

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.6
(183 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
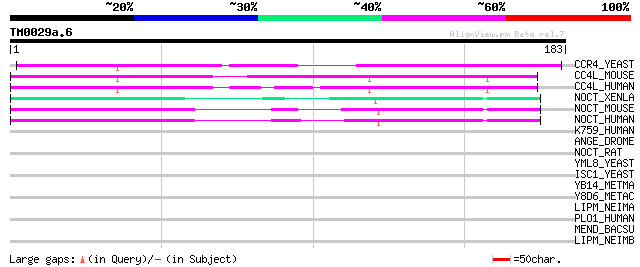
Score E
Sequences producing significant alignments: (bits) Value
CCR4_YEAST (P31384) Glucose-repressible alcohol dehydrogenase tr... 68 1e-11
CC4L_MOUSE (Q8K3P5) Carbon catabolite repressor protein 4 homolo... 62 6e-10
CC4L_HUMAN (Q9ULM6) Carbon catabolite repressor protein 4 homolo... 62 7e-10
NOCT_XENLA (P79942) Nocturnin (Rhythmic message 1) (RM1) 54 2e-07
NOCT_MOUSE (O35710) Nocturnin (CCR4 protein homolog) 53 5e-07
NOCT_HUMAN (Q9UK39) Nocturnin (CCR4 protein homolog) 53 5e-07
K759_HUMAN (Q9UNK9) Protein KIAA0759 41 0.001
ANGE_DROME (Q24239) Angel protein (Angel 39) (ANG39) 38 0.015
NOCT_RAT (Q9ET55) Nocturnin (CCR4 protein homolog) (Fragment) 37 0.020
YML8_YEAST (Q03210) Hypothetical 57.7 kDa protein in NDI1-ATR1 i... 35 0.13
ISC1_YEAST (P40015) Inositol phosphosphingolipids phospholipase ... 31 1.4
YB14_METMA (Q8PXV2) Hypothetical UPF0264 protein MM1114 30 3.1
Y8D6_METAC (Q8THS6) Hypothetical UPF0264 protein MA4436 30 3.1
LIPM_NEIMA (P57037) Capsule polysaccharide modification protein ... 29 7.0
PLO1_HUMAN (Q02809) Procollagen-lysine,2-oxoglutarate 5-dioxygen... 28 9.1
MEND_BACSU (P23970) Menaquinone biosynthesis protein menD [Inclu... 28 9.1
LIPM_NEIMB (Q05013) Capsule polysaccharide modification protein ... 28 9.1
>CCR4_YEAST (P31384) Glucose-repressible alcohol dehydrogenase
transcriptional effector (EC 3.1.-.-) (Cytoplasmic
deadenylase) (Carbon catabolite repressor protein 4)
Length = 837
Score = 68.2 bits (165), Expect = 1e-11
Identities = 56/190 (29%), Positives = 77/190 (40%), Gaps = 31/190 (16%)
Query: 3 THVNVHQDLKDVKLWQVHTLLKGLEKIAASAD----------IPMLVCGDFNSNPGSAPH 52
TH++ DVK +QV LL LE + P+L+CGDFNS SA +
Sbjct: 664 THLHWDPKFNDVKTFQVGVLLDHLETLLKEETSHNFRQDIKKFPVLICGDFNSYINSAVY 723
Query: 53 ALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSA 112
L+ G+V D H L L S+Y+
Sbjct: 724 ELINTGRVQIHQEGNGRD--FGYMSEKNFSHNLALKSSYNCIG----------------- 764
Query: 113 TNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALL 172
E FTN T F +DYI+++ +L V LL +D E K P+ ++ SDHI LL
Sbjct: 765 --ELPFTNFTPSFTDVIDYIWFSTHALRVRGLLGEVDPEYESKFIGFPNDKFPSDHIPLL 822
Query: 173 AEFRCCKNRS 182
A F K +
Sbjct: 823 ARFEFMKTNT 832
>CC4L_MOUSE (Q8K3P5) Carbon catabolite repressor protein 4 homolog
(EC 3.1.-.-) (Cytoplasmic deadenylase) (CCR4 carbon
catabolite repression 4-like) (Nocturnin homolog)
Length = 557
Score = 62.4 bits (150), Expect = 6e-10
Identities = 55/191 (28%), Positives = 82/191 (42%), Gaps = 28/191 (14%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASAD-------------IPMLVCGDFNSNP 47
AN H++ + DVKL Q L ++ I A IP+++C D NS P
Sbjct: 358 ANAHMHWDPEYSDVKLVQTMMFLSEVKNIIDKASRSLKSSVLGECGTIPLVLCADLNSLP 417
Query: 48 GSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKR 107
S L+ G V+ +H D +L + L + + + G H
Sbjct: 418 DSGVVEYLSTGGVETNHKDF-----------KELRYNESLTNFSCNGKNGMTNGRITHGF 466
Query: 108 RLDSATNEPL--FTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKD--TALPSPE 163
+L SA L +TN T DF G +DYIFY+ L ++L LD L ++ + P P
Sbjct: 467 KLKSAYENGLMPYTNYTFDFKGIIDYIFYSKPQLNTLAILGPLDHHWLVENNISGCPHPL 526
Query: 164 WSSDHIALLAE 174
SDH +L A+
Sbjct: 527 IPSDHFSLFAQ 537
>CC4L_HUMAN (Q9ULM6) Carbon catabolite repressor protein 4 homolog
(EC 3.1.-.-) (Cytoplasmic deadenylase) (CCR4 carbon
catabolite repression 4-like) (Nocturnin homolog)
Length = 557
Score = 62.0 bits (149), Expect = 7e-10
Identities = 58/191 (30%), Positives = 82/191 (42%), Gaps = 28/191 (14%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASAD-------------IPMLVCGDFNSNP 47
AN H++ + DVKL Q L ++ I A IP+++C D NS P
Sbjct: 358 ANAHMHWDPEYSDVKLVQTMMFLSEVKNIIDKASRNLKSSVLGEFGTIPLVLCADLNSLP 417
Query: 48 GSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKR 107
S L+ G V+ +H D LR + L + S + T G H
Sbjct: 418 DSGVVEYLSTGGVETNHKDF-----KELRYNESLTN----FSCHGKNGTTNGR--ITHGF 466
Query: 108 RLDSATNEPL--FTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKD--TALPSPE 163
+L SA L +TN T DF G +DYIFY+ L +L LD L ++ + P P
Sbjct: 467 KLQSAYESGLMPYTNYTFDFKGIIDYIFYSKPQLNTLGILGPLDHHWLVENNISGCPHPL 526
Query: 164 WSSDHIALLAE 174
SDH +L A+
Sbjct: 527 IPSDHFSLFAQ 537
>NOCT_XENLA (P79942) Nocturnin (Rhythmic message 1) (RM1)
Length = 388
Score = 54.3 bits (129), Expect = 2e-07
Identities = 48/179 (26%), Positives = 73/179 (39%), Gaps = 44/179 (24%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
A TH+ + +L Q LL LE I A +P+++CGDFN++P + A
Sbjct: 240 AVTHLKARTGWERFRLAQGSDLLDNLESITQGATVPLIICGDFNADPTEEVYKRFASS-- 297
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFT- 119
L L SAY K + +EP +T
Sbjct: 298 -----------------------SLNLNSAY--------------KLLSEDGESEPPYTT 320
Query: 120 ---NVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEF 175
T + LDYI+Y+ +L V + L L EE + + LPS + SDH++L+ +F
Sbjct: 321 WKIRTTGESCHTLDYIWYSQHALRVNAALGLPTEEQIGPN-RLPSFNYPSDHLSLVCDF 378
>NOCT_MOUSE (O35710) Nocturnin (CCR4 protein homolog)
Length = 429
Score = 52.8 bits (125), Expect = 5e-07
Identities = 47/179 (26%), Positives = 74/179 (41%), Gaps = 44/179 (24%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
A TH+ + + Q LL+ L+ I A IP++VCGDFN+ P + A +
Sbjct: 281 AVTHLKARTGWERFRSAQGCDLLQNLQNITQGAKIPLIVCGDFNAEPTEEVYKHFASSSL 340
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTN 120
+ L SAY + +EP +T
Sbjct: 341 N-------------------------LNSAYKLLS--------------PDGQSEPPYTT 361
Query: 121 ----VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEF 175
+ + LDYI+Y+ +L V S L+LL EE + + LPS + SDH++L+ +F
Sbjct: 362 WKIRTSGECRHTLDYIWYSRHALSVTSALDLLTEEQIGPN-RLPSFHYPSDHLSLVCDF 419
>NOCT_HUMAN (Q9UK39) Nocturnin (CCR4 protein homolog)
Length = 431
Score = 52.8 bits (125), Expect = 5e-07
Identities = 47/179 (26%), Positives = 74/179 (41%), Gaps = 44/179 (24%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
A TH+ + + Q LL+ L+ I A IP++VCGDFN+ P + A +
Sbjct: 283 AVTHLKARTGWERFRSAQGCDLLQNLQNITQGAKIPLIVCGDFNAEPTEEVYKHFASSNL 342
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTN 120
+ L SAY + +EP +T
Sbjct: 343 N-------------------------LNSAYKLLSA--------------DGQSEPPYTT 363
Query: 121 ----VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEF 175
+ + LDYI+Y+ +L V S L+LL EE + + LPS + SDH++L+ +F
Sbjct: 364 WKIRTSGECRHTLDYIWYSKHALNVRSALDLLTEEQIGPN-RLPSFNYPSDHLSLVCDF 421
>K759_HUMAN (Q9UNK9) Protein KIAA0759
Length = 670
Score = 41.2 bits (95), Expect = 0.001
Identities = 23/63 (36%), Positives = 37/63 (58%), Gaps = 3/63 (4%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASAD---IPMLVCGDFNSNPGSAPHALLAM 57
ANTH+ + DVKL Q+ LL ++K+A +D P+++CGD NS P S + +
Sbjct: 389 ANTHILYNPRRGDVKLAQMAILLAEVDKVARLSDGSHCPIILCGDLNSVPDSPLYNFIRD 448
Query: 58 GKV 60
G++
Sbjct: 449 GEL 451
Score = 39.3 bits (90), Expect = 0.005
Identities = 33/103 (32%), Positives = 42/103 (40%), Gaps = 19/103 (18%)
Query: 83 HQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFI----------GALDYI 132
H L L S Y+ F +R T PL +T D+I D+
Sbjct: 570 HCLHLTSVYTHFLP---------QRGRPEVTTMPLGLGMTVDYIFFSAESCENGNRTDHR 620
Query: 133 FYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEF 175
Y +L + L LL EE L LP+P SSDH+ LLA F
Sbjct: 621 LYRDGTLKLLGRLSLLSEEILWAANGLPNPFCSSDHLCLLASF 663
>ANGE_DROME (Q24239) Angel protein (Angel 39) (ANG39)
Length = 354
Score = 37.7 bits (86), Expect = 0.015
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 2/51 (3%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAP 51
A TH+ + DV+ QV +L+ L+ S D P+++ GDFNS P S+P
Sbjct: 210 ATTHLLFNTKRSDVRCAQVERILEELQSF--STDTPIVLTGDFNSLPDSSP 258
>NOCT_RAT (Q9ET55) Nocturnin (CCR4 protein homolog) (Fragment)
Length = 253
Score = 37.4 bits (85), Expect = 0.020
Identities = 18/47 (38%), Positives = 26/47 (55%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNP 47
A TH+ + + Q LL+ L+ I A IP++VCGDFN+ P
Sbjct: 207 AVTHLKARTGWERFRSAQGCDLLQSLQNITEGAKIPLIVCGDFNAEP 253
>YML8_YEAST (Q03210) Hypothetical 57.7 kDa protein in NDI1-ATR1
intergenic region
Length = 505
Score = 34.7 bits (78), Expect = 0.13
Identities = 50/244 (20%), Positives = 89/244 (35%), Gaps = 68/244 (27%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIP---MLVCGDFNSNPGSAPHALLAM 57
ANTH+ H +L Q + +L+ +++I A + L+ GDFN+ P P+ +
Sbjct: 250 ANTHLFWHPFGVFERLRQSYLVLQKIQEIKACSKYNGWHSLLMGDFNTEPEEPPYLAITK 309
Query: 58 GKVDPSHPDLAIDPLSILRPHSK------------------------------------- 80
+ P A+ S+ +SK
Sbjct: 310 RPLILKGPIRAMVECSLAYRYSKKRNGEESDQDDEECDEKSRGEGHSDQPQNPKPESFTA 369
Query: 81 ------LVHQLPLV--SAYSSFARTIGLGYEQ-HKRRLDSATNEPLFTNVTRDFIGALDY 131
LV+QL + S + G+GY + H + + EP +N + G LDY
Sbjct: 370 TKEEKALVNQLVALHNSLHVKGVSLYGIGYGKVHPENANGSHGEPGLSNWANTWCGLLDY 429
Query: 132 IFYTA-------------------DSLVVESLLELLDEESLRKDTALPSPEWSSDHIALL 172
IFY +++ + L + + + K + E++SDHI+L+
Sbjct: 430 IFYIEGDHNQDTRQKEPLNAFEGNNNVKIIGYLRMPCAQEMPKHSQPFEGEYASDHISLM 489
Query: 173 AEFR 176
+ R
Sbjct: 490 CQIR 493
>ISC1_YEAST (P40015) Inositol phosphosphingolipids phospholipase C
(EC 3.1.4.-) (IPS phospholipase C) (IPS-PLC) (Neutral
sphingomyelinase) (N-SMase) (nSMase)
Length = 477
Score = 31.2 bits (69), Expect = 1.4
Identities = 17/38 (44%), Positives = 20/38 (51%), Gaps = 2/38 (5%)
Query: 26 LEKIAASADIPMLVCGDFNSNPGSAPHALLAM--GKVD 61
L K+ A ++V GD NS PGS PH L G VD
Sbjct: 215 LIKLYRQAGYAVIVVGDLNSRPGSLPHKFLTQEAGLVD 252
>YB14_METMA (Q8PXV2) Hypothetical UPF0264 protein MM1114
Length = 234
Score = 30.0 bits (66), Expect = 3.1
Identities = 32/143 (22%), Positives = 61/143 (42%), Gaps = 12/143 (8%)
Query: 18 QVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRP 77
+V ++ G + I+A+ GDFN PG+A A L + + + +
Sbjct: 44 EVKAVVNGRQPISATI-------GDFNYKPGTAALAALGAAVAGADYIKVGLYDIQTESQ 96
Query: 78 HSKLVHQLP-LVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTA 136
+L+ ++ V Y+ + + GY +K R++S + PL G +D +
Sbjct: 97 ALELLTKITRAVKDYNPLKKVVASGYSDYK-RINSIS--PLLLPAVAAEAG-VDVVMVDT 152
Query: 137 DSLVVESLLELLDEESLRKDTAL 159
+S E +DE+ L++ T L
Sbjct: 153 GVKDGKSTFEFMDEKELKEFTDL 175
>Y8D6_METAC (Q8THS6) Hypothetical UPF0264 protein MA4436
Length = 234
Score = 30.0 bits (66), Expect = 3.1
Identities = 29/120 (24%), Positives = 51/120 (42%), Gaps = 5/120 (4%)
Query: 41 GDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPL-VSAYSSFARTIG 99
GDFN PG+A A L + + + + +L+ ++ L V Y + +
Sbjct: 60 GDFNYKPGTASLAALGAAVAGADYIKVGLYDIQTEAQALELLTKITLAVKDYDPSKKVVA 119
Query: 100 LGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTAL 159
GY +K R++S + PL G +D + +S E +DE+ L++ T L
Sbjct: 120 SGYSDYK-RINSIS--PLLLPAVAAEAG-VDVVMVDTGIKDGKSTFEFMDEQELKEFTDL 175
>LIPM_NEIMA (P57037) Capsule polysaccharide modification protein
lipA
Length = 704
Score = 28.9 bits (63), Expect = 7.0
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 10/92 (10%)
Query: 83 HQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVE 142
HQLP ++ F R++GLG + + ++ + TR L+ + AD++ E
Sbjct: 75 HQLPFIALEDGFLRSLGLGVSGYPPYSIVYDDIGIYYDTTRP--SRLEQLILAADTMPSE 132
Query: 143 SL------LELLDEESLRKDTALPSPEWSSDH 168
+L ++ + + L K +PE S DH
Sbjct: 133 TLAQARQAMDFILQHHLSKYN--HAPELSDDH 162
>PLO1_HUMAN (Q02809) Procollagen-lysine,2-oxoglutarate 5-dioxygenase
1 precursor (EC 1.14.11.4) (Lysyl hydroxylase 1) (LH1)
Length = 727
Score = 28.5 bits (62), Expect = 9.1
Identities = 28/115 (24%), Positives = 49/115 (42%), Gaps = 10/115 (8%)
Query: 18 QVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRP 77
+V L K LEK A D+ +L ++ S P LL K + + ++ P
Sbjct: 77 KVRLLKKALEKHADKEDLVILFTDSYDVLFASGPRELLK--KFRQARSQVVFSAEELIYP 134
Query: 78 HSKLVHQLPLVSAYSSFARTIG-LGYEQHKRRL-------DSATNEPLFTNVTRD 124
+L + P+VS F + G +GY + +L DS +++ +T + D
Sbjct: 135 DRRLETKYPVVSDGKRFLGSGGFIGYAPNLSKLVAEWEGQDSDSDQLFYTKIFLD 189
>MEND_BACSU (P23970) Menaquinone biosynthesis protein menD
[Includes:
2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate
synthase (EC 2.5.1.64) (SHCHC synthase); 2-oxoglutarate
decarboxylase (EC 4.1.1.71) (Alpha-ketoglutarate
decarboxylase) (KDC)]
Length = 580
Score = 28.5 bits (62), Expect = 9.1
Identities = 23/74 (31%), Positives = 42/74 (56%), Gaps = 6/74 (8%)
Query: 23 LKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLV 82
L + ++ A A+ M+VCG+ +S+ + ++A+ K +P LA DPLS LR + +
Sbjct: 209 LSDVAEMLAEAEKGMIVCGELHSD--ADKENIIALSKA-LQYPILA-DPLSNLR--NGVH 262
Query: 83 HQLPLVSAYSSFAR 96
+ ++ AY SF +
Sbjct: 263 DKSTVIDAYDSFLK 276
>LIPM_NEIMB (Q05013) Capsule polysaccharide modification protein
lipA
Length = 704
Score = 28.5 bits (62), Expect = 9.1
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 10/92 (10%)
Query: 83 HQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVE 142
HQLP ++ F R++GLG + + ++ + TR L+ + AD++ E
Sbjct: 75 HQLPFIALEDGFLRSLGLGVAGYPPYSIVYDDIGIYYDTTRP--SRLEQLILAADTMPSE 132
Query: 143 SL------LELLDEESLRKDTALPSPEWSSDH 168
+L ++ + + L K +PE S DH
Sbjct: 133 TLAQAQQAMDFILQHHLSKYN--HAPELSDDH 162
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,669,719
Number of Sequences: 164201
Number of extensions: 843020
Number of successful extensions: 1983
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1955
Number of HSP's gapped (non-prelim): 24
length of query: 183
length of database: 59,974,054
effective HSP length: 103
effective length of query: 80
effective length of database: 43,061,351
effective search space: 3444908080
effective search space used: 3444908080
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0029a.6


