

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.5
(1389 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
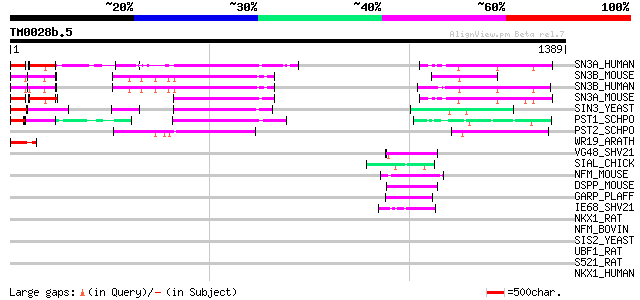
Score E
Sequences producing significant alignments: (bits) Value
SN3A_HUMAN (Q96ST3) Paired amphipathic helix protein Sin3a 212 5e-54
SN3B_MOUSE (Q62141) Paired amphipathic helix protein Sin3b 198 1e-49
SN3B_HUMAN (O75182) Paired amphipathic helix protein Sin3b 197 2e-49
SN3A_MOUSE (Q60520) Paired amphipathic helix protein Sin3a 183 3e-45
SIN3_YEAST (P22579) Paired amphipathic helix protein SIN3 139 7e-32
PST1_SCHPO (Q09750) Paired amphipathic helix protein pst1 (SIN3 ... 132 7e-30
PST2_SCHPO (O13919) Paired amphipathic helix protein pst2 (SIN3 ... 121 1e-26
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 64 3e-09
VG48_SHV21 (Q01033) Hypothetical gene 48 protein 55 1e-06
SIAL_CHICK (P79780) Bone sialoprotein II precursor (BSP II) (Cel... 50 6e-05
NFM_MOUSE (P08553) Neurofilament triplet M protein (160 kDa neur... 50 6e-05
DSPP_MOUSE (P97399) Dentin sialophosphoprotein precursor (Dentin... 47 3e-04
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 46 6e-04
IE68_SHV21 (Q01042) Immediate-early protein 46 8e-04
NKX1_RAT (Q9QZM6) Sodium/potassium/calcium exchanger 1 precursor... 45 0.001
NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa neur... 45 0.001
SIS2_YEAST (P36024) SIS2 protein (Halotolerance protein HAL3) 45 0.001
UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream bin... 44 0.002
S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding ... 44 0.002
NKX1_HUMAN (O60721) Sodium/potassium/calcium exchanger 1 precurs... 44 0.002
>SN3A_HUMAN (Q96ST3) Paired amphipathic helix protein Sin3a
Length = 1273
Score = 212 bits (540), Expect = 5e-54
Identities = 145/463 (31%), Positives = 233/463 (50%), Gaps = 79/463 (17%)
Query: 266 EFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLMEEFNEFL 325
E F DKV++ LR+ + Y+ FL+CL I+++E+I++ EL LV LGK+P+L FN F
Sbjct: 463 ESLFFDKVRKALRSAEAYENFLRCLVIFNQEVISRAELVQLVSPFLGKFPEL---FNWF- 518
Query: 326 VQAEKNDGGFLAGVMNKKSLWIEGPGLKPMKAEDRDRDRDRDRYRDDGMKERDRECRERD 385
KN G+ K+S+ +E
Sbjct: 519 ----KNFLGY------KESVHLE------------------------------------- 531
Query: 386 RSTVVSNKEVSGLKIKDKHFLKPINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAK 445
T + G+ + E+D ++C R SYR LPK+Y P + ++ + +
Sbjct: 532 --TYPKERATEGIAM----------EIDYASCKRLGSSYRALPKSYQQPKCTGRTPLCKE 579
Query: 446 VLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEK 505
VLND WVS S SED +F +K QYEE ++RCED+RFELD++LE+ T + +E + +K
Sbjct: 580 VLNDTWVSFPSWSEDSTFVSSKKTQYEEHIYRCEDERFELDVVLETNLATIRVLEAIQKK 639
Query: 506 INNNVIKGDRPVRIDEHL----TALNLRCIERLYGDHGLDVMEVLRKNAPLALPVILTRL 561
++ + R+D L ++ + ++R+Y D D+++ LRKN +A+P++L RL
Sbjct: 640 LSRLSAEEQAKFRLDNTLGGTSEVIHRKALQRIYADKAADIIDGLRKNPSIAVPIVLKRL 699
Query: 562 KQKQDEWARCRADFSKVWAEIYAKNYHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEK 621
K K++EW + F+KVW E K Y KSLDH+ FKQ DTK L +K+LL E++ I ++
Sbjct: 700 KMKEEEWREAQRGFNKVWREQNEKYYLKSLDHQGINFKQNDTKVLRSKSLLNEIESIYDE 759
Query: 622 KHKEDDVLLAIAAGSRQPIIPNLEFHYPDLDIHEDLYQLVKYSCGELCTAEQLD--KVMK 679
+ ++ A + P+ P+L Y D I ED L+ + ++ D K+ +
Sbjct: 760 RQEQ-----ATEENAGVPVGPHLSLAYEDKQILEDAAALIIHHVKRQTGIQKEDKYKIKQ 814
Query: 680 VWTTFLEPMLCVPSRAQGAEDTEDVVKAKNTEDVVNTENNSVK 722
+ F+ P L R D DV + + E V+ +VK
Sbjct: 815 IMHHFI-PDLLFAQRG----DLSDVEEEEEEEMDVDEATGAVK 852
Score = 108 bits (271), Expect = 8e-23
Identities = 99/390 (25%), Positives = 172/390 (43%), Gaps = 73/390 (18%)
Query: 1025 DVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVK 1084
D+ ++ D + EE E+ + D EA G V H+ G G S S+
Sbjct: 822 DLLFAQRGDLSDVEEEEEEEMDVD-------EATGAVKKHN--GVGGSPPKSKLLFSNT- 871
Query: 1085 PLAKYVPAALHEKDRNSQVFYGSDSFYVLFRLHQTL-----------------------Y 1121
L D +FY ++++Y+ RLHQ L +
Sbjct: 872 -----AAQKLRGMDEVYNLFYVNNNWYIFMRLHQILCLRLLRICSQAERQIEEENREREW 926
Query: 1122 ER----IQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFEDDCRAI 1177
ER I+ K +S + + + K D D Y F++ + SLLDG+ D++++ED R +
Sbjct: 927 EREVLGIKRDKSDSPAIQLRLKEPMDVDVEDYYPAFLDMVRSLLDGNIDSSQYEDSLREM 986
Query: 1178 IGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYE----------KSRKLGKFI 1227
+Y+ FT+DKLI +V+QLQ I S+E+ ++ LY E ++ +
Sbjct: 987 FTIHAYIAFTMDKLIQSIVRQLQHIVSDEICVQVTDLYLAENNNGATGGQLNTQNSRSLL 1046
Query: 1228 DIVYHENACVILHEENIYRIEY--SPGPKKLSIQLMDCGHDKPE--VTAVSVDPKFSTYL 1283
+ Y A ++ +EN +++ + S G +L+I+L+D + + V A Y+
Sbjct: 1047 ESTYQRKAEQLMSDENCFKLMFIQSQGQVQLTIELLDTEEENSDDPVEAERWSDYVERYM 1106
Query: 1284 HNDFLSVVPDKKE---KSRLFLKRNKHRY------------AGSSEFSSQAMEGLQVFNG 1328
++D S P+ +E + +FL RN R G S + ME + +
Sbjct: 1107 NSDTTS--PELREHLAQKPVFLPRNLRRIRKCQRGREQQEKEGKEGNSKKTMENVDSLDK 1164
Query: 1329 LECKIACSSSKVSYVLDTEDVLYRKKSKRR 1358
LEC+ +S K+ YV+ +ED +YR+ + R
Sbjct: 1165 LECRFKLNSYKMVYVIKSEDYMYRRTALLR 1194
Score = 85.5 bits (210), Expect = 9e-16
Identities = 81/295 (27%), Positives = 121/295 (40%), Gaps = 88/295 (29%)
Query: 47 KPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENK-------------CINEVY 93
+PVEF AIN+VNKIK RF+G +YK+FL+IL+ Y+KE + EVY
Sbjct: 300 QPVEFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTEQEVY 359
Query: 94 HEVAVLFQGHADLLDEFTHFLPDASAAASSYYMSARNSVLRDRRSA--MPTVRQAHVVKR 151
+VA LF+ DLL EF FLPDA+ +SVL + +A + +VR H
Sbjct: 360 AQVARLFKNQEDLLSEFGQFLPDAN-----------SSVLLSKTTAEKVDSVRNDHGGTV 408
Query: 152 ERTTVSHCDRDPSVDRPD-PDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGRDHEH 210
++ +++ + PS + H G K++ + + + D + H
Sbjct: 409 KKPQLNNKPQRPSQNGCQIRRHPTGTTPPVKKKPKLLNLKDSSMADASK----------H 458
Query: 211 DGGQYRERLSHKRKSDRKAEDFGAEPLLDADEFFGTRPMSSTCDDKKSLKTKYPEEFAFC 270
GG RK+ R AE + E F C
Sbjct: 459 GGGTESLFFDKVRKALRSAEAY--------------------------------ENFLRC 486
Query: 271 DKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLMEEFNEFL 325
L I+++E+I++ EL LV LGK+P+L F FL
Sbjct: 487 -------------------LVIFNQEVISRAELVQLVSPFLGKFPELFNWFKNFL 522
Score = 58.5 bits (140), Expect = 1e-07
Identities = 26/40 (65%), Positives = 32/40 (80%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLED 40
IDT GVI+RV +LF+GH DLI+GFNTFLP GY+I + D
Sbjct: 159 IDTPGVISRVSQLFKGHPDLIMGFNTFLPPGYKIEVQTND 198
Score = 49.7 bits (117), Expect = 6e-05
Identities = 22/67 (32%), Positives = 41/67 (60%)
Query: 49 VEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLD 108
++ E+A+++++++K +F VY FLDI+ ++ ++ V V+ LF+GH DL+
Sbjct: 121 LKVEDALSYLDQVKLQFGSQPQVYNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDLIM 180
Query: 109 EFTHFLP 115
F FLP
Sbjct: 181 GFNTFLP 187
Score = 44.7 bits (104), Expect = 0.002
Identities = 30/102 (29%), Positives = 45/102 (43%), Gaps = 18/102 (17%)
Query: 248 PMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRN-----PDGYQEFLKCLHIYSKE------ 296
P++ + SL+ P EF K++N PD Y+ FL+ LH Y KE
Sbjct: 285 PVTISLGTAPSLQNNQPVEFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKE 344
Query: 297 -------IITQQELQSLVGDLLGKYPDLMEEFNEFLVQAEKN 331
+T+QE+ + V L DL+ EF +FL A +
Sbjct: 345 AGGNYTPALTEQEVYAQVARLFKNQEDLLSEFGQFLPDANSS 386
Score = 33.5 bits (75), Expect = 4.2
Identities = 22/91 (24%), Positives = 43/91 (47%), Gaps = 6/91 (6%)
Query: 248 PMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRN-PDGYQEFLKCLHIYSKEIITQQELQSL 306
P++ ++ + K + ++ D+VK + + P Y +FL + + + I + S
Sbjct: 108 PVAPVQGQQQFQRLKVEDALSYLDQVKLQFGSQPQVYNDFLDIMKEFKSQSIDTPGVISR 167
Query: 307 VGDLLGKYPDLMEEFNEFL-----VQAEKND 332
V L +PDL+ FN FL ++ + ND
Sbjct: 168 VSQLFKGHPDLIMGFNTFLPPGYKIEVQTND 198
>SN3B_MOUSE (Q62141) Paired amphipathic helix protein Sin3b
Length = 954
Score = 198 bits (503), Expect = 1e-49
Identities = 152/490 (31%), Positives = 233/490 (47%), Gaps = 90/490 (18%)
Query: 257 KSLKTKYPEEFAFCDKVKEK-LRNPDGYQEFLKCLHIYSKEII----------TQQELQS 305
+S ++ ++ +K+K + L +P+ Y+ FL+ LH Y KE + +++E+ +
Sbjct: 148 ESDSVEFNNAISYVNKIKTRFLDHPEIYRSFLEILHTYQKEQLHTKGRPFRGMSEEEVFT 207
Query: 306 LVGDLLGKYPDLMEEFNEFLVQA----------------EKNDGGFLAGVMNKK----SL 345
V +L DL+ EF +FL +A +KN+ L NKK SL
Sbjct: 208 EVANLFRGQEDLLSEFGQFLPEAKRSLFTGNGSCEMNSGQKNEEKSLEH--NKKRSRPSL 265
Query: 346 W--IEGPGLKPMKAEDRDR------------------DRDRDRYRDDGMKERDRECRERD 385
+ P K MK D+ R + + E C
Sbjct: 266 LRPVSAPAKKKMKLRGTKDLSIAAVGKYGTLQEFSFFDKVRRVLKSQEVYENFLRCIALF 325
Query: 386 RSTVVSNKEV-------------------SGLKIKDKHFLKPIN---------ELDLSNC 417
+VS E+ S L +K+ F P++ E+D ++C
Sbjct: 326 NQELVSGSELLQLVSPFLGKFPELFAQFKSFLGVKELSFAPPMSDRSGDGISREIDYASC 385
Query: 418 DRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFR 477
R SYR LPK Y P S ++ I +VLND WVS S SED +F +K YEE L R
Sbjct: 386 KRIGSSYRALPKTYQQPKCSGRTAICKEVLNDTWVSFPSWSEDSTFVSSKKTPYEEQLHR 445
Query: 478 CEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHL----TALNLRCIER 533
CED+RFELD++LE+ T + +E + +K++ + +R+D+ L + R I R
Sbjct: 446 CEDERFELDVVLETNLATIRVLESVQKKLSRMAPEDQEKLRLDDCLGGTSEVIQRRAIHR 505
Query: 534 LYGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYAKNYHKSLDH 593
+YGD +V+E L++N A+PV+L RLK K++EW + F+K+W E Y K Y KSLDH
Sbjct: 506 IYGDKAPEVIESLKRNPATAVPVVLKRLKAKEEEWREAQQGFNKIWREQYEKAYLKSLDH 565
Query: 594 RSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEFHYPDLDI 653
++ FKQ DTK L +K+LL E++ + + +H+E A S P+L F Y D I
Sbjct: 566 QAVNFKQNDTKALRSKSLLNEIESVYD-EHQEQHSEGRSAPSSE----PHLIFVYEDRQI 620
Query: 654 HEDLYQLVKY 663
ED L+ Y
Sbjct: 621 LEDAAALISY 630
Score = 99.0 bits (245), Expect = 8e-20
Identities = 65/161 (40%), Positives = 79/161 (48%), Gaps = 44/161 (27%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPL---------------------- 38
IDT GVI RV +LF H DLI+GFN FLP GY I +P
Sbjct: 70 IDTPGVIRRVSQLFHEHPDLIVGFNAFLPLGYRIDIPKNGKLNIQSPLSSQDNSHSHGDC 129
Query: 39 ----------ED--EQPPAKKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKE- 85
ED + P VEF AI++VNKIK RF +Y+SFL+IL+ Y+KE
Sbjct: 130 GEDFKQMSYKEDRGQVPLESDSVEFNNAISYVNKIKTRFLDHPEIYRSFLEILHTYQKEQ 189
Query: 86 ---------NKCINEVYHEVAVLFQGHADLLDEFTHFLPDA 117
EV+ EVA LF+G DLL EF FLP+A
Sbjct: 190 LHTKGRPFRGMSEEEVFTEVANLFRGQEDLLSEFGQFLPEA 230
Score = 67.0 bits (162), Expect = 3e-10
Identities = 49/194 (25%), Positives = 85/194 (43%), Gaps = 29/194 (14%)
Query: 1055 GEAEGMVDAHDVEGDGMSLTISERFLQTVKPLAKYVPAALHEKDRNSQVFYGSDSFYVLF 1114
G ++ D D + D S R KP A P D +F+ ++++Y
Sbjct: 665 GTSDDSADERDRDRD--SAEPERRRPTDEKPPADASPEPPKVLDDVYSLFFANNNWYFFL 722
Query: 1115 RLHQTLYERI---------------------------QSAKFNSSSAEKKWKASHDTGST 1147
RLHQTL R+ + K + E + K +
Sbjct: 723 RLHQTLCARLLKIYRQAQKQLLEHRREQEREQLLCEGRREKAADPAMELRLKQPSEVELE 782
Query: 1148 DQYDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEM 1207
+ Y F++ + SLL+GS D T++ED R + +Y+ FT+DKL+ + +QL + S+++
Sbjct: 783 EYYPAFLDMVRSLLEGSIDPTQYEDTLREMFTIHAYIGFTMDKLVQNIARQLHHLVSDDV 842
Query: 1208 DNKLLQLYAYEKSR 1221
K+++LY E+ R
Sbjct: 843 CLKVVELYLNEQQR 856
Score = 50.8 bits (120), Expect = 3e-05
Identities = 24/70 (34%), Positives = 39/70 (55%)
Query: 46 KKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHAD 105
K PV E+A+ +++++K RF D Y FL+I+ ++ ++ V V+ LF H D
Sbjct: 29 KLPVHVEDALTYLDQVKIRFGSDPATYNGFLEIMKEFKSQSIDTPGVIRRVSQLFHEHPD 88
Query: 106 LLDEFTHFLP 115
L+ F FLP
Sbjct: 89 LIVGFNAFLP 98
>SN3B_HUMAN (O75182) Paired amphipathic helix protein Sin3b
Length = 1130
Score = 197 bits (501), Expect = 2e-49
Identities = 148/489 (30%), Positives = 229/489 (46%), Gaps = 87/489 (17%)
Query: 257 KSLKTKYPEEFAFCDKVKEK-LRNPDGYQEFLKCLHIYSKEII----------TQQELQS 305
+S ++ ++ +K+K + L +P+ Y+ FL+ LH Y KE + +++E+ +
Sbjct: 156 ESDSVEFNNAISYVNKIKTRFLDHPEIYRSFLEILHTYQKEQLNTRGRPFRGMSEEEVFT 215
Query: 306 LVGDLLGKYPDLMEEFNEFLVQA----------------EKNDGGFLAGVMNKKSLW--- 346
V +L DL+ EF +FL +A +KN+ K+S
Sbjct: 216 EVANLFRGQEDLLSEFGQFLPEAKRSLFTGNGPCEMHSVQKNEHDKTPEHSRKRSRPSLL 275
Query: 347 --IEGPGLKPMKAEDRDR------------------DRDRDRYRDDGMKERDRECRERDR 386
+ P K MK D+ R + + E C
Sbjct: 276 RPVSAPAKKKMKLRGTKDLSIAAVGKYGTLQEFSFFDKVRRVLKSQEVYENFLRCIALFN 335
Query: 387 STVVSNKEV-------------------SGLKIKDKHFLKPIN---------ELDLSNCD 418
+VS E+ S L +K+ F P++ E+D ++C
Sbjct: 336 QELVSGSELLQLVSPFLGKFPELFAQFKSFLGVKELSFAPPMSDRSGDGISREIDYASCK 395
Query: 419 RCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRC 478
R SYR LPK Y P S ++ I +VLND WVS S SED +F +K YEE L RC
Sbjct: 396 RIGSSYRALPKTYQQPKCSGRTAICKEVLNDTWVSFPSWSEDSTFVSSKKTPYEEQLHRC 455
Query: 479 EDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHL----TALNLRCIERL 534
ED+RFELD++LE+ T + +E + +K++ + R+D+ L + R I R+
Sbjct: 456 EDERFELDVVLETNLATIRVLESVQKKLSRMAPEDQEKFRLDDSLGGTSEVIQRRAIYRI 515
Query: 535 YGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYAKNYHKSLDHR 594
YGD +++E L+KN A+PV+L RLK K++EW + F+K+W E Y K Y KSLDH+
Sbjct: 516 YGDKAPEIIESLKKNPVTAVPVVLKRLKAKEEEWREAQQGFNKIWREQYEKAYLKSLDHQ 575
Query: 595 SFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEFHYPDLDIH 654
+ FKQ DTK L +K+LL E++ + + +H+E A S P+L F Y D I
Sbjct: 576 AVNFKQNDTKALRSKSLLNEIESVYD-EHQEQHSEGRSAPSSE----PHLIFVYEDRQIL 630
Query: 655 EDLYQLVKY 663
ED L+ Y
Sbjct: 631 EDAAALISY 639
Score = 100 bits (249), Expect = 3e-20
Identities = 69/162 (42%), Positives = 83/162 (50%), Gaps = 45/162 (27%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLP----------------------- 37
IDT GVI RV +LF H DLI+GFN FLP GY I +P
Sbjct: 77 IDTPGVIRRVSQLFHEHPDLIVGFNAFLPLGYRIDIPKNGKLNIQSPLTSQENSHNHGDG 136
Query: 38 LED--EQPPAK--KP--------VEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKE 85
ED +Q P K KP VEF AI++VNKIK RF +Y+SFL+IL+ Y+KE
Sbjct: 137 AEDFKQQVPYKEDKPQVPLESDSVEFNNAISYVNKIKTRFLDHPEIYRSFLEILHTYQKE 196
Query: 86 ----------NKCINEVYHEVAVLFQGHADLLDEFTHFLPDA 117
EV+ EVA LF+G DLL EF FLP+A
Sbjct: 197 QLNTRGRPFRGMSEEEVFTEVANLFRGQEDLLSEFGQFLPEA 238
Score = 82.4 bits (202), Expect = 8e-15
Identities = 85/389 (21%), Positives = 168/389 (42%), Gaps = 67/389 (17%)
Query: 1021 SENVDVSGSE-SADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEGDGMSLTISERF 1079
S+ +D+ SE SAD + S + + K + G + GD +
Sbjct: 669 SQQLDLGASEESADEDRDSPQGQTTDPSERKKPAPGPHSSPPEEKGAFGDAPA------- 721
Query: 1080 LQTVKPLAKYVPAALHEK-DRNSQVFYGSDSFYVLFRLHQTLYERI-------------- 1124
T +P P A H+ D +F+ ++++Y RLHQTL R+
Sbjct: 722 --TEQP--PLPPPAPHKPLDDVYSLFFANNNWYFFLRLHQTLCSRLLKIYRQAQKQLLEY 777
Query: 1125 -------------QSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFE 1171
+ K + + E + K + + Y F++ + SLL+GS D T++E
Sbjct: 778 RTEKEREKLLCEGRREKGSDPAMELRLKQPSEVELEEYYPAFLDMVRSLLEGSIDPTQYE 837
Query: 1172 DDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEK----------SR 1221
D R + +Y+ FT+DKL+ + +QL + S+++ K+++LY EK SR
Sbjct: 838 DTLREMFTIHAYVGFTMDKLVQNIARQLHHLVSDDVCLKVVELYLNEKKRGAAGGNLSSR 897
Query: 1222 KLGKFIDIVYHENACVILHEENIYRIEY--SPGPKKLSIQLMDCGHDKPE--VTAVSVDP 1277
+ + Y A + +EN +++ + G ++I+L+D + E V +
Sbjct: 898 CVRAARETSYQWKAERCMADENCFKVMFLQRKGQVIMTIELLDTEEAQTEDPVEVQHLAR 957
Query: 1278 KFSTYLHNDFLSVVPDKKEKSR-LFLKRNKHRY------------AGSSEFSSQAMEGLQ 1324
Y+ + S P + + +FL+RN ++ G + S + + G++
Sbjct: 958 YVEQYVGTEGASSSPTEGFLLKPVFLQRNLKKFRRRWQSEQARALRGEARSSWKRLVGVE 1017
Query: 1325 VFNGLECKIACSSSKVSYVLDTEDVLYRK 1353
++C+ S+ K+ +++++ED +YR+
Sbjct: 1018 SACDVDCRFKLSTHKMVFIVNSEDYMYRR 1046
Score = 50.8 bits (120), Expect = 3e-05
Identities = 24/70 (34%), Positives = 39/70 (55%)
Query: 46 KKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHAD 105
K PV E+A+ +++++K RF D Y FL+I+ ++ ++ V V+ LF H D
Sbjct: 36 KLPVHVEDALTYLDQVKIRFGSDPATYNGFLEIMKEFKSQSIDTPGVIRRVSQLFHEHPD 95
Query: 106 LLDEFTHFLP 115
L+ F FLP
Sbjct: 96 LIVGFNAFLP 105
>SN3A_MOUSE (Q60520) Paired amphipathic helix protein Sin3a
Length = 1282
Score = 183 bits (464), Expect = 3e-45
Identities = 96/255 (37%), Positives = 153/255 (59%), Gaps = 9/255 (3%)
Query: 411 ELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRKNQ 470
E+D ++C R SYR LPK+Y P + ++ + +VLND WVS S SED +F +K Q
Sbjct: 546 EIDYASCKRLGSSYRALPKSYQQPKCTGRTPLCKEVLNDTWVSFPSWSEDSTFVSSKKTQ 605
Query: 471 YEESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHL----TAL 526
YEE ++RCED+RFELD++LE+ T + +E + +K++ + R+D L +
Sbjct: 606 YEEHIYRCEDERFELDVVLETNLATIRVLEAIQKKLSRLSAEEQAKFRLDNTLGGTSEVI 665
Query: 527 NLRCIERLYGDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIYAKN 586
+ + ++R+Y D D+++ LRKN +A+P++L RLK K++EW + F+KVW + K
Sbjct: 666 HRKALQRIYADKAADIIDGLRKNPSIAVPIVLKRLKMKEEEWREAQRGFNKVWRDENEKY 725
Query: 587 YHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEF 646
Y KSLDH+ FKQ DTK L +K+LL E++ I +++ ++ A + P+ P+L
Sbjct: 726 YLKSLDHQGINFKQNDTKVLRSKSLLNEIESIYDERQEQ-----ATEENAGVPVGPHLSL 780
Query: 647 HYPDLDIHEDLYQLV 661
Y D I ED L+
Sbjct: 781 AYEDKQILEDAAALI 795
Score = 105 bits (262), Expect = 9e-22
Identities = 96/396 (24%), Positives = 173/396 (43%), Gaps = 77/396 (19%)
Query: 1025 DVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVK 1084
D+ ++ D + EE E+ + D EA G H+ G G S S+
Sbjct: 823 DLLFAQRGDLSDVEEEEEEEMDVD-------EATGAPKKHN--GVGGSPPKSKLLFSNT- 872
Query: 1085 PLAKYVPAALHEKDRNSQVFYGSDSFYVLFRLHQTL-----------------------Y 1121
L D +FY ++++Y+ RLHQ L +
Sbjct: 873 -----AAQKLRGMDEVYNLFYVNNNWYIFMRLHQILCLRLLRICSQAERQIEEENREREW 927
Query: 1122 ER----IQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFEDDCRAI 1177
ER I+ K +S + + + K D D Y F++ + LLDG+ D++++ED R +
Sbjct: 928 EREVLGIKRDKSDSPAIQLRLKEPMDVDVEDYYPAFLDMVRQLLDGNIDSSQYEDSLREM 987
Query: 1178 IGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYE----------KSRKLGKFI 1227
+Y+ FT+DKLI +V+QLQ I S+E+ ++ LY E S+ +
Sbjct: 988 FTIHAYIAFTMDKLIQSIVRQLQHIVSDEVCVQVTDLYLAENNNGATGGQLNSQTSRSLV 1047
Query: 1228 DIVYHENACVILHEENIYRIEY--SPGPKKLSIQLMDCGHD-KPEVTAVSVDPKFSTYLH 1284
+ Y A ++ +EN +++ + S G +L+++L+D + + V ++S Y
Sbjct: 1048 ESAYQRKAEQLMSDENCFKLMFIQSQGQVQLTVELLDTEEENSDDPVEAEVWTRWSDYRW 1107
Query: 1285 NDFL-------SVVPDKKE---KSRLFLKRNKHRY------------AGSSEFSSQAMEG 1322
+D++ + P+ +E + +FL RN R G S + ME
Sbjct: 1108 SDYVERYMSSDTTSPELREHLAQKPVFLPRNLRRIRKCQRGREQQEKEGKEGNSKKTMEN 1167
Query: 1323 LQVFNGLECKIACSSSKVSYVLDTEDVLYRKKSKRR 1358
++ + LEC+ +S K+ YV+ +ED +YR+ + R
Sbjct: 1168 VESLDKLECRFKLNSYKMVYVIKSEDYMYRRTALLR 1203
Score = 76.3 bits (186), Expect = 6e-13
Identities = 40/87 (45%), Positives = 54/87 (61%), Gaps = 13/87 (14%)
Query: 47 KPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENK-------------CINEVY 93
+PV F AIN+VNKIK RF+G +YK+FL+IL+ Y+KE + EVY
Sbjct: 300 QPVHFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTEQEVY 359
Query: 94 HEVAVLFQGHADLLDEFTHFLPDASAA 120
+VA LF+ DLL EF FLPDA+++
Sbjct: 360 AQVARLFKNQEDLLSEFGQFLPDANSS 386
Score = 58.5 bits (140), Expect = 1e-07
Identities = 26/40 (65%), Positives = 32/40 (80%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLED 40
IDT GVI+RV +LF+GH DLI+GFNTFLP GY+I + D
Sbjct: 159 IDTPGVISRVSQLFKGHPDLIMGFNTFLPPGYKIEVQTND 198
Score = 47.8 bits (112), Expect = 2e-04
Identities = 21/67 (31%), Positives = 41/67 (60%)
Query: 49 VEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLD 108
++ E+A+++++++K +F V+ FLDI+ ++ ++ V V+ LF+GH DL+
Sbjct: 121 LKVEDALSYLDQVKLQFGSQPQVHNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDLIM 180
Query: 109 EFTHFLP 115
F FLP
Sbjct: 181 GFNTFLP 187
Score = 42.7 bits (99), Expect = 0.007
Identities = 29/102 (28%), Positives = 44/102 (42%), Gaps = 18/102 (17%)
Query: 248 PMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRN-----PDGYQEFLKCLHIYSKE------ 296
P++ + SL+ P F K++N PD Y+ FL+ LH Y KE
Sbjct: 285 PVTISLGTAPSLQNNQPVHFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKE 344
Query: 297 -------IITQQELQSLVGDLLGKYPDLMEEFNEFLVQAEKN 331
+T+QE+ + V L DL+ EF +FL A +
Sbjct: 345 AGGNYTPALTEQEVYAQVARLFKNQEDLLSEFGQFLPDANSS 386
>SIN3_YEAST (P22579) Paired amphipathic helix protein SIN3
Length = 1536
Score = 139 bits (349), Expect = 7e-32
Identities = 89/254 (35%), Positives = 138/254 (54%), Gaps = 12/254 (4%)
Query: 410 NELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVS-VTSGSEDYSFKHMRK 468
+ LDL C+ PSY+ LPK+ S + + +VLND WV SED F RK
Sbjct: 741 HRLDLDLCEAFGPSYKRLPKSDTFMPCSGRDDMCWEVLNDEWVGHPVWASEDSGFIAHRK 800
Query: 469 NQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHL--TAL 526
NQYEE+LF+ E++R E D +ES T + +E + KI N ++ L T++
Sbjct: 801 NQYEETLFKIEEERHEYDFYIESNLRTIQCLETIVNKIENMTENEKANFKLPPGLGHTSM 860
Query: 527 NL--RCIERLYG-DHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAEIY 583
+ + I ++Y + G ++++ L ++ + PV+L RLKQK +EW R + +++KVW E+
Sbjct: 861 TIYKKVIRKVYDKERGFEIIDALHEHPAVTAPVVLKRLKQKDEEWRRAQREWNKVWRELE 920
Query: 584 AKNYHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPIIPN 643
K + KSLDH FKQ D K L+TK L++E+ I K D I + +P
Sbjct: 921 QKVFFKSLDHLGLTFKQADKKLLTTKQLISEISSI-----KVDQTNKKIHWLTPKP-KSQ 974
Query: 644 LEFHYPDLDIHEDL 657
L+F +PD +I D+
Sbjct: 975 LDFDFPDKNIFYDI 988
Score = 89.7 bits (221), Expect = 5e-17
Identities = 49/107 (45%), Positives = 65/107 (59%), Gaps = 2/107 (1%)
Query: 42 QPPAKK--PVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVL 99
Q AKK VEF +AI++VNKIK RF +YK FL+IL Y++E K INEVY +V L
Sbjct: 397 QEDAKKNVDVEFSQAISYVNKIKTRFADQPDIYKHFLEILQTYQREQKPINEVYAQVTHL 456
Query: 100 FQGHADLLDEFTHFLPDASAAASSYYMSARNSVLRDRRSAMPTVRQA 146
FQ DLL++F FLPD+SA+A+ A+ + + M QA
Sbjct: 457 FQNAPDLLEDFKKFLPDSSASANQQVQHAQQHAQQQHEAQMHAQAQA 503
Score = 49.3 bits (116), Expect = 7e-05
Identities = 53/286 (18%), Positives = 113/286 (38%), Gaps = 30/286 (10%)
Query: 1004 DEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDA 1063
+E S ++ +S + + ++ + +E S + R K + + +G V
Sbjct: 1030 EESLYSHKQNVSESSGSDDGSSIASRKRPYQQEMSLLDILHRSRYQKLKRSNDEDGKVPQ 1089
Query: 1064 HDVEGDGMSLTISERFLQTVKPLAKYVPAALHEKD------RNSQVF--YGSDSFYVLFR 1115
+ TI E L + ++ L E+ +N +F + + + Y+ FR
Sbjct: 1090 LSEPPEEEPNTIEEEELIDEEAKNPWLTGNLVEEANSQGIIQNRSIFNLFANTNIYIFFR 1149
Query: 1116 LHQTLYERIQSAKFNSSSAEKKWKASH--------------------DTGSTDQYDRFMN 1155
T+YER+ K + K+ D D Y + +
Sbjct: 1150 HWTTIYERLLEIKQMNERVTKEINTRSTVTFAKDLDLLSSQLSEMGLDFVGEDAYKQVLR 1209
Query: 1156 ALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLY 1215
L++G ++ FE+ R +++ L+T+DK+ LVK + ++ +++ L+
Sbjct: 1210 LSRRLINGDLEHQWFEESLRQAYNNKAFKLYTIDKVTQSLVKHAHTLMTDAKTAEIMALF 1269
Query: 1216 AYEKSRKLGKFID-IVYH-ENACVILHEENIYRIEYSPGPKKLSIQ 1259
+++ D I+Y + + + EN++RIE+ +SIQ
Sbjct: 1270 VKDRNASTTSAKDQIIYRLQVRSHMSNTENMFRIEFDKRTLHVSIQ 1315
Score = 48.9 bits (115), Expect = 1e-04
Identities = 24/43 (55%), Positives = 27/43 (61%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQP 43
IDT GVI RV LF G+ LI GFNTFLP+GY I + P
Sbjct: 257 IDTPGVIERVSTLFRGYPILIQGFNTFLPQGYRIECSSNPDDP 299
Score = 45.8 bits (107), Expect = 8e-04
Identities = 23/73 (31%), Positives = 42/73 (57%), Gaps = 1/73 (1%)
Query: 254 DDKKSLKTKYPEEFAFCDKVKEKLRN-PDGYQEFLKCLHIYSKEIITQQELQSLVGDLLG 312
D KK++ ++ + ++ +K+K + + PD Y+ FL+ L Y +E E+ + V L
Sbjct: 399 DAKKNVDVEFSQAISYVNKIKTRFADQPDIYKHFLEILQTYQREQKPINEVYAQVTHLFQ 458
Query: 313 KYPDLMEEFNEFL 325
PDL+E+F +FL
Sbjct: 459 NAPDLLEDFKKFL 471
Score = 43.9 bits (102), Expect = 0.003
Identities = 19/69 (27%), Positives = 39/69 (55%)
Query: 47 KPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADL 106
+P+ ++A++++ ++K +F +Y FLDI+ ++ + V V+ LF+G+ L
Sbjct: 217 RPLNVKDALSYLEQVKFQFSSRPDIYNLFLDIMKDFKSQAIDTPGVIERVSTLFRGYPIL 276
Query: 107 LDEFTHFLP 115
+ F FLP
Sbjct: 277 IQGFNTFLP 285
Score = 43.1 bits (100), Expect = 0.005
Identities = 24/66 (36%), Positives = 34/66 (51%)
Query: 265 EEFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLMEEFNEF 324
EE F +K K + N Y EFLK L++YS++I+ +L V LG +L F F
Sbjct: 664 EEVTFFEKAKRYIGNKHLYTEFLKILNLYSQDILDLDDLVEKVDFYLGSNKELFTWFKNF 723
Query: 325 LVQAEK 330
+ EK
Sbjct: 724 VGYQEK 729
Score = 38.5 bits (88), Expect = 0.13
Identities = 19/71 (26%), Positives = 37/71 (51%), Gaps = 1/71 (1%)
Query: 44 PAKKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGH 103
P + + E + F K K R+ G+ H+Y FL ILN+Y ++ ++++ +V +
Sbjct: 655 PIENNISLNEEVTFFEKAK-RYIGNKHLYTEFLKILNLYSQDILDLDDLVEKVDFYLGSN 713
Query: 104 ADLLDEFTHFL 114
+L F +F+
Sbjct: 714 KELFTWFKNFV 724
>PST1_SCHPO (Q09750) Paired amphipathic helix protein pst1 (SIN3
homolog 1)
Length = 1522
Score = 132 bits (332), Expect = 7e-30
Identities = 88/293 (30%), Positives = 152/293 (51%), Gaps = 15/293 (5%)
Query: 408 PINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSV-TSGSEDYSFKHM 466
P +DL+ C C PSYRLLPK + S + + +LND WVS T SED F
Sbjct: 584 PRPRVDLTQCKSCGPSYRLLPKIELLLPCSGRDDLCWTILNDAWVSFPTLASEDSGFIAH 643
Query: 467 RKNQYEESLFRCEDDRFELDMLLES----VNLTTKRVEELFEKINNNVIKGDRPVRIDEH 522
RKNQ+EE+L + E++R+E D + + + L +++ + P +
Sbjct: 644 RKNQFEENLHKLEEERYEYDRHIGANMRFIELLQIHADKMLKMSEVEKANWTLPSNLGGK 703
Query: 523 LTALNLRCIERLYG-DHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRADFSKVWAE 581
++ + I+++YG +H ++E L+KN + +P++L RLK+K EW + ++++W +
Sbjct: 704 SVSIYHKVIKKVYGKEHAQQIIENLQKNPSVTIPIVLERLKKKDREWRSLQNHWNELWHD 763
Query: 582 IYAKNYHKSLDHRSFYFKQQDTKRLSTKALLAEVKEISEKKHKEDDVLLAIAAGSRQPII 641
I KN+++SLDH+ FK D K + K L++E++ +++++ E ++ G P
Sbjct: 764 IEEKNFYRSLDHQGVSFKSVDKKSTTPKFLISELRNLAQQQKVE------LSEGKVTPSH 817
Query: 642 PNLEFHYPDLDIHEDLYQL--VKYSCGELCTAEQLDKVMKVWTTFLEPMLCVP 692
L F Y D +I D+ +L V G +AE +K+ +FL VP
Sbjct: 818 QFL-FSYKDPNIITDIARLFGVFLIHGSTHSAEDNEKMSNFLRSFLSLFFDVP 869
Score = 78.2 bits (191), Expect = 1e-13
Identities = 66/271 (24%), Positives = 102/271 (37%), Gaps = 54/271 (19%)
Query: 37 PLEDEQPPA----KKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEV 92
P PP K+ + ++AINFVN +K RF Y SFLDIL Y+ + + I V
Sbjct: 331 PAAPSYPPVDASVKQAADLDQAINFVNNVKNRFSHKPEAYNSFLDILKSYQHDQRPIQLV 390
Query: 93 YHEVAVLFQGHADLLDEFTHFLPDASAAASSYYMSARNSVLRDRRSAMPTVRQAHVVKRE 152
Y +V+ LF DLL+EF FLPD S A + +D+ + +P KR
Sbjct: 391 YFQVSQLFAEAPDLLEEFKRFLPDVSVNAPAE--------TQDKSTVVPQESATATPKRS 442
Query: 153 RTTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGRDHEHDG 212
+ P P + EK + + ++H +
Sbjct: 443 PSATPTSALPPIGKFAPPTTAKAQPAPEKRRGEPAVQTRNHSK----------------- 485
Query: 213 GQYRERLSHKRKSDRKAEDFGAEPLLDADEFFGTRPMSSTCDDKKSLKTKYPEEFAFCDK 272
R R + + F P++ + K P E F +
Sbjct: 486 ---RTRTATSSVEETTPRAFNV-------------PIA---------QNKNPSELEFLEH 520
Query: 273 VKEKLRNPDGYQEFLKCLHIYSKEIITQQEL 303
++ L N Y EF+K L +YS+E+ + L
Sbjct: 521 ARQYLANESKYNEFIKLLELYSQEVFDKNAL 551
Score = 67.4 bits (163), Expect = 3e-10
Identities = 86/386 (22%), Positives = 154/386 (39%), Gaps = 68/386 (17%)
Query: 1011 HRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEGDG 1070
+RSS + SE+ + + S E S E D E + S GE V+ + DG
Sbjct: 931 NRSSVKEDYVSESTERTPDASEIDEHISEHEENDDE-SSSVFSTGEV--WVNCKFTDTDG 987
Query: 1071 MSLTISERFLQTVKPLAKYVPAALHEKDRNSQVFYGSDSFYVLFRLHQTLYERIQSAKFN 1130
L + DR+ +G+ S Y FRL TLY R++ K
Sbjct: 988 SLLDDGTKL-----------------SDRSVYNLFGNMSLYCFFRLFHTLYSRLEEIKNL 1030
Query: 1131 SSSAEKKWKASHDTGSTDQ------------------------YDRFMNALYSLLDGSSD 1166
A K HD S Y++ + L++G D
Sbjct: 1031 EQMAYSK---QHDVKSNPVAVELGLVRHPSERLGFALPTADTVYEQAIQLCERLMEGEID 1087
Query: 1167 NTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEK----SRK 1222
FED R + G ++ L+T++KL+ ++KQL ++ + + +L Q++ Y + R+
Sbjct: 1088 QNGFEDALRCLYGIHAFRLYTVEKLVTSIIKQLHSVTT---NRRLAQVFMYYEKDRVQRR 1144
Query: 1223 LGKFIDIVYH-ENACVILHEENIYRIEYSPGPKKLSIQLMDCGHDKPEVTAVSVDPKFST 1281
I+Y + +EN+ I+++ ++ +I+LM G + + + D +
Sbjct: 1145 TSPRQQIMYRIQTETAFGPDENLCCIDWNSQTRQSAIRLM--GREDLTMGTLKSDAEKWC 1202
Query: 1282 YLHNDFLSVVPDK---KEKSRL-FLKR-------NKHRYAGSSEFSSQAMEGLQVFNGLE 1330
Y ++ P + E R+ FL++ N+ + S S+ A+ + +GL
Sbjct: 1203 YYIGSYIMSSPTEGILPEHVRIPFLRKCLPSDEGNEDDESSSVVKSANAIITSFLESGLA 1262
Query: 1331 CKIACSSSKVSYVLDTEDVLYRKKSK 1356
I ++ K+ Y TEDV R +
Sbjct: 1263 LTIPINTVKIRYENGTEDVFARNSEQ 1288
Score = 46.2 bits (108), Expect = 6e-04
Identities = 23/85 (27%), Positives = 45/85 (52%), Gaps = 5/85 (5%)
Query: 36 LPLEDEQPPAKKPVEFEE-----AINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCIN 90
+P ++ P P E+ + A+++++ +K +F + +Y FLDI+ ++ +
Sbjct: 162 VPNQNPSAPPPPPQEYRQLNVTDALSYLDLVKLQFHQEPEIYNEFLDIMKEFKSQAIETP 221
Query: 91 EVYHEVAVLFQGHADLLDEFTHFLP 115
EV V+ LF G+ +L+ F FLP
Sbjct: 222 EVITRVSKLFAGYPNLIQGFNTFLP 246
Score = 45.8 bits (107), Expect = 8e-04
Identities = 22/40 (55%), Positives = 27/40 (67%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLED 40
I+T VI RV +LF G+ +LI GFNTFLP GY I + D
Sbjct: 218 IETPEVITRVSKLFAGYPNLIQGFNTFLPPGYSIEISSAD 257
Score = 39.7 bits (91), Expect = 0.058
Identities = 24/81 (29%), Positives = 41/81 (49%), Gaps = 6/81 (7%)
Query: 265 EEFAFCDKVKEKL-RNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLMEEFNE 323
+ ++ D VK + + P+ Y EFL + + + I E+ + V L YP+L++ FN
Sbjct: 184 DALSYLDLVKLQFHQEPEIYNEFLDIMKEFKSQAIETPEVITRVSKLFAGYPNLIQGFNT 243
Query: 324 FL-----VQAEKNDGGFLAGV 339
FL ++ D G LAG+
Sbjct: 244 FLPPGYSIEISSADPGSLAGI 264
Score = 35.4 bits (80), Expect = 1.1
Identities = 21/64 (32%), Positives = 29/64 (44%), Gaps = 1/64 (1%)
Query: 269 FCDKVKEKLRN-PDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLMEEFNEFLVQ 327
F + VK + + P+ Y FL L Y + Q + V L + PDL+EEF FL
Sbjct: 355 FVNNVKNRFSHKPEAYNSFLDILKSYQHDQRPIQLVYFQVSQLFAEAPDLLEEFKRFLPD 414
Query: 328 AEKN 331
N
Sbjct: 415 VSVN 418
>PST2_SCHPO (O13919) Paired amphipathic helix protein pst2 (SIN3
homolog 2)
Length = 1075
Score = 121 bits (304), Expect = 1e-26
Identities = 108/401 (26%), Positives = 176/401 (42%), Gaps = 46/401 (11%)
Query: 259 LKTKYPEEFAFCDKVKEKL-RNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDL 317
L Y F +V+ L NP+ + + L + + ELQ++V LL ++P L
Sbjct: 138 LPCTYHRAIGFVSRVRRALLSNPEQFFKLQDSLRKFKNSECSLSELQTIVTSLLAEHPSL 197
Query: 318 MEEFNEFLVQA-----EKNDGGF-LAGVMNKKSLWIEGPGLKPMKAEDRDR------DRD 365
EF+ FL + + G F L G+ + + L D +
Sbjct: 198 AHEFHNFLPSSIFFGSKPPLGSFPLRGIQSSQFTLSNISDLLSQSRPDNLSPFSHLSNES 257
Query: 366 RDRYRDDGMKERDRECRER-------------DRSTVVSN-----KEVSGL--------K 399
D +++ D E DR+ +VS K SGL
Sbjct: 258 SDFFKNVKNVLTDVETYHEFLKLLNLYVQGIIDRNILVSRGFGFLKSNSGLWRSFLSLTS 317
Query: 400 IKDKHFLKPINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVS-VTSGS 458
+ + FL N S+ C PSYRLLP S + +LND WVS T S
Sbjct: 318 LSPEEFLSVYNSA-CSDFPECGPSYRLLPVEERNISCSGRDDFAWGILNDDWVSHPTWAS 376
Query: 459 EDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVR 518
E+ F RK YEE++ + E++R+E D +E+ + T K ++++ +IN +
Sbjct: 377 EESGFIVQRKTPYEEAMTKLEEERYEFDRHIEATSWTIKSLKKIQNRINELPEEERETYT 436
Query: 519 IDEHL----TALNLRCIERLY-GDHGLDVMEVLRKNAPLALPVILTRLKQKQDEWARCRA 573
++E L ++ + I+ +Y +H ++ + L + L LP++++RL++K +EW +
Sbjct: 437 LEEGLGLPSKSIYKKTIKLVYTSEHAEEMFKALERMPCLTLPLVISRLEEKNEEWKSVKR 496
Query: 574 DFSKVWAEIYAKNYHKSLDHRSFYFKQQDTKRLSTKALLAE 614
W I KNY KSLD + YFK +D K +S+K LLAE
Sbjct: 497 SLQPGWRSIEFKNYDKSLDSQCVYFKARDKKNVSSKFLLAE 537
Score = 57.0 bits (136), Expect = 4e-07
Identities = 63/273 (23%), Positives = 112/273 (40%), Gaps = 29/273 (10%)
Query: 1105 YGSDSFYVLFRLHQTLYERIQSAKFNS---------------SSAEKKWK-----ASHDT 1144
+G S YV FRL LYER+ + S +K W+ S
Sbjct: 744 FGYSSMYVFFRLFNLLYERLYELQRLEDQVSIIQQRIIPNPVSQKQKIWRDRWNDLSDVP 803
Query: 1145 GSTDQYDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIAS 1204
Y+ + L+ G D + FED R G ++Y ++T+DKL++ KQ+ I S
Sbjct: 804 DEKTHYENTYVMILRLIYGIVDQSAFEDYLRFYYGNKAYKIYTIDKLVWSAAKQVHHIVS 863
Query: 1205 EEMDNKLLQLYAYEKSRKLGK-FIDIVYHENACVILH-EENIYRIEYSPGPKKLSIQLMD 1262
+ + L S K + D +Y +L+ +E ++R + K I++M
Sbjct: 864 DGKYKFVTSLVEQNSSASPKKNYDDFLYRLEIEKLLNPDEILFRFCWINKFKSFGIKIMK 923
Query: 1263 CGHDKPEVTAVSVDPKFSTYLHNDFLSVVPDK---KEKSRLFLKRN--KHRYAGS--SEF 1315
+ + + + + Y+ N + + ++ K FL RN K R S
Sbjct: 924 RANLIVDQSLDTQRRVWKKYVQNYRIQKLTEEISYKNYRCPFLCRNIEKERTVEQLVSRL 983
Query: 1316 SSQAMEGLQVFNGLECKIACSSSKVSYVLDTED 1348
++ + ++ +GL+ K+ S K+ Y+ TED
Sbjct: 984 QTKLLRSAELVSGLQAKLCLDSFKLLYLPRTED 1016
Score = 43.1 bits (100), Expect = 0.005
Identities = 31/137 (22%), Positives = 52/137 (37%), Gaps = 20/137 (14%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGY-------------EITLPLEDEQPPA-- 45
+D G I R+ + + DL+ N FLP Y + T P P+
Sbjct: 72 VDFPGFIERISVILRDYPDLLEYLNIFLPSSYKYLLSNSGANFTLQFTTPSGPVSTPSTY 131
Query: 46 -----KKPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLF 100
P + AI FV++++ + + D L ++ ++E+ V L
Sbjct: 132 VATYNDLPCTYHRAIGFVSRVRRALLSNPEQFFKLQDSLRKFKNSECSLSELQTIVTSLL 191
Query: 101 QGHADLLDEFTHFLPDA 117
H L EF +FLP +
Sbjct: 192 AEHPSLAHEFHNFLPSS 208
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 63.9 bits (154), Expect = 3e-09
Identities = 31/66 (46%), Positives = 47/66 (70%), Gaps = 11/66 (16%)
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+DT+GVIAR+K+LF+GH DL+LGFNT+L K Y+IT+ ED+ P I+F++K
Sbjct: 341 VDTSGVIARLKDLFKGHDDLLLGFNTYLSKEYQITILPEDDFP-----------IDFLDK 389
Query: 61 IKARFE 66
++ +E
Sbjct: 390 VEGPYE 395
Score = 39.7 bits (91), Expect = 0.058
Identities = 21/62 (33%), Positives = 36/62 (57%), Gaps = 1/62 (1%)
Query: 53 EAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTH 112
+A+N++ +K +FE D Y +FL++LN + + + V + LF+GH DLL F
Sbjct: 308 DALNYLKAVKDKFE-DSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 113 FL 114
+L
Sbjct: 367 YL 368
>VG48_SHV21 (Q01033) Hypothetical gene 48 protein
Length = 797
Score = 55.1 bits (131), Expect = 1e-06
Identities = 39/133 (29%), Positives = 62/133 (46%), Gaps = 4/133 (3%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKG-GDMSQKYQSRHGEQVCCEAKGEN 998
K E +EG+ GD ED + + G D G +G+ GD + G++ E + E
Sbjct: 517 KDEGDEGDEGDEGDEGEDEWEDEGDEGEDEGDEGEDEGDEGEDEGEDEGDEGEDEGEDEG 576
Query: 999 DVDADDEGEESPHRSSDDSENASENVDVSGSESAD-GEECSREEHEDREHDNKAESEG-E 1056
D + +DEGE+ D+ E+ + + G + D G+E E E + ++ E EG E
Sbjct: 577 D-EGEDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGDEGEDEGDEGEDEGDEGEDEGDE 635
Query: 1057 AEGMVDAHDVEGD 1069
E D + EGD
Sbjct: 636 GEDEGDEGEDEGD 648
Score = 47.8 bits (112), Expect = 2e-04
Identities = 37/132 (28%), Positives = 55/132 (41%), Gaps = 8/132 (6%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGEND 999
+ E +EGE D ED E+ G D G +G+ + + GE E + E D
Sbjct: 573 EDEGDEGE-----DEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGDEGEDEGDEGEDEGD 627
Query: 1000 V--DADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEA 1057
D DEGE+ D+ + + D E +G+E E E + ++ E EG+
Sbjct: 628 EGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGDEGEDEGDEGEDEGDEGEDEGDE 687
Query: 1058 EGMVDAHDVEGD 1069
D D EGD
Sbjct: 688 GDEGDEGD-EGD 698
Score = 46.6 bits (109), Expect = 5e-04
Identities = 36/126 (28%), Positives = 57/126 (44%), Gaps = 10/126 (7%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDAD 1003
+EG+ G+ E D + G D G +G+ + + GE E GE++ D
Sbjct: 607 DEGDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDE--GEDEGDEG 664
Query: 1004 DEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDA 1063
DEGE+ D+ E+ + + G E +G+E E E +++ E EGE EG
Sbjct: 665 DEGEDE----GDEGEDEGDEGEDEGDEGDEGDEGD----EGDEGEDEGEDEGEDEGDEGT 716
Query: 1064 HDVEGD 1069
D EG+
Sbjct: 717 KDKEGN 722
Score = 46.6 bits (109), Expect = 5e-04
Identities = 39/136 (28%), Positives = 56/136 (40%), Gaps = 12/136 (8%)
Query: 942 EREEGE--------LSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCE 993
E +EGE D +D E+ G D G +G GD + ++ E
Sbjct: 417 EEDEGEDEEDEKDEKEEGEDEGDDGEDEGEDEGEDEGDEGDEGDEGEDEGEDEDDE---E 473
Query: 994 AKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAES 1053
+GE++ D DEGE+ D+ E+ + D E +G+E E E E D E
Sbjct: 474 DEGEDEGDEGDEGEDEGD-EGDEGEDEGDEGDEGKDEGDEGDEGKDEGDEGDEGDEGDEG 532
Query: 1054 EGEAEGMVDAHDVEGD 1069
E E E D + EGD
Sbjct: 533 EDEWEDEGDEGEDEGD 548
Score = 46.6 bits (109), Expect = 5e-04
Identities = 35/126 (27%), Positives = 53/126 (41%), Gaps = 8/126 (6%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDAD 1003
+EG+ G+ E D + G D G +G G K + G++ E ++ D
Sbjct: 478 DEGDEGDEGEDEGDE----GDEGEDEGDEGDEG----KDEGDEGDEGKDEGDEGDEGDEG 529
Query: 1004 DEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDA 1063
DEGE+ D+ E+ + + G E D E +E ED D E E E E D
Sbjct: 530 DEGEDEWEDEGDEGEDEGDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDE 589
Query: 1064 HDVEGD 1069
+ EG+
Sbjct: 590 GEDEGE 595
Score = 45.8 bits (107), Expect = 8e-04
Identities = 38/132 (28%), Positives = 56/132 (41%), Gaps = 8/132 (6%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGL--DAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDV- 1000
+EG+ G E D ++ G D G +G G+ + + GE E + E D
Sbjct: 498 DEGDEGDEGKDEGDEGDEGKDEGDEGDEGDEGDEGEDEWEDEGDEGEDEGDEGEDEGDEG 557
Query: 1001 --DADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAE-SEGEA 1057
+ +DEG+E D+ + + + G E D E +E ED D E EGE
Sbjct: 558 EDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGDEGED 617
Query: 1058 EGMVDAHDVEGD 1069
EG D + EGD
Sbjct: 618 EG--DEGEDEGD 627
Score = 45.4 bits (106), Expect = 0.001
Identities = 42/143 (29%), Positives = 60/143 (41%), Gaps = 17/143 (11%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKY------QSRHGEQVCCE 993
K E+EEGE GD ED E+ G D G +G GD + + GE E
Sbjct: 428 KDEKEEGE--DEGDDGEDEG---EDEGEDEGDEGDEGDEGEDEGEDEDDEEDEGEDEGDE 482
Query: 994 A-KGENDVDADDEGEESPHRSS---DDSENASENVDVS--GSESADGEECSREEHEDREH 1047
+GE++ D DEGE+ D+ + E D G E +G+E E ++ +
Sbjct: 483 GDEGEDEGDEGDEGEDEGDEGDEGKDEGDEGDEGKDEGDEGDEGDEGDEGEDEWEDEGDE 542
Query: 1048 DNKAESEGEAEGMVDAHDVEGDG 1070
EGE EG + E +G
Sbjct: 543 GEDEGDEGEDEGDEGEDEGEDEG 565
Score = 44.3 bits (103), Expect = 0.002
Identities = 35/132 (26%), Positives = 61/132 (45%), Gaps = 6/132 (4%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEA-KGEN 998
+ E +EG+ G+ E ++ E+ G D G +G G+ + + GE E +G++
Sbjct: 450 EDEGDEGDEGDEGEDEGEDEDDEEDEGEDEGDEGDEGE-DEGDEGDEGEDEGDEGDEGKD 508
Query: 999 DVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAE 1058
+ D DEG++ D+ + E D E +GE+ E E + ++ E EGE E
Sbjct: 509 EGDEGDEGKDEGDEG-DEGDEGDEGEDEWEDEGDEGED---EGDEGEDEGDEGEDEGEDE 564
Query: 1059 GMVDAHDVEGDG 1070
G + E +G
Sbjct: 565 GDEGEDEGEDEG 576
Score = 43.1 bits (100), Expect = 0.005
Identities = 34/122 (27%), Positives = 52/122 (41%), Gaps = 10/122 (8%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGEND 999
+ E +EGE GD ED + G D G +G+ + + GE E G+
Sbjct: 616 EDEGDEGE--DEGDEGEDE----GDEGEDEGDEGEDEGDEGEDEGDEGEDEGDE--GDEG 667
Query: 1000 VDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEG 1059
D DEGE+ D+ + E + G E +GE+ +E ED + + EG A
Sbjct: 668 EDEGDEGEDEGDEGEDEGDEGDEGDE--GDEGDEGEDEGEDEGEDEGDEGTKDKEGNANK 725
Query: 1060 MV 1061
+V
Sbjct: 726 VV 727
Score = 42.7 bits (99), Expect = 0.007
Identities = 36/130 (27%), Positives = 52/130 (39%), Gaps = 15/130 (11%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEA----- 994
+ E +EGE D ED E+ G D G +G G+ G++ E
Sbjct: 584 EDEGDEGE-----DEGEDEGDEGEDEGEDEGDEGDEGEDEGDEGEDEGDEGEDEGDEGED 638
Query: 995 KGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHD-----N 1049
+G+ D DEGE+ D+ + E D +G+E E E E D +
Sbjct: 639 EGDEGEDEGDEGEDEGDEGEDEGDEGDEGEDEGDEGEDEGDEGEDEGDEGDEGDEGDEGD 698
Query: 1050 KAESEGEAEG 1059
+ E EGE EG
Sbjct: 699 EGEDEGEDEG 708
Score = 36.2 bits (82), Expect = 0.64
Identities = 27/94 (28%), Positives = 39/94 (40%), Gaps = 9/94 (9%)
Query: 977 DMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEE 1036
D S R ++ E + N + +DEGE D E+ + + E DGE+
Sbjct: 392 DKSDDRDDRDKDEYELENEEYNRDEEEDEGE--------DEEDEKDEKEEGEDEGDDGED 443
Query: 1037 CSREEHEDREHDNKAESEGEAEGMVDAHDVEGDG 1070
+E ED + EGE EG D D E +G
Sbjct: 444 EGEDEGEDEGDEGDEGDEGEDEG-EDEDDEEDEG 476
Score = 35.4 bits (80), Expect = 1.1
Identities = 30/122 (24%), Positives = 54/122 (43%), Gaps = 16/122 (13%)
Query: 942 EREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVD 1001
+++E EL + EE N E+ G D + +K + GE + + E + +
Sbjct: 401 DKDEYEL----ENEEYNRDEEEDEGED--------EEDEKDEKEEGEDEGDDGEDEGEDE 448
Query: 1002 ADDEGEESPH--RSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAE--SEGEA 1057
+DEG+E D+ E+ + D E +G+E E E E +++ + EG+
Sbjct: 449 GEDEGDEGDEGDEGEDEGEDEDDEEDEGEDEGDEGDEGEDEGDEGDEGEDEGDEGDEGKD 508
Query: 1058 EG 1059
EG
Sbjct: 509 EG 510
Score = 32.3 bits (72), Expect = 9.2
Identities = 26/84 (30%), Positives = 35/84 (40%), Gaps = 3/84 (3%)
Query: 162 DPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHRE---DGKRDWERDGRDHEHDGGQYRER 218
D S DR D D D L E+ R E E + E D K + E +G D E +G E
Sbjct: 392 DKSDDRDDRDKDEYELENEEYNRDEEEDEGEDEEDEKDEKEEGEDEGDDGEDEGEDEGED 451
Query: 219 LSHKRKSDRKAEDFGAEPLLDADE 242
+ + ED G + + DE
Sbjct: 452 EGDEGDEGDEGEDEGEDEDDEEDE 475
>SIAL_CHICK (P79780) Bone sialoprotein II precursor (BSP II)
(Cell-binding sialoprotein) (Integrin-binding
sialoprotein)
Length = 276
Score = 49.7 bits (117), Expect = 6e-05
Identities = 44/190 (23%), Positives = 71/190 (37%), Gaps = 20/190 (10%)
Query: 892 LMILVVDISSRFQIRQGGDSTRPGTFTNGATTGGTE---VHRYQEQSIRHFKSEREEGEL 948
L+ ++ +S F +R R G A ++RY + +K E
Sbjct: 6 LLACLLATASAFSVRSWLRRARAGDSEENAVLKSRHRYYLYRYAYPPLHRYKGSDSSEEE 65
Query: 949 SPNGDMEEDNFAVYE------NSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDA 1002
+ EE+ A GL G G GGD + +Q G Q +G++ +
Sbjct: 66 GDGSEEEEEGGAPSHAGTQAAGEGLTLGDVGPGGDAASAHQDCKGGQK--GTRGDSGDED 123
Query: 1003 DDEGEESPHRSSDDSENASENVDVSGSESADGEE---------CSREEHEDREHDNKAES 1053
DE EE ++ E ++V V+G+ + E EE +D E + + E
Sbjct: 124 SDEEEEEEEEEEEEEEVEEQDVSVNGTSTNTTAETPHGNNTVAAEEEEDDDEEEEEEEEE 183
Query: 1054 EGEAEGMVDA 1063
E EAE A
Sbjct: 184 EEEAEATTAA 193
>NFM_MOUSE (P08553) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium polypeptide)
(NF-M)
Length = 848
Score = 49.7 bits (117), Expect = 6e-05
Identities = 40/159 (25%), Positives = 66/159 (41%), Gaps = 9/159 (5%)
Query: 929 HRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGH-KGKGGDMSQKYQSRHG 987
H++ E+ I K E E+ E MEE A+ E A K + + ++ ++
Sbjct: 449 HKFVEEIIEETKVEDEKSE------MEETLTAIAEELAASAKEEKEEAEEKEEEPEAEKS 502
Query: 988 EQVCCEAKGENDVDADDEGEESPHRSSDDSENA-SENVDVSGSESADGEECSREEHEDRE 1046
EAK E + +E EE ++ E S+ + GSE E E E+ E
Sbjct: 503 PVKSPEAKEEEEEGEKEEEEEGQEEEEEEDEGVKSDQAEEGGSEKEGSSEKDEGEQEEEE 562
Query: 1047 HDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKP 1085
+ +AE EGE + +EG + + E ++ KP
Sbjct: 563 GETEAEGEGEEAEAKEEKKIEGKVEEVAVKEE-IKVEKP 600
Score = 33.5 bits (75), Expect = 4.2
Identities = 25/120 (20%), Positives = 58/120 (47%), Gaps = 17/120 (14%)
Query: 169 DPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGR----DHEHDGGQYRERLSHKRK 224
+P+ ++ +++ + + E EK+ E+G+ + E + D +GG +E S K +
Sbjct: 496 EPEAEKSPVKSPEAKEEEEEGEKEEEEEGQEEEEEEDEGVKSDQAEEGGSEKEGSSEKDE 555
Query: 225 SDRKAEDFGAEPLLDADEFFGTRPMSSTCDDKKSLKTKYPEEFAFCDKVK----EKLRNP 280
+++ E+ E + +E + ++K ++ K EE A +++K EK ++P
Sbjct: 556 GEQEEEEGETEAEGEGEE--------AEAKEEKKIEGKV-EEVAVKEEIKVEKPEKAKSP 606
>DSPP_MOUSE (P97399) Dentin sialophosphoprotein precursor (Dentin
matrix protein-3) (DMP-3) [Contains: Dentin
phosphoprotein (Dentin phosphophoryn) (DPP); Dentin
sialoprotein (DSP)]
Length = 934
Score = 47.4 bits (111), Expect = 3e-04
Identities = 28/130 (21%), Positives = 53/130 (40%), Gaps = 4/130 (3%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDAD 1003
+ + S + D + + + + D+ D S S + + D+
Sbjct: 805 DSSDSSNSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSS 864
Query: 1004 DEGEESPHRSSDDSENASENVDV----SGSESADGEECSREEHEDREHDNKAESEGEAEG 1059
D + S S DS N+S++ D S S+S+DG+ S + D D+ ++S+ ++EG
Sbjct: 865 DSSDSSDSSDSSDSSNSSDSSDSDSKDSSSDSSDGDSKSGNGNSDSNSDSNSDSDSDSEG 924
Query: 1060 MVDAHDVEGD 1069
H D
Sbjct: 925 SDSNHSTSDD 934
Score = 43.1 bits (100), Expect = 0.005
Identities = 38/170 (22%), Positives = 69/170 (40%), Gaps = 12/170 (7%)
Query: 897 VDISSRFQIRQGGDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDMEE 956
V++ R +QG GT A R S E + S GD
Sbjct: 396 VELDKRNSPKQGESDKPQGTAEKSAAHSNLGHSRIGSSSNSDGHDSYEFDDESMQGD--- 452
Query: 957 DNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDD 1016
D + E++G D D + + + G + +A +D +DD+ + H DD
Sbjct: 453 DPKSSDESNGSDES------DTNSESANESGSRG--DASYTSDESSDDDNDSDSHAGEDD 504
Query: 1017 S-ENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDAHD 1065
S +++S + D + D E ++E + +HDN ++SE +++ + D
Sbjct: 505 SSDDSSGDGDSDSNGDGDSESEDKDESDSSDHDNSSDSESKSDSSDSSDD 554
Score = 40.4 bits (93), Expect = 0.034
Identities = 24/121 (19%), Positives = 45/121 (36%), Gaps = 1/121 (0%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDAD 1003
+ + S + D + + + + D+ D S S C + + D+
Sbjct: 560 DSSDSSDSSDSSDSSDSSDSSDSSDSNSSSDSSDSSGSSDSSDSSDTCDSSDSSDSSDSS 619
Query: 1004 DEGEESPHRSSDDSENASENVDVSGS-ESADGEECSREEHEDREHDNKAESEGEAEGMVD 1062
D + S S DS ++S++ D S S +S+D CS D+ S+ D
Sbjct: 620 DSSDSSDSSDSSDSSDSSDSSDSSSSSDSSDSSSCSDSSDSSDSSDSSDSSDSSDSSSSD 679
Query: 1063 A 1063
+
Sbjct: 680 S 680
Score = 39.3 bits (90), Expect = 0.076
Identities = 37/172 (21%), Positives = 66/172 (37%), Gaps = 20/172 (11%)
Query: 909 GDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLD 968
GD + +NG+ T E R S + S + D + D+ A ++S D
Sbjct: 451 GDDPKSSDESNGSDESDTNSESANESGSRGDASYTSDE--SSDDDNDSDSHAGEDDSSDD 508
Query: 969 AGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVS- 1027
+ G G+++ + DE + S H +S DSE+ S++ D S
Sbjct: 509 SSGDGDSDS---------------NGDGDSESEDKDESDSSDHDNSSDSESKSDSSDSSD 553
Query: 1028 -GSESADGEECS-REEHEDREHDNKAESEGEAEGMVDAHDVEGDGMSLTISE 1077
S+S+D + S + D + + ++ D+ D G S S+
Sbjct: 554 DSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSNSSSDSSDSSGSSDSSDSSD 605
Score = 38.5 bits (88), Expect = 0.13
Identities = 43/231 (18%), Positives = 85/231 (36%), Gaps = 3/231 (1%)
Query: 945 EGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDV-DAD 1003
+G+ NGD + ++ E+ D + S S ++ +D D+
Sbjct: 512 DGDSDSNGDGDSESEDKDESDSSDHDNSSDSESKSDSSDSSDDSSDSSDSSDSSDSSDSS 571
Query: 1004 DEGEESPHRSSDDSENASENVDVSGS-ESADGEE-CSREEHEDREHDNKAESEGEAEGMV 1061
D + S S DS ++S++ D SGS +S+D + C + D + + ++
Sbjct: 572 DSSDSSDSSDSSDSNSSSDSSDSSGSSDSSDSSDTCDSSDSSDSSDSSDSSDSSDSSDSS 631
Query: 1062 DAHDVEGDGMSLTISERFLQTVKPLAKYVPAALHEKDRNSQVFYGSDSFYVLFRLHQTLY 1121
D+ D S + S+ + + + D + S +
Sbjct: 632 DSSDSSDSSDSSSSSDSSDSSSCSDSSDSSDSSDSSDSSDSSDSSSSDSSSSSNSSDSSD 691
Query: 1122 ERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFED 1172
S+ +SS + +S +GS+D D ++ S SSD++ D
Sbjct: 692 SSDSSSSSDSSDSSDSSDSSDSSGSSDSSDSSASSDSSSSSDSSDSSSSSD 742
Score = 38.1 bits (87), Expect = 0.17
Identities = 89/409 (21%), Positives = 145/409 (34%), Gaps = 66/409 (16%)
Query: 715 NTENNSVKS--GIVSVAESGDSKAWQLNGD---SGVREDRCLDSDHTVYKT----ETAGN 765
N+ N ++S GIV AE +S G SG D + D T+ + E +
Sbjct: 130 NSTANGIRSQVGIVENAEEAESSVHGQAGQNTKSGGASDVSQNGDATLVQENEPPEASIK 189
Query: 766 NTQHGKTNIIGF------TPDELSGVNKQNQSSE---------RLVNANVSPAS------ 804
N+ + + I G T + G+ +NQ +E L + + SP+
Sbjct: 190 NSTNHEAGIHGSGVATHETTPQREGLGSENQGTEVTPSIGEDAGLDDTDGSPSGNGVEED 249
Query: 805 ---------GMEQSNGRTKIDNTSGHTATPSGNGNTSVEGGLELPSSEPILFQTRKFPSN 855
G E +GR D T G G GNT G + + + + FP+
Sbjct: 250 EDTGSGDGEGAEAGDGRESHDGTKGQGGQSHG-GNTDHRGQSSVSTEDDDSKEQEGFPNG 308
Query: 856 YSIFSLMLDILINFGLHLHECILQRILFVKHPSYGWLMILVVDISSRFQIRQGGDSTRPG 915
++ + + + G + L K I+ S+ + G S G
Sbjct: 309 HNGDNSSEENGVEEGDSTQATQDKEKLSPKDTRDAEGGII-----SQSEACPSGKSQDQG 363
Query: 916 TFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNG----DMEEDNFAVYENSGLDAGH 971
T G G +SI +S + G NG ++++ N S G
Sbjct: 364 IETEGPNKGN--------KSIITKESGKLSGSKDSNGHQGVELDKRNSPKQGESDKPQGT 415
Query: 972 KGKGGDMSQKYQSRHGEQVCCEAKGENDVD-----ADDEGEESPHRSSDDSENASENVDV 1026
K S SR G + + D DD SD+S+ SE+ +
Sbjct: 416 AEKSAAHSNLGHSRIGSSSNSDGHDSYEFDDESMQGDDPKSSDESNGSDESDTNSESANE 475
Query: 1027 SGSE---SADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEGDGMS 1072
SGS S +E S ++++ H + +S ++ G D+ D GDG S
Sbjct: 476 SGSRGDASYTSDESSDDDNDSDSHAGEDDSSDDSSGDGDS-DSNGDGDS 523
Score = 37.7 bits (86), Expect = 0.22
Identities = 30/131 (22%), Positives = 50/131 (37%), Gaps = 5/131 (3%)
Query: 945 EGELSPNGDMEEDNFAVYENSGLDAG-----HKGKGGDMSQKYQSRHGEQVCCEAKGEND 999
+G S NG E+++ + G +AG H G G Q + + E+D
Sbjct: 238 DGSPSGNGVEEDEDTGSGDGEGAEAGDGRESHDGTKGQGGQSHGGNTDHRGQSSVSTEDD 297
Query: 1000 VDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEG 1059
+ EG + H + SE S + D E+ S ++ D E ++SE G
Sbjct: 298 DSKEQEGFPNGHNGDNSSEENGVEEGDSTQATQDKEKLSPKDTRDAEGGIISQSEACPSG 357
Query: 1060 MVDAHDVEGDG 1070
+E +G
Sbjct: 358 KSQDQGIETEG 368
Score = 33.5 bits (75), Expect = 4.2
Identities = 57/380 (15%), Positives = 123/380 (32%), Gaps = 43/380 (11%)
Query: 693 SRAQGAEDTEDVVKAKNTEDVVNTENNSVKSGIVSVAESGDSKAWQLNGDSGVREDRCLD 752
S + + D+ D + ++ D ++ ++S SG ++S D+ + DS D
Sbjct: 565 SDSSDSSDSSDSSDSSDSSDSNSSSDSSDSSGSSDSSDSSDTCDSSDSSDSSDSSDSSDS 624
Query: 753 SDHTVYKTETAGNNTQHGKTNIIGFTPDELSGVNKQNQSSERLVNANVSPASGMEQSNGR 812
SD + + +++ ++ S + + SS+ +++ S +S + S+
Sbjct: 625 SDSSDSSDSSDSSDSSDSSSSSDSSDSSSCSDSSDSSDSSDSSDSSDSSDSSSSDSSSSS 684
Query: 813 TKIDNTSGHTATPSGNGNTSVEGGLELPSSEPILFQTRKFPSNYSIFSLMLDILINFGLH 872
D++ ++ S + + S + SS S+ S S D +
Sbjct: 685 NSSDSSDSSDSSSSSDSSDSSDSSDSSDSSGSSDSSDSSASSDSSSSSDSSDSSSSS--- 741
Query: 873 LHECILQRILFVKHPSYGWLMILVVDISSRFQIRQGGDSTRPGTFTNGATTGGTEVHRYQ 932
D S DS+ +N + + +
Sbjct: 742 -------------------------DSSDSSDSSDSSDSSESSDSSNSSDSSDSS----- 771
Query: 933 EQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCC 992
+ + S + D + + + + D+ D S S
Sbjct: 772 --------DSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDSSDSSDSSD 823
Query: 993 EAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAE 1052
+ + D+ D + S SSD S+++ + S+S+D + S D+
Sbjct: 824 SSDSSDSSDSSDSSDSSD--SSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSNS 881
Query: 1053 SEGEAEGMVDAHDVEGDGMS 1072
S+ D+ DG S
Sbjct: 882 SDSSDSDSKDSSSDSSDGDS 901
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 46.2 bits (108), Expect = 6e-04
Identities = 26/118 (22%), Positives = 57/118 (48%), Gaps = 1/118 (0%)
Query: 942 EREEGELSPNGDMEEDNFAVYENSG-LDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDV 1000
++EE + + +++E++ V E+ ++ + + + ++ + E+ E + E +
Sbjct: 546 DKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 605
Query: 1001 DADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAE 1058
D D+E E+ DD+E ++ + E D EE E+ ++ E D + E E E E
Sbjct: 606 DEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEEEEEEEE 663
Score = 38.1 bits (87), Expect = 0.17
Identities = 28/122 (22%), Positives = 52/122 (41%), Gaps = 10/122 (8%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDAD 1003
EE EL ++++ E+ + K D + + E+ E + E + + +
Sbjct: 535 EEAELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEE---EEEEEEEEEEE 591
Query: 1004 DEGEESPH-------RSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGE 1056
+E EE +D ++A E+ D + + D EE EE +D E D++ E E E
Sbjct: 592 EEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDE 651
Query: 1057 AE 1058
+
Sbjct: 652 ED 653
Score = 36.6 bits (83), Expect = 0.49
Identities = 48/273 (17%), Positives = 112/273 (40%), Gaps = 12/273 (4%)
Query: 134 RDRRSAMPTVRQAHVVKRERTTVSHCDRDPS--VDRPDPDHDRGLLRAEKEQRRRVEKEK 191
+D + +PT ++ DP+ ++ D D++ + +++ +KEK
Sbjct: 73 KDEKDDVPTTSNDNLKNAHNNNEISSSTDPTNIINVNDKDNENSV------DKKKDKKEK 126
Query: 192 DHREDGKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAEDFGAEPLLDADEFFGTRPMSS 251
H++D K E+ + + D + + + K K D+K ++ ++ + +P ++
Sbjct: 127 KHKKDKKEKKEKKDKKEKKDKKEKKHKKEKKHKKDKKKKE--NSEVMSLYKTGQHKPKNA 184
Query: 252 TCDDKKSLKTKYPEEFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLL 311
T +++L + E + L +P Y+E C I S T + + + +
Sbjct: 185 TEHGEENLDEEMVSEINNNAQGGLLLSSPYQYREQGGC-GIISSVHETSNDTKDNDKENI 243
Query: 312 GKYPDLMEEFNEFLVQAEKNDGGFLAGVMNKKSLWIEGPGLKPMKAEDRDRDRDRDRYRD 371
+ + E L +K + M K+ IE K + E + ++++R +
Sbjct: 244 SEDKKEDHQQEEMLKTLDKKERKQKEKEM-KEQEKIEKKKKKQEEKEKKKQEKERKKQEK 302
Query: 372 DGMKERDRECRERDRSTVVSNKEVSGLKIKDKH 404
K++++E +++ + K+ K K KH
Sbjct: 303 KERKQKEKEMKKQKKIEKERKKKEEKEKKKKKH 335
Score = 35.0 bits (79), Expect = 1.4
Identities = 26/124 (20%), Positives = 48/124 (37%), Gaps = 5/124 (4%)
Query: 927 EVHRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRH 986
E + + + ++ E +E E D EE+ E + + + + ++ +
Sbjct: 549 EDKKEESKEVQEESKEVQEDEEEVEEDEEEE-----EEEEEEEEEEEEEEEEEEEEEEEE 603
Query: 987 GEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDRE 1046
E E + + + D DD E+ DD E + D E D E+ EE E+ E
Sbjct: 604 EEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEEEEEEEE 663
Query: 1047 HDNK 1050
+ K
Sbjct: 664 SEKK 667
Score = 35.0 bits (79), Expect = 1.4
Identities = 20/80 (25%), Positives = 35/80 (43%), Gaps = 3/80 (3%)
Query: 993 EAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEE---CSREEHEDREHDN 1049
E + E + + ++E EE ++ E E D + D EE + E+ +D E D+
Sbjct: 574 EDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDD 633
Query: 1050 KAESEGEAEGMVDAHDVEGD 1069
E + E + D + E D
Sbjct: 634 DEEDDDEEDDDEDEDEDEED 653
Score = 34.3 bits (77), Expect = 2.4
Identities = 29/121 (23%), Positives = 47/121 (37%), Gaps = 11/121 (9%)
Query: 950 PNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAK-GENDVDADDEGEE 1008
P D + A E + L K + +K +S+ ++ E + E +V+ D+E EE
Sbjct: 522 PRRDNHKKKMAKIEEAELQK-QKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEE 580
Query: 1009 SPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEG 1068
++ E E + EE EE ++ E D E E + D D E
Sbjct: 581 EEEEEEEEEEEEEEEEE---------EEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEE 631
Query: 1069 D 1069
D
Sbjct: 632 D 632
>IE68_SHV21 (Q01042) Immediate-early protein
Length = 407
Score = 45.8 bits (107), Expect = 8e-04
Identities = 32/142 (22%), Positives = 66/142 (45%), Gaps = 6/142 (4%)
Query: 924 GGTEVHRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQ 983
G +++ +Q + +R E E+ G+ E + E G + G +G+G + +++ +
Sbjct: 44 GDDDINTTHQQQAALTEEQRRE-EVEEEGE-ERERRGEEEREG-EGGEEGEGREEAEEEE 100
Query: 984 SRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHE 1043
+ E E + + + + E EE+ +++ E E + +E A+ EE E E
Sbjct: 101 AEEKE---AEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEE 157
Query: 1044 DREHDNKAESEGEAEGMVDAHD 1065
E + + E+E EAE +A +
Sbjct: 158 AEEEEAEEEAEEEAEEAEEAEE 179
Score = 41.6 bits (96), Expect = 0.015
Identities = 38/160 (23%), Positives = 65/160 (39%), Gaps = 19/160 (11%)
Query: 927 EVHRYQEQSIRHFKSERE-----EGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQK 981
EV E+ R + ERE EGE + EE E + + + +++
Sbjct: 66 EVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEE 125
Query: 982 YQSRHGEQVCCEAKGENDVDA-------------DDEGEESPHRSSDDSENASENVDVSG 1028
++ E EA+ E +A ++E EE ++++E A E +
Sbjct: 126 AEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEA 185
Query: 1029 SESADGEECSREEHEDREHDNKA-ESEGEAEGMVDAHDVE 1067
E+ + EE E E E + +A E+E EAE +A + E
Sbjct: 186 EEAEEAEEAEEAEEEAEEAEEEAEEAEEEAEEAEEAEEAE 225
Score = 38.5 bits (88), Expect = 0.13
Identities = 33/147 (22%), Positives = 62/147 (41%), Gaps = 11/147 (7%)
Query: 932 QEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVC 991
+E+ ++E EE E + + EE+ E +A + + +++ + E+
Sbjct: 97 EEEEAEEKEAEEEEAEEAEE-EAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAE 155
Query: 992 CEAKGEN-DVDADDEGEESPHRSSDDSENASENVDVSGSESAD---------GEECSREE 1041
EA+ E + +A++E EE+ + E A E + +E A+ EE E
Sbjct: 156 EEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEAEEAEEEA 215
Query: 1042 HEDREHDNKAESEGEAEGMVDAHDVEG 1068
E E + E+E EAE + + G
Sbjct: 216 EEAEEAEEAEEAEEEAEEAEEEEEEAG 242
>NKX1_RAT (Q9QZM6) Sodium/potassium/calcium exchanger 1 precursor
(Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod
Na-Ca+K exchanger)
Length = 1181
Score = 45.4 bits (106), Expect = 0.001
Identities = 42/155 (27%), Positives = 66/155 (42%), Gaps = 21/155 (13%)
Query: 924 GGTEVHRY---QEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQ 980
G TE R QE+ E++EGE G E++ E G +A H+G+ +
Sbjct: 741 GETEAERKEDGQEEETETKGKEKQEGETESEGKDEQEGET--EAEGKEADHEGET-EAEG 797
Query: 981 KYQSRHGEQVC----------CEAKG---ENDVDADDEGEESPHRSSDDSENASENVDVS 1027
K GE EA+G E + + + EG+E H ++E N +
Sbjct: 798 KEVEHEGETEAEGTEDEQEGETEAEGKEVEQEGETEAEGKEVEHEVETEAERKETNHE-- 855
Query: 1028 GSESADGEECSREEHEDREHDNKAESEGEAEGMVD 1062
G A+G+E E + E + + + E EAEG V+
Sbjct: 856 GETEAEGKEADHEGETEAEGNVEHQGETEAEGKVE 890
Score = 40.4 bits (93), Expect = 0.034
Identities = 42/144 (29%), Positives = 65/144 (44%), Gaps = 16/144 (11%)
Query: 925 GTEV-HRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKG---GDMSQ 980
G EV H + ++ R K EGE G E D+ E G + H+G+ G +
Sbjct: 836 GKEVEHEVETEAER--KETNHEGETEAEGK-EADHEGETEAEG-NVEHQGETEAEGKVEH 891
Query: 981 KYQSRHGEQVCCEAKGENDVDADD----EGE-ESPHRSSDDSENASENVDVSGSESADGE 1035
+ ++ GE+ E +G+++ ADD +GE E+ + D E A G +DG
Sbjct: 892 EGETEAGEKD--EHEGQSETQADDTEVKDGEGEAEANAEDQCETAQGEKGADGGGGSDGG 949
Query: 1036 ECSREE-HEDREHDNKAESEGEAE 1058
+ EE ED E + + E E E E
Sbjct: 950 DSEEEEDEEDEEEEEEEEEEEEEE 973
Score = 40.0 bits (92), Expect = 0.044
Identities = 37/175 (21%), Positives = 68/175 (38%), Gaps = 22/175 (12%)
Query: 904 QIRQGGDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDMEEDNF-AVY 962
++ G++ GT E +++ + + E E+ + +E N
Sbjct: 799 EVEHEGETEAEGTEDEQEGETEAEGKEVEQEGETEAEGKEVEHEVETEAERKETNHEGET 858
Query: 963 ENSGLDAGHKGKG-GDMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSE--- 1018
E G +A H+G+ + + ++Q + E +GE + DE E +DD+E
Sbjct: 859 EAEGKEADHEGETEAEGNVEHQGETEAEGKVEHEGETEAGEKDEHEGQSETQADDTEVKD 918
Query: 1019 -------NASENVDVS----------GSESADGEECSREEHEDREHDNKAESEGE 1056
NA + + + GS+ D EE EE E+ E + + E E E
Sbjct: 919 GEGEAEANAEDQCETAQGEKGADGGGGSDGGDSEEEEDEEDEEEEEEEEEEEEEE 973
Score = 40.0 bits (92), Expect = 0.044
Identities = 30/120 (25%), Positives = 50/120 (41%), Gaps = 13/120 (10%)
Query: 938 HFKSEREEGELSPNGDMEEDNFAVYEN---SGLDAGHKGKGGDMSQKYQSRHGEQVCCEA 994
H EG + G+ E + +E +G H+G+ + + + GE
Sbjct: 867 HEGETEAEGNVEHQGETEAEGKVEHEGETEAGEKDEHEGQSETQADDTEVKDGE------ 920
Query: 995 KGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESE 1054
GE + +A+D+ E + D S+ D SE + EE EE E+ E + + ESE
Sbjct: 921 -GEAEANAEDQCETAQGEKGADGGGGSDGGD---SEEEEDEEDEEEEEEEEEEEEEEESE 976
Score = 38.9 bits (89), Expect = 0.099
Identities = 36/135 (26%), Positives = 64/135 (46%), Gaps = 21/135 (15%)
Query: 952 GDMEEDNFAVYENSGLDAGHKGK-GGDMSQKYQSRHGEQVCCEAKGENDVDADDEGE--- 1007
GD EED + +A H+G G+ ++ G++ + +GE + + ++G+
Sbjct: 701 GDQEEDPGCQEDVD--EAEHRGDMTGEEGERETEAEGKK---DEEGETEAERKEDGQEEE 755
Query: 1008 -ESPHRSSDDSENASENVD-VSGSESADGEECSRE-----EHEDREHDNKAESEG---EA 1057
E+ + + E SE D G A+G+E E E ++ EH+ + E+EG E
Sbjct: 756 TETKGKEKQEGETESEGKDEQEGETEAEGKEADHEGETEAEGKEVEHEGETEAEGTEDEQ 815
Query: 1058 EGMVDAH--DVEGDG 1070
EG +A +VE +G
Sbjct: 816 EGETEAEGKEVEQEG 830
Score = 37.0 bits (84), Expect = 0.38
Identities = 33/133 (24%), Positives = 55/133 (40%), Gaps = 22/133 (16%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDAD 1003
EEGE + ++D E + G + + ++ Q E E K E + + +
Sbjct: 725 EEGERETEAEGKKDEEGETEAERKEDGQEEETETKGKEKQEGETES---EGKDEQEGETE 781
Query: 1004 DEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEG-------- 1055
EG+E+ H ++E + V+ G A+G E E + + E+EG
Sbjct: 782 AEGKEADHEGETEAEG--KEVEHEGETEAEGTE--------DEQEGETEAEGKEVEQEGE 831
Query: 1056 -EAEGMVDAHDVE 1067
EAEG H+VE
Sbjct: 832 TEAEGKEVEHEVE 844
Score = 34.7 bits (78), Expect = 1.9
Identities = 25/99 (25%), Positives = 42/99 (42%), Gaps = 14/99 (14%)
Query: 993 EAKGENDVD--ADDEGEESPHRSSDDSENASENVDVSGSESADGE-ECSREE-------- 1041
E KG+ + D ++ +E+ HR E + G + +GE E R+E
Sbjct: 698 EDKGDQEEDPGCQEDVDEAEHRGDMTGEEGERETEAEGKKDEEGETEAERKEDGQEEETE 757
Query: 1042 ---HEDREHDNKAESEGEAEGMVDAHDVEGDGMSLTISE 1077
E +E + ++E + E EG +A E D T +E
Sbjct: 758 TKGKEKQEGETESEGKDEQEGETEAEGKEADHEGETEAE 796
>NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium polypeptide)
(NF-M) (Fragment)
Length = 810
Score = 45.4 bits (106), Expect = 0.001
Identities = 34/137 (24%), Positives = 58/137 (41%), Gaps = 13/137 (9%)
Query: 929 HRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGE 988
H++ E+ I K E E+ E MEE A+ E + + K + +K + E
Sbjct: 335 HKFVEEIIEETKVEDEKSE------MEEALTAITEELAVSVKEEVKEEEAEEKEEKEEAE 388
Query: 989 QVCCEAKGENDVDA------DDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEH 1042
+ AK ++ V A ++EGE+ ++ E E +E E+ E
Sbjct: 389 EEVVAAK-KSPVKATAPELKEEEGEKEEEEGQEEEEEEEEAAKSDQAEEGGSEKEGSSEK 447
Query: 1043 EDREHDNKAESEGEAEG 1059
E+ E + + E+E E EG
Sbjct: 448 EEGEQEEEGETEAEGEG 464
>SIS2_YEAST (P36024) SIS2 protein (Halotolerance protein HAL3)
Length = 562
Score = 45.1 bits (105), Expect = 0.001
Identities = 19/67 (28%), Positives = 37/67 (54%)
Query: 998 NDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEGEA 1057
N+ + DDE EE +D+E+ +EN + + D ++ ++ +D + D++ E E E
Sbjct: 495 NEEEDDDEDEEEDDDEEEDTEDKNENNNDDDDDDDDDDDDDDDDDDDDDDDDEDEDEAET 554
Query: 1058 EGMVDAH 1064
G++D H
Sbjct: 555 PGIIDKH 561
>UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream binding
factor 1) (UBF-1)
Length = 764
Score = 44.3 bits (103), Expect = 0.002
Identities = 26/79 (32%), Positives = 40/79 (49%), Gaps = 2/79 (2%)
Query: 993 EAKGENDVDADDEGEESPHRSSDDSENASENVDVSG-SESADGEEC-SREEHEDREHDNK 1050
E E D D DDE EE + D SE+ ++ + S ES DG+E ++ ED + D+
Sbjct: 679 EDDDEEDDDDDDEEEEEDDENGDSSEDGGDSSESSSEDESEDGDENEDDDDDEDDDEDDD 738
Query: 1051 AESEGEAEGMVDAHDVEGD 1069
+ + E+EG + GD
Sbjct: 739 EDEDNESEGSSSSSSSSGD 757
Score = 40.4 bits (93), Expect = 0.034
Identities = 22/88 (25%), Positives = 41/88 (46%), Gaps = 1/88 (1%)
Query: 978 MSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEEC 1037
+ K +S + + + + + DDE +S D SE++SE+ G E+ D ++
Sbjct: 671 LQSKSESEEDDDEEDDDDDDEEEEEDDENGDSSEDGGDSSESSSEDESEDGDENEDDDD- 729
Query: 1038 SREEHEDREHDNKAESEGEAEGMVDAHD 1065
++ ED + D ESEG + + D
Sbjct: 730 DEDDDEDDDEDEDNESEGSSSSSSSSGD 757
Score = 38.9 bits (89), Expect = 0.099
Identities = 19/65 (29%), Positives = 32/65 (49%)
Query: 993 EAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAE 1052
E EN ++D G+ S S D+SE+ EN D E D ++ E++E + +
Sbjct: 694 EEDDENGDSSEDGGDSSESSSEDESEDGDENEDDDDDEDDDEDDDEDEDNESEGSSSSSS 753
Query: 1053 SEGEA 1057
S G++
Sbjct: 754 SSGDS 758
Score = 35.0 bits (79), Expect = 1.4
Identities = 24/92 (26%), Positives = 46/92 (49%), Gaps = 2/92 (2%)
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGEND 999
KSE EE + + D +++ + +G + G + S + +S G++ + E+D
Sbjct: 674 KSESEEDDDEEDDDDDDEEEEEDDENGDSSEDGGDSSESSSEDESEDGDENEDDDDDEDD 733
Query: 1000 VDADDEGEESPHRSSDDSENASENVDVSGSES 1031
+ DDE E+ + S S ++S + D S S+S
Sbjct: 734 DEDDDEDED--NESEGSSSSSSSSGDSSDSDS 763
Score = 32.3 bits (72), Expect = 9.2
Identities = 22/83 (26%), Positives = 36/83 (42%), Gaps = 6/83 (7%)
Query: 1000 VDADDEGEESPHRSSDDSENASENV-DVSGSESADGEECSREEHEDR-----EHDNKAES 1053
+ + E EE DD ++ E D +G S DG + S ED E+++ +
Sbjct: 671 LQSKSESEEDDDEEDDDDDDEEEEEDDENGDSSEDGGDSSESSSEDESEDGDENEDDDDD 730
Query: 1054 EGEAEGMVDAHDVEGDGMSLTIS 1076
E + E + D E +G S + S
Sbjct: 731 EDDDEDDDEDEDNESEGSSSSSS 753
>S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding
protein)
Length = 738
Score = 44.3 bits (103), Expect = 0.002
Identities = 38/158 (24%), Positives = 63/158 (39%), Gaps = 25/158 (15%)
Query: 919 NGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDM 978
+G+ G EV R ++ R +S +EEG G E + G + ++ +GG
Sbjct: 150 DGSERIGLEVDR---RASRSSQSSKEEGNSEEYGSDHETGSSASSEQGNNTENEEEGG-- 204
Query: 979 SQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECS 1038
E DV+ D+E +E + E+A E D E + EE
Sbjct: 205 ------------------EEDVEEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEEDEEEE 246
Query: 1039 REEHEDREHDNKAESE--GEAEGMVDAHDVEGDGMSLT 1074
EE E+ E D + + E + + +A D + +S T
Sbjct: 247 EEEEEEYEQDERDQKEEGNDYDTRSEASDSGSESVSFT 284
Score = 35.0 bits (79), Expect = 1.4
Identities = 24/101 (23%), Positives = 44/101 (42%), Gaps = 11/101 (10%)
Query: 160 DRDPSVDRP---DPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGRDHEHDGGQYR 216
+ D VD D + D E E+ E++++ E+ + ++E+D RD + +G Y
Sbjct: 209 EEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEEDEEEEEEEEEEYEQDERDQKEEGNDYD 268
Query: 217 ERLSHKRKSDRKAEDFGAEPLLDADEFFGTRPMSSTCDDKK 257
R +A D G+E + D + + D+KK
Sbjct: 269 TR--------SEASDSGSESVSFTDGSVRSGSGTDGSDEKK 301
>NKX1_HUMAN (O60721) Sodium/potassium/calcium exchanger 1 precursor
(Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod
Na-Ca+K exchanger)
Length = 1099
Score = 44.3 bits (103), Expect = 0.002
Identities = 39/135 (28%), Positives = 57/135 (41%), Gaps = 30/135 (22%)
Query: 944 EEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQ-KYQSRHGEQVCCEAKGENDVDA 1002
EEGE + G+ EE SG + +G+G +Q K + E EA+G+ D +
Sbjct: 759 EEGETAGEGETEE-------KSGGETQPEGEGETETQGKGEECEDEN---EAEGKGDNEG 808
Query: 1003 DDEGE---ESPHRSSDDSENASENV----------------DVSGSESADGEECSREEHE 1043
+DEGE E ++ E S+ + D GS+ D EE EE E
Sbjct: 809 EDEGEIHAEDGEMKGNEGETESQELSAENHGEAKNDEKGVEDGGGSDGGDSEEEEEEEEE 868
Query: 1044 DREHDNKAESEGEAE 1058
E + + E E E E
Sbjct: 869 QEEEEEEEEQEEEEE 883
Score = 34.3 bits (77), Expect = 2.4
Identities = 36/150 (24%), Positives = 57/150 (38%), Gaps = 27/150 (18%)
Query: 906 RQGGDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDME-EDNFAVYEN 964
+ GG++ G G TE E+ +E E GD E ED ++
Sbjct: 772 KSGGETQPEG-------EGETETQGKGEEC-------EDENEAEGKGDNEGEDEGEIHAE 817
Query: 965 SGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENV 1024
G G++G+ + + HGE +ND ++G S S++ E E
Sbjct: 818 DGEMKGNEGE-TESQELSAENHGE-------AKNDEKGVEDGGGSDGGDSEEEEEEEEEQ 869
Query: 1025 DVSGSESADGEECSREEHEDREHDNKAESE 1054
+ E + EE EE E+ E + K E
Sbjct: 870 E----EEEEEEEQEEEEEEEEEEEEKGNEE 895
Score = 33.9 bits (76), Expect = 3.2
Identities = 38/173 (21%), Positives = 64/173 (36%), Gaps = 14/173 (8%)
Query: 942 EREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQ----------KYQSRHGEQVC 991
E+ GE P G+ E + E + +GKG + + + + GE
Sbjct: 771 EKSGGETQPEGEGETETQGKGEECEDENEAEGKGDNEGEDEGEIHAEDGEMKGNEGETES 830
Query: 992 CEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKA 1051
E EN +A ++ + D ++ E + + + EE +EE E+ E +
Sbjct: 831 QELSAENHGEAKNDEKGVEDGGGSDGGDSEEEEEEEEEQEEEEEEEEQEEEEEEEEEE-- 888
Query: 1052 ESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKPLAKYVPAALHEKDRNSQVF 1104
E +G E + + D I L V PL VP ++ R VF
Sbjct: 889 EEKGNEEPL--SLDWPETRQKQAIYLFLLPIVFPLWLTVPDVRRQESRKFFVF 939
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 170,905,495
Number of Sequences: 164201
Number of extensions: 7869478
Number of successful extensions: 24743
Number of sequences better than 10.0: 339
Number of HSP's better than 10.0 without gapping: 114
Number of HSP's successfully gapped in prelim test: 229
Number of HSP's that attempted gapping in prelim test: 20971
Number of HSP's gapped (non-prelim): 2304
length of query: 1389
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1266
effective length of database: 39,777,331
effective search space: 50358101046
effective search space used: 50358101046
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0028b.5


