK+P + A + LAK + +EEK ++ G A E DG+
Sbjct: 136 PYGATFGPKAQRKKPRVAAASLEDLAKATDSDSQKYEEKKELDSTLGLMAATEQEDGWSQ 195
Query: 112 LLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMM 171
+ +F KGQ+K I ELYKVIDSSDVV+ VLDARDP GTRC +E+++K+ +KH++
Sbjct: 196 VAKEAIFHKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHLI 255
Query: 172 LLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAI 231
+LNKCDL+P W W++ LSK+ PTLAFHASI SFGKGSL+ +LRQF++L D+Q I
Sbjct: 256 YVLNKCDLVPTWLAAAWVKHLSKDRPTLAFHASITNSFGKGSLIQLLRQFSQLHKDRQQI 315
Query: 232 SVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQN-K 290
SVGFIGYPN GKSS+INTLR K VC+VAPIPGETKVWQYITL KRIFLIDCPG+V + K
Sbjct: 316 SVGFIGYPNTGKSSIINTLRKKKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPSAK 375
Query: 291 DSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTG 350
D+E D++ +GVVRV ++ I VL+R + HL R Y+I W + +F+ L + G
Sbjct: 376 DTEEDILFRGVVRVEHVSHPEQYIPAVLRRCKRHHLERTYEISGWADATEFIEMLARKQG 435
Query: 351 KLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSE 390
+LLKGGEPD AK VL+D+ RGKIP+FV PP ++ E
Sbjct: 436 RLLKGGEPDETGVAKQVLNDFNRGKIPWFVSPPDRDPQPE 475
>NOG2_CANGA (Q6FWS1) Nucleolar GTP-binding protein 2
Length = 494
Score = 363 bits (933), Expect = e-100
Identities = 180/338 (53%), Positives = 237/338 (69%), Gaps = 6/338 (1%)
Query: 55 FTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKY----AAGASGTAEANEADGFI 110
F FGPK +RK+P + A + L + + +EEK G G E +E +G+
Sbjct: 137 FDQTFGPKAQRKKPRVAASSLEELVQATEEENKTYEEKEELKATLGLMGKQE-DEENGWT 195
Query: 111 DLLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHM 170
+ +F KGQ+K I ELYKVIDSSDVV+ VLDARDP GTRC +E+++K+ +KH+
Sbjct: 196 QVTKEAIFSKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHL 255
Query: 171 MLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQA 230
+ +LNKCDL+P W W++ LSK+ PTLAFHASI SFGKGSL+ +LRQF++L +D++
Sbjct: 256 IYVLNKCDLVPTWVAAAWVKHLSKDRPTLAFHASITNSFGKGSLIQLLRQFSQLHTDRKQ 315
Query: 231 ISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVV-YQN 289
ISVGFIGYPN GKSS+INTLR K VC+VAPIPGETKVWQYITL KRIFLIDCPG+V
Sbjct: 316 ISVGFIGYPNTGKSSIINTLRKKKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPST 375
Query: 290 KDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLT 349
KDSE D++ +GVVRV ++ I VLKR +HL R Y+I W++ DF+ L +
Sbjct: 376 KDSEEDILFRGVVRVEHVSHPEQYIPGVLKRCQTKHLERTYEISGWKDATDFIEMLARKQ 435
Query: 350 GKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQED 387
G+LLKGGEPD +K +L+D+ RGKIP+FV PP++E+
Sbjct: 436 GRLLKGGEPDESGVSKQILNDFNRGKIPWFVIPPEKEE 473
>NOG2_YEAST (P53742) Nucleolar GTP-binding protein 2
Length = 486
Score = 363 bits (932), Expect = e-100
Identities = 185/375 (49%), Positives = 253/375 (67%), Gaps = 9/375 (2%)
Query: 16 SFSAMVKKKESSGKSKQSLDANGNNDAKMEQSNSRVQRPFTPVFGPKTKRKRPCLLAFDY 75
++ ++++ + + DA+ + A++ + S + FGPK +RKRP L A +
Sbjct: 103 TYQVLLRRNKLPMSLLEEKDADESPKARILDTES-----YADAFGPKAQRKRPRLAASNL 157
Query: 76 QSLAKKAHVAQEAFEEKYAAGASGTAEANEAD---GFIDLLGHTMFQKGQTKHISGELYK 132
+ L K + +EEK A+ N+ D G+ +F KGQ+K I ELYK
Sbjct: 158 EDLVKATNEDITKYEEKQVLDATLGLMGNQEDKENGWTSAAKEAIFSKGQSKRIWNELYK 217
Query: 133 VIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVL 192
VIDSSDVV+ VLDARDP GTRC +E+++K+ +KH++ +LNKCDL+P W W++ L
Sbjct: 218 VIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHLIYVLNKCDLVPTWVAAAWVKHL 277
Query: 193 SKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAISVGFIGYPNVGKSSVINTLRT 252
SKE PTLAFHASI SFGKGSL+ +LRQF++L +D++ ISVGFIGYPN GKSS+INTLR
Sbjct: 278 SKERPTLAFHASITNSFGKGSLIQLLRQFSQLHTDRKQISVGFIGYPNTGKSSIINTLRK 337
Query: 253 KNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVV-YQNKDSEADVVLKGVVRVTNLKDAA 311
K VC+VAPIPGETKVWQYITL KRIFLIDCPG+V +KDSE D++ +GVVRV ++
Sbjct: 338 KKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPSSKDSEEDILFRGVVRVEHVTHPE 397
Query: 312 DRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDW 371
I VLKR +HL R Y+I W++ +F+ L + G+LLKGGEPD +K +L+D+
Sbjct: 398 QYIPGVLKRCQVKHLERTYEISGWKDATEFIEILARKQGRLLKGGEPDESGVSKQILNDF 457
Query: 372 QRGKIPFFVPPPQQE 386
RGKIP+FV PP++E
Sbjct: 458 NRGKIPWFVLPPEKE 472
>NOG2_PNECA (Q9C3Z4) Nucleolar GTP-binding protein 2
(Binding-inducible GTPase)
Length = 483
Score = 353 bits (906), Expect = 5e-97
Identities = 181/360 (50%), Positives = 243/360 (67%), Gaps = 9/360 (2%)
Query: 43 KMEQSNSRVQRPFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGASGTAE 102
K ++N PF FG K+ RKR L A ++L+ A + E + +K +E
Sbjct: 112 KTRKANIIDIEPFDDTFGKKSXRKRAKLYASSIENLSNFAFESYENYIKK-------NSE 164
Query: 103 ANEADGFIDLLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLK 162
D I +F KG +K I ELYK IDSSDV++Q+LDAR+P GTRC H+E++LK
Sbjct: 165 YENVDKNIQKSFEAIFSKGTSKRIWNELYKXIDSSDVIIQLLDARNPLGTRCKHVEEYLK 224
Query: 163 ENCKYKHMMLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFA 222
+ +KHM+LLLNKCDLIP W T+ W++ LSKEYPTLAFHASIN FGKGSL+ +LRQF+
Sbjct: 225 KEKPHKHMILLLNKCDLIPTWCTREWIKQLSKEYPTLAFHASINNPFGKGSLIQLLRQFS 284
Query: 223 RLKSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDC 282
+L S+++ ISVGFIGYPN GKSSVINTLR+K VC APIPGETKVWQY+ +T +IF+IDC
Sbjct: 285 KLHSNRRQISVGFIGYPNTGKSSVINTLRSKKVCNTAPIPGETKVWQYVRMTSKIFMIDC 344
Query: 283 PGVVYQN-KDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENEN-D 340
PG+V N DSE ++++KG +R+ + + I +L +HL R Y+I WEN++
Sbjct: 345 PGIVPPNSNDSETEIIIKGALRIEKVSNPEQYIHAILNLCETKHLERTYQISGWENDSTK 404
Query: 341 FLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSEEPIVNGLDVD 400
F+ L + TGKLLKGGE D + AKMV++D+ RGKIP+F+ P Q+ D S + N V+
Sbjct: 405 FIELLARKTGKLLKGGEVDESSIAKMVINDFIRGKIPWFIAPAQENDPSNIKLSNNTLVE 464
>NOG2_NEUCR (Q7SHR8) Nucleolar GTP-binding protein 2
Length = 619
Score = 344 bits (882), Expect = 3e-94
Identities = 199/469 (42%), Positives = 280/469 (59%), Gaps = 24/469 (5%)
Query: 27 SGKSKQSLDANGNNDA-KMEQSNSRVQ-RPFTPVFGPKTKRKRPCLLAFDYQSLAKKAHV 84
S K SL +G DA K Q+ ++ PF+ FGPK +RKRP L L + +
Sbjct: 109 SNKLPMSLIRDGPKDALKKHQAKMTIESEPFSQTFGPKAQRKRPKLSFNTIGDLTEHSEK 168
Query: 85 AQEAF-----EEKYAAGASGTA------EANEADGFIDLLGHTMFQKGQTKHISGELYKV 133
+ + + E K +GASG + E D + +F KGQ+K I ELYKV
Sbjct: 169 SMDTYQARLEEIKLLSGASGYGGGLADDDVQEEDFSVATAKEAIFTKGQSKRIWNELYKV 228
Query: 134 IDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVLS 193
IDSSDV++ V+DARDP GTRC H+EK+L +KH++ +LNK DL+P+ W+RVL
Sbjct: 229 IDSSDVILHVIDARDPLGTRCRHVEKYLATEAPHKHLIFVLNKIDLVPSKTAAAWIRVLQ 288
Query: 194 KEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAISVGFIGYPNVGKSSVINTLRTK 253
K++PT A +SI FG+GSL+ +LRQF+ L D++ ISVG +GYPNVGKSS+IN LR K
Sbjct: 289 KDHPTCAMRSSIKNPFGRGSLIDLLRQFSILHKDRKQISVGLVGYPNVGKSSIINALRGK 348
Query: 254 NVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNK-DSEADVVLKGVVRVTNLKDAAD 312
V KVAPIPGETKVWQY+TL +RI+LIDCPG+V N+ D+ D++L+GVVRV N+ +
Sbjct: 349 PVAKVAPIPGETKVWQYVTLMRRIYLIDCPGIVPPNQNDTPQDLLLRGVVRVENVDNPEQ 408
Query: 313 RIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQ 372
I VL +V H+ R Y++K W++ FL L + G+LLKGGEPD+ AKMVL+D+
Sbjct: 409 YIPAVLNKVKPHHMERTYELKGWKDHIHFLEMLARKGGRLLKGGEPDVDGVAKMVLNDFM 468
Query: 373 RGKIPFFVPPPQQEDLSEEPIVNGLDVDDGV--------DSNQASAAIKAIANVLSSQQQ 424
RGKIP+F P P++E+ E + G + G + + A A A A + ++
Sbjct: 469 RGKIPWFTPAPEKEE-GETDTMEGREGRYGEMSKKRKRDEDDSAPATTPASAGEDAKEED 527
Query: 425 SSVPVQKDLFTENELEEEADNLLLISGDSPDGDVLDSDSDTSEQDPDTE 473
D +++E+EE A+ + D LD+ SD E+D + E
Sbjct: 528 PENFAGFDSDSDSEVEEAAEEKGEEKSTAEDMIPLDASSD-EEEDGEEE 575
>NOG2_CRYBA (Q6TGJ8) Nucleolar GTP-binding protein 2
Length = 731
Score = 343 bits (879), Expect = 7e-94
Identities = 199/424 (46%), Positives = 258/424 (59%), Gaps = 43/424 (10%)
Query: 54 PFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAG-ASGTAE-ANEADGFID 111
PF FGPK +RKRP L + L + + A A E A A+GTA+ A+
Sbjct: 148 PFGDTFGPKAQRKRPRLDIGSIEELGESSAAAATAAEAATAESQANGTADLADIYHPTTS 207
Query: 112 LLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMM 171
++ KG ++ I GELYKV+DSSDVV+ VLDARDP GTRC + ++L++ +KH++
Sbjct: 208 TAREPIYAKGTSRRIWGELYKVLDSSDVVIHVLDARDPLGTRCKPVVEYLRKEKAHKHLV 267
Query: 172 LLLNKCDLIPAWATKG------------WLRVLSKEYPTLAFHASINKSFGKGSLLSVLR 219
+LNK DL+P W T G W++ LS PT+AFHASIN SFGKGSL+ +LR
Sbjct: 268 YVLNKVDLVPTWVTSGPYAYAYANGPARWVKHLSLSAPTIAFHASINNSFGKGSLIQLLR 327
Query: 220 QFARLKSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFL 279
QF+ L SDK+ ISVGFIGYPN GKSS+INTL+ K VC VAPIPGETKVWQYITL +RI+L
Sbjct: 328 QFSVLHSDKKQISVGFIGYPNTGKSSIINTLKKKKVCTVAPIPGETKVWQYITLMRRIYL 387
Query: 280 IDCPGVV-YQNKDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKE---- 334
IDCPG+V KDS+ D VLKGVVRV NL A+ I +L+RV E+L R Y ++
Sbjct: 388 IDCPGIVPVSAKDSDTDTVLKGVVRVENLATPAEHIPALLERVRPEYLERTYNLEHVEGG 447
Query: 335 WENEND---FLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSEE 391
W E L + K +GKLLKGGEPD AAKMVL+DW RGKIPFFV PP +
Sbjct: 448 WHGEQGATVILTAIAKKSGKLLKGGEPDQEAAAKMVLNDWIRGKIPFFVAPPAKS----- 502
Query: 392 PIVNGLDVDDGVDSNQASAAIKAIANVLSSQQQSSVPVQKDLFTENE----LEEEADNLL 447
+ +AA A A ++++ + +++L E E LEE+ +L
Sbjct: 503 ------------EPGAPTAATAAAATTTEAEKKKTEAEEQELAEERETMEILEEQERSLG 550
Query: 448 LISG 451
+ G
Sbjct: 551 KVLG 554
>NOG2_CRYNV (Q8J109) Nucleolar GTP-binding protein 2
Length = 708
Score = 333 bits (855), Expect = 4e-91
Identities = 197/458 (43%), Positives = 259/458 (56%), Gaps = 61/458 (13%)
Query: 54 PFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGASGTAEANEADGFIDLL 113
PF FGPK +RKRP L A A + T+ A E
Sbjct: 146 PFGNTFGPKAQRKRPRLDIGKGNGTADLADIYHPT-----------TSTARE-------- 186
Query: 114 GHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLL 173
++ KG ++ I GELYKV+DSSDVV+ VLDARDP GTRC + ++L++ +KH++ +
Sbjct: 187 --PIYAKGTSRRIWGELYKVLDSSDVVIHVLDARDPLGTRCKPVVEYLRKEKAHKHLVYV 244
Query: 174 LNKCDLIPAWATKG---------------WLRVLSKEYPTLAFHASINKSFGKGSLLSVL 218
LNK DL+P W T G W++ LS PT+AFHASIN SFGKGSL+ +L
Sbjct: 245 LNKVDLVPTWVTSGPYAYPYANGPHFGARWVKHLSLSAPTIAFHASINNSFGKGSLIQLL 304
Query: 219 RQFARLKSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIF 278
RQF+ L SDK+ ISVGFIGYPN GKSS+INTL+ K VC VAPIPGETKVWQYITL +RI+
Sbjct: 305 RQFSVLHSDKKQISVGFIGYPNTGKSSIINTLKKKKVCTVAPIPGETKVWQYITLMRRIY 364
Query: 279 LIDCPGVV-YQNKDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKE--- 334
LIDCPG+V KDS+ D VLKGVVRV NL A+ I +L+RV E+L R Y ++
Sbjct: 365 LIDCPGIVPVSAKDSDTDTVLKGVVRVENLATPAEHIPALLERVRPEYLERTYGLEHVEG 424
Query: 335 -WENEND---FLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSE 390
W E L + K +GKLLKGGEPD AAKMVL+DW RGK+PFFV PP + +
Sbjct: 425 GWHGEEGATFVLTAIAKKSGKLLKGGEPDQEAAAKMVLNDWIRGKVPFFVAPPTKPESGA 484
Query: 391 EPIVNGLDVDDGVDSNQASAAIKAIANVLSSQQQSSV--------------PVQKDLFTE 436
+ V+ + + Q A + + +Q+ S+ P+ K +
Sbjct: 485 DAHVSSATTEKVQEQEQKELAEEKETKEMLEEQERSLGKVLGIKRVKGVEQPISKIVTMT 544
Query: 437 NELEEEADNLL---LISGDSPDGDVLDSDSDTSEQDPD 471
L ++A + + D D ++ + D D E D D
Sbjct: 545 KFLGDDARRYVEEEEVDVDDVDKEMAEEDEDEDEDDDD 582
>NUG1_YEAST (P40010) Nuclear GTP-binding protein NUG1 (Nuclear
GTPase 1)
Length = 520
Score = 139 bits (351), Expect = 1e-32
Identities = 115/395 (29%), Positives = 191/395 (48%), Gaps = 50/395 (12%)
Query: 74 DYQSLAKKAHVAQEAFEEKYAAGASGTAEANEADGFIDLLGHTMFQKGQTKHISGELYK- 132
D LA AQ+A E Y S A+ + + +D++ + + G+ EL K
Sbjct: 105 DENGLAALVESAQQAAAE-YEGTPSNDADVRDDE--LDVIDYNIDFYGEDVEGESELEKS 161
Query: 133 ----------VIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPA 182
VID+SDV++ VLDARDP+ TR +E+ + ++ + K ++L+LNK DLIP
Sbjct: 162 RKAYDKIFKSVIDASDVILYVLDARDPESTRSRKVEEAVLQS-QGKRLILILNKVDLIPP 220
Query: 183 WATKGWLRVLSKEYPTLAFHAS------------INKSFGKGSLLSVLRQFARLKSDKQA 230
+ WL L +PT+ AS ++++ +LL L+ ++ + K++
Sbjct: 221 HVLEQWLNYLKSSFPTIPLRASSGAVNGTSFNRKLSQTTTASALLESLKTYSNNSNLKRS 280
Query: 231 ISVGFIGYPNVGKSSVINTLRTK-----NVCKVAPIPGETKVWQYITLTKRIFLIDCPGV 285
I VG IGYPNVGKSSVIN L + C V G T + I + ++ ++D PG+
Sbjct: 281 IVVGVIGYPNVGKSSVINALLARRGGQSKACPVGNEAGVTTSLREIKIDNKLKILDSPGI 340
Query: 286 VY--QNK-----DSEADVVLKGVVRVTNLKDAADRIGEVLKRV-----MKEHLHRAYKIK 333
+ +NK + EA++ L + ++ D + ++KR+ M E + Y+I
Sbjct: 341 CFPSENKKRSKVEHEAELALLNALPAKHIVDPYPAVLMLVKRLAKSDEMTESFKKLYEIP 400
Query: 334 EW-ENEND-----FLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQED 387
N+ D FL+ + + G+L KGG P+L +A VL+DW+ GKI +V P
Sbjct: 401 PIPANDADTFTKHFLIHVARKRGRLGKGGIPNLASAGLSVLNDWRDGKILGWVLPNTSAA 460
Query: 388 LSEEPIVNGLDVDDGVDSNQASAAIKAIANVLSSQ 422
S++ N ++ G +A I + S +
Sbjct: 461 ASQQDKQNLSTINTGTKQAPIAANESTIVSEWSKE 495
>YE64_METJA (Q58859) Hypothetical GTP-binding protein MJ1464
Length = 373
Score = 130 bits (327), Expect = 8e-30
Identities = 85/255 (33%), Positives = 134/255 (52%), Gaps = 20/255 (7%)
Query: 132 KVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRV 191
K+ID DV++ VLDARDP+ TR LEK +K K K ++ +LNK DL+P + W V
Sbjct: 19 KIIDECDVILLVLDARDPEMTRNRELEKKIK--AKGKKLIYVLNKADLVPKDILEKWKEV 76
Query: 192 LSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAISVGFIGYPNVGKSSVINTLR 251
+ S + G L +++Q + K+ VG +GYPNVGKSS+IN L
Sbjct: 77 FGEN----TVFVSAKRRLGTKILREMIKQSLKEMGKKEG-KVGIVGYPNVGKSSIINALT 131
Query: 252 TKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNKDSEADVVLKGVVRVTNLKDAA 311
K + G TK Q++ LTK I L+D PGV+ E D+V+ G +R+ +++
Sbjct: 132 GKRKALTGSVAGLTKGEQWVRLTKNIKLMDTPGVLEMR--DEDDLVISGALRLEKVENPI 189
Query: 312 DRIGEVLKR-------VMKEHLHRAYKIKEWENENDFLLQLCKLTGKLLKGGEPDLMTAA 364
++L R ++KE+ Y+ E + + L ++ L KGGE DL+ A
Sbjct: 190 PPALKILSRINNFDNSIIKEYFGVDYE----EVDEELLKKIGNKRSYLTKGGEVDLVRTA 245
Query: 365 KMVLHDWQRGKIPFF 379
K ++ ++Q GK+ ++
Sbjct: 246 KTIIKEYQDGKLNYY 260
>YAWG_SCHPO (Q10190) Hypothetical GTP-binding protein C3F10.16c in
chromosome I
Length = 616
Score = 122 bits (306), Expect = 2e-27
Identities = 99/341 (29%), Positives = 159/341 (46%), Gaps = 56/341 (16%)
Query: 101 AEANEADGFIDLLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKH 160
A+ + +GFI T F++ I +L++VI+ SDVVVQ++DAR+P R HLE++
Sbjct: 141 AQLQDVEGFIV----TPFERNL--EIWRQLWRVIERSDVVVQIVDARNPLFFRSAHLEQY 194
Query: 161 LKENCKYKHMMLLLNKCDLIPAWATKGWLRVLSKE-YPTLAFHASINKSFGKGS------ 213
+KE K LL+NK D++ W ++ P L F A + +
Sbjct: 195 VKEVGPSKKNFLLVNKADMLTEEQRNYWSSYFNENNIPFLFFSARMAAEANERGEDLETY 254
Query: 214 -----------------------------LLSVLRQFARLKSD-KQAISVGFIGYPNVGK 243
L + +FA D K ++ G +GYPNVGK
Sbjct: 255 ESTSSNEIPESLQADENDVHSSRIATLKVLEGIFEKFASTLPDGKTKMTFGLVGYPNVGK 314
Query: 244 SSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQN-KDSEADVVLKGVV 302
SS IN L V+ PG+TK +Q I L++++ L+DCPG+V+ + ++AD+VL GV+
Sbjct: 315 SSTINALVGSKKVSVSSTPGKTKHFQTINLSEKVSLLDCPGLVFPSFATTQADLVLDGVL 374
Query: 303 RVTNLKDAADRIGEVLKRVMKEHLHRAY----KIKEWE-------NENDFLLQLCKLTGK 351
+ L++ + +R+ KE L Y +IK E + + L + G
Sbjct: 375 PIDQLREYTGPSALMAERIPKEVLETLYTIRIRIKPIEEGGTGVPSAQEVLFPFARSRGF 434
Query: 352 L-LKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQEDLSEE 391
+ G PD AA+++L D+ GK+ + PPP + E
Sbjct: 435 MRAHHGTPDDSRAARILLKDYVNGKLLYVHPPPNYPNSGSE 475
>YGJ9_YEAST (P53145) Hypothetical GTP-binding protein in SEH1-PRP20
intergenic region
Length = 640
Score = 119 bits (298), Expect = 2e-26
Identities = 87/318 (27%), Positives = 147/318 (45%), Gaps = 59/318 (18%)
Query: 129 ELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGW 188
+L++V++ SD+VVQ++DAR+P R LE+++KE+ K +LL+NK DL+ W
Sbjct: 190 QLWRVVERSDLVVQIVDARNPLLFRSVDLERYVKESDDRKANLLLVNKADLLTKKQRIAW 249
Query: 189 LRVLSKEYPTLAFHASIN--------------------------------KSFGKGSLLS 216
+ + + F++++ K K +LS
Sbjct: 250 AKYFISKNISFTFYSALRANQLLEKQKEMGEDYREQDFEEADKEGFDADEKVMEKVKILS 309
Query: 217 V--LRQFARLKSDKQA-----------ISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPG 263
+ L + K+ + I++G +GYPNVGKSS IN+L V+ PG
Sbjct: 310 IDQLEELFLSKAPNEPLLPPLPGQPPLINIGLVGYPNVGKSSTINSLVGAKKVSVSSTPG 369
Query: 264 ETKVWQYITLTKRIFLIDCPGVVYQN-KDSEADVVLKGVVRVTNLKDAADRIGEVLKRVM 322
+TK +Q I L+ + L DCPG+V+ N ++ ++V GV+ + L+D G V +R+
Sbjct: 370 KTKHFQTIKLSDSVMLCDCPGLVFPNFAYNKGELVCNGVLPIDQLRDYIGPAGLVAERIP 429
Query: 323 KEHLHRAYKIKEWENEND------------FLLQLCKLTGKLLKG-GEPDLMTAAKMVLH 369
K ++ Y I D L+ + G + +G G D A++ +L
Sbjct: 430 KYYIEAIYGIHIQTKSRDEGGNGDIPTAQELLVAYARARGYMTQGYGSADEPRASRYILK 489
Query: 370 DWQRGKIPFFVPPPQQED 387
D+ GK+ + PPP ED
Sbjct: 490 DYVNGKLLYVNPPPHLED 507
>Y442_MYCGE (Q49435) Hypothetical protein MG442
Length = 270
Score = 66.6 bits (161), Expect = 1e-10
Identities = 76/270 (28%), Positives = 119/270 (43%), Gaps = 37/270 (13%)
Query: 121 GQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLI 180
G K I +L K+ D +++++DAR P T H E Y LNK LI
Sbjct: 14 GHMKKIHDQLKKLSSQIDGIIEIVDARAPTLT-------HNSEIISY-----FLNKPKLI 61
Query: 181 PAWATKGWLRVLSKEYPTLAF-HASINKSFG--KGSLLSVLRQFARLKSDKQA------- 230
A T L++ P S+ + F K L ++ FA + +A
Sbjct: 62 LALKTD-----LAQYKPNKKILFGSLKEPFKLKKKVLKTLTTLFANKRQQLKAKGLLIKQ 116
Query: 231 ISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNK 290
+ IG PNVGKSS+IN L KN KVA G TK +I ++ + L D PGV +
Sbjct: 117 FRLAVIGMPNVGKSSLINLLINKNHLKVANRAGITKSLNWIQISPELLLSDTPGVFLKRI 176
Query: 291 DS---EADVVLKGVVR--VTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFLLQL 345
D +VL V+R V N+++ +G +K+H + E ++ +FL +
Sbjct: 177 DEIQIGYKLVLTNVIRREVVNIEE----VGMFAFNYLKKHYKQLLPF-EADSFINFLEKF 231
Query: 346 CKLTGKLLKGGEPDLMTAAKMVLHDWQRGK 375
K+ G + K E + A ++ +++ GK
Sbjct: 232 AKVRGLIKKANELNTNLACEIFINELINGK 261
>GNL1_MOUSE (P36916) Guanine nucleotide-binding protein-like 1
(GTP-binding protein MMR1)
Length = 430
Score = 65.1 bits (157), Expect = 4e-10
Identities = 31/91 (34%), Positives = 53/91 (58%)
Query: 231 ISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNK 290
+++G IG+PNVGKSS+IN L + V V+ PG T+ +Q LT + L DCPG+++ +
Sbjct: 184 VTIGCIGFPNVGKSSLINGLVGRKVVSVSRTPGHTRYFQTYFLTPSVKLCDCPGLIFPSL 243
Query: 291 DSEADVVLKGVVRVTNLKDAADRIGEVLKRV 321
VL G+ + +++ +G + R+
Sbjct: 244 LPRQLQVLAGIYPIAQIQEPYTSVGYLASRI 274
>GNL1_HUMAN (P36915) Guanine nucleotide-binding protein-like 1
(GTP-binding protein HSR1)
Length = 430
Score = 64.7 bits (156), Expect = 5e-10
Identities = 30/91 (32%), Positives = 53/91 (57%)
Query: 231 ISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNK 290
+++G +G+PNVGKSS+IN L + V V+ PG T+ +Q LT + L DCPG+++ +
Sbjct: 184 VTIGCVGFPNVGKSSLINGLVGRKVVSVSRTPGHTRYFQTYFLTPSVKLCDCPGLIFPSL 243
Query: 291 DSEADVVLKGVVRVTNLKDAADRIGEVLKRV 321
VL G+ + +++ +G + R+
Sbjct: 244 LPRQLQVLAGIYPIAQIQEPYTAVGYLASRI 274
Score = 30.8 bits (68), Expect = 8.1
Identities = 22/77 (28%), Positives = 35/77 (44%), Gaps = 3/77 (3%)
Query: 133 VIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVL 192
V++ SD+V+ + D R P L +++ ++L+LNK DL P W
Sbjct: 7 VLEMSDIVLLITDIRHPVVNFPPALYEYVTGELGLA-LVLVLNKVDLAPRRLVVAWKHYF 65
Query: 193 SKEYPTLAFHASINKSF 209
+ YP L H + SF
Sbjct: 66 HQHYPQL--HVVLFTSF 80
>Y442_MYCPN (P75135) Hypothetical protein MG442 homolog (K05_orf271)
Length = 271
Score = 63.9 bits (154), Expect = 9e-10
Identities = 60/197 (30%), Positives = 86/197 (43%), Gaps = 32/197 (16%)
Query: 121 GQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLI 180
G K +L K+ S D V++V+DAR P T+ + + K + L K DL
Sbjct: 15 GHMKKTHDQLKKLASSLDGVIEVVDARAPTLTQNPEITAYFTNKPK----LTLALKADLA 70
Query: 181 PAWATKGWL-----------RVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQ 229
A L R++ K+ TL F A N+ KG L+ R
Sbjct: 71 QTVANSNILWGTLKQGLQLKRLVIKKLQTL-FQAKKNQLKAKGLLVHQFR---------- 119
Query: 230 AISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQN 289
+ IG PNVGKSS+IN L KN +VA G TK + ++ +L D PGV ++
Sbjct: 120 ---LAVIGMPNVGKSSLINLLLNKNHLQVANRAGVTKSMSWNQISSEFYLSDTPGVFFKR 176
Query: 290 KDSEA---DVVLKGVVR 303
D A +VL V++
Sbjct: 177 IDEMAVGYKLVLTNVIK 193
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,537,569
Number of Sequences: 164201
Number of extensions: 2379239
Number of successful extensions: 6499
Number of sequences better than 10.0: 393
Number of HSP's better than 10.0 without gapping: 245
Number of HSP's successfully gapped in prelim test: 148
Number of HSP's that attempted gapping in prelim test: 5976
Number of HSP's gapped (non-prelim): 569
length of query: 482
length of database: 59,974,054
effective HSP length: 114
effective length of query: 368
effective length of database: 41,255,140
effective search space: 15181891520
effective search space used: 15181891520
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0030.10


