

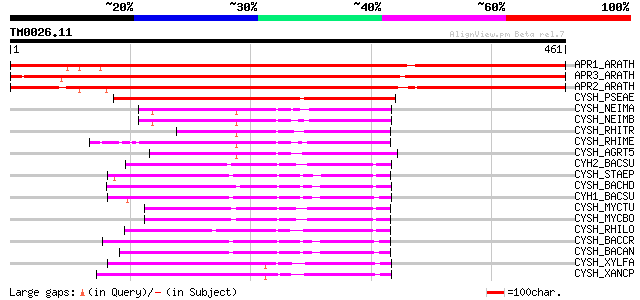
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.11
(461 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
APR1_ARATH (P92979) 5'-adenylylsulfate reductase 1, chloroplast ... 684 0.0
APR3_ARATH (P92980) 5'-adenylylsulfate reductase 3, chloroplast ... 683 0.0
APR2_ARATH (P92981) 5'-adenylylsulfate reductase 2, chloroplast ... 660 0.0
CYSH_PSEAE (O05927) Phosphoadenosine phosphosulfate reductase (E... 290 5e-78
CYSH_NEIMA (Q9JUD5) Phosphoadenosine phosphosulfate reductase (E... 171 4e-42
CYSH_NEIMB (Q9JRT1) Phosphoadenosine phosphosulfate reductase (E... 167 5e-41
CYSH_RHITR (O33579) Phosphoadenosine phosphosulfate reductase (E... 159 1e-38
CYSH_RHIME (P56891) Phosphoadenosine phosphosulfate reductase (E... 159 2e-38
CYSH_AGRT5 (Q8UH67) Phosphoadenosine phosphosulfate reductase (E... 158 3e-38
CYH2_BACSU (O06737) Probable phosphoadenosine phosphosulfate red... 122 1e-27
CYSH_STAEP (Q8CMX3) Phosphoadenosine phosphosulfate reductase (E... 114 5e-25
CYSH_BACHD (Q9KCT3) Phosphoadenosine phosphosulfate reductase (E... 109 1e-23
CYH1_BACSU (P94498) Phosphoadenosine phosphosulfate reductase (E... 109 1e-23
CYSH_MYCTU (P65668) Probable phosphoadenosine phosphosulfate red... 109 2e-23
CYSH_MYCBO (P65669) Probable phosphoadenosine phosphosulfate red... 109 2e-23
CYSH_RHILO (Q98GP9) Phosphoadenosine phosphosulfate reductase (E... 108 3e-23
CYSH_BACCR (Q81FZ1) Phosphoadenosine phosphosulfate reductase (E... 101 4e-21
CYSH_BACAN (Q81T49) Phosphoadenosine phosphosulfate reductase (E... 99 2e-20
CYSH_XYLFA (Q9PD82) Phosphoadenosine phosphosulfate reductase (E... 97 1e-19
CYSH_XANCP (Q8P607) Phosphoadenosine phosphosulfate reductase (E... 94 6e-19
>APR1_ARATH (P92979) 5'-adenylylsulfate reductase 1, chloroplast
precursor (EC 1.8.4.9) (Adenosine 5'-phosphosulfate
5'-adenylylsulfate sulfotransferase 1) (APS
sulfotransferase 1) (Thioredoxin independent APS
reductase 1) (3'-phosphoadenosine-5'-phosphos
Length = 465
Score = 684 bits (1766), Expect = 0.0
Identities = 342/471 (72%), Positives = 400/471 (84%), Gaps = 16/471 (3%)
Query: 1 MALAVTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVN---ASQRRSFVKP- 56
MA++V S S SS +S S +PK SQI S+R+ + H V+ + +R S VKP
Sbjct: 1 MAMSVNVSSSSSSGIINSRFGVSLEPKVSQIGSLRLLDRVHVAPVSLNLSGKRSSSVKPL 60
Query: 57 --PQRSRDSIAPLAATIVAS---DVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHI 111
+++DS+ PLAAT+VA +V E +++++LA LENASPL+IMD ALEK+GN I
Sbjct: 61 NAEPKTKDSMIPLAATMVAEIAEEVEVVEIEDFEELAKKLENASPLEIMDKALEKYGNDI 120
Query: 112 AIAFSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAV 171
AIAFSGAEDVALIEYA LTGRPFRVFSLDTGRLNPETYRFFD VEKHYGI IEYMFPD+V
Sbjct: 121 AIAFSGAEDVALIEYAHLTGRPFRVFSLDTGRLNPETYRFFDAVEKHYGIRIEYMFPDSV 180
Query: 172 EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVV 231
EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGL+AWITGQRKDQSPGTRSE+PVV
Sbjct: 181 EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVV 240
Query: 232 QVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRA 291
QVDPVFEG+DGGVGSLVKWNPVANV+G+D+W+FLRTM+VPVN+LH+ GYISIGCEPCT+A
Sbjct: 241 QVDPVFEGLDGGVGSLVKWNPVANVEGNDVWNFLRTMDVPVNTLHAAGYISIGCEPCTKA 300
Query: 292 VLPGQHEREGRWWWEDAKAKECGLHKGNVKQDA-EAELNGNGVANTNGTATVADIFNTQN 350
VLPGQHEREGRWWWEDAKAKECGLHKGNVK+++ +A++NG + VADIF ++N
Sbjct: 301 VLPGQHEREGRWWWEDAKAKECGLHKGNVKENSDDAKVNG------ESKSAVADIFKSEN 354
Query: 351 VVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRA 410
+V+LSR GIENL KLENRKEPW+VVLYAPWCP+CQAME SY +LADKLA SG+KV KFRA
Sbjct: 355 LVTLSRQGIENLMKLENRKEPWIVVLYAPWCPFCQAMEASYDELADKLAGSGIKVAKFRA 414
Query: 411 DGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNALR 461
DG+QKEFAK+EL LGSFPTI+ FPK+SSRPIKYPSE RDV+SL +F+N +R
Sbjct: 415 DGDQKEFAKQELQLGSFPTILVFPKNSSRPIKYPSEKRDVESLTSFLNLVR 465
>APR3_ARATH (P92980) 5'-adenylylsulfate reductase 3, chloroplast
precursor (EC 1.8.4.9) (Adenosine 5'-phosphosulfate
5'-adenylylsulfate sulfotransferase 3) (APS
sulfotransferase 3) (Thioredoxin independent APS
reductase 3) (3'-phosphoadenosine-5'-phosphos
Length = 458
Score = 683 bits (1763), Expect = 0.0
Identities = 338/463 (73%), Positives = 391/463 (84%), Gaps = 7/463 (1%)
Query: 1 MALAVTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEIPH--GGGVNASQRRSFVKPPQ 58
MALA+ S S SSS+ SSS+F SS K ++I S+R+ + ++ S +RS VK
Sbjct: 1 MALAINVSSS-SSSAISSSSFPSSDLKVTKIGSLRLLNRTNVSAASLSLSGKRSSVKALN 59
Query: 59 RSRDSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFSGA 118
+ + A+ V + E +++++LA LENASPL+IMD ALEKFGN IAIAFSGA
Sbjct: 60 VQSITKESIVASEVTEKLDVVEVEDFEELAKRLENASPLEIMDKALEKFGNDIAIAFSGA 119
Query: 119 EDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQGLVR 178
EDVALIEYA LTGRP+RVFSLDTGRLNPETYR FD VEKHYGI IEYMFPDAVEVQ LVR
Sbjct: 120 EDVALIEYAHLTGRPYRVFSLDTGRLNPETYRLFDTVEKHYGIRIEYMFPDAVEVQALVR 179
Query: 179 SKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFE 238
+KGLFSFYEDGHQECCR+RKVRPLRRALKGLRAWITGQRKDQSPGTRSE+PVVQVDPVFE
Sbjct: 180 NKGLFSFYEDGHQECCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFE 239
Query: 239 GVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHE 298
G+DGGVGSLVKWNPVANV+G+D+W+FLRTM+VPVN+LH+ GY+SIGCEPCTRAVLPGQHE
Sbjct: 240 GLDGGVGSLVKWNPVANVEGNDVWNFLRTMDVPVNTLHAAGYVSIGCEPCTRAVLPGQHE 299
Query: 299 REGRWWWEDAKAKECGLHKGNVKQDAEAELNGNGVANTNGTATVADIFNTQNVVSLSRTG 358
REGRWWWEDAKAKECGLHKGN+K++ NGN AN NGTA+VADIFN++NVV+LSR G
Sbjct: 300 REGRWWWEDAKAKECGLHKGNIKENT----NGNATANVNGTASVADIFNSENVVNLSRQG 355
Query: 359 IENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFA 418
IENL KLENRKE W+VVLYAPWCP+CQAME S+ +LADKL SGVKV KFRADG+QK+FA
Sbjct: 356 IENLMKLENRKEAWIVVLYAPWCPFCQAMEASFDELADKLGGSGVKVAKFRADGDQKDFA 415
Query: 419 KRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNALR 461
K+EL LGSFPTI+ FPK+SSRPIKYPSE RDVDSL +F+N +R
Sbjct: 416 KKELQLGSFPTILVFPKNSSRPIKYPSEKRDVDSLTSFLNLVR 458
>APR2_ARATH (P92981) 5'-adenylylsulfate reductase 2, chloroplast
precursor (EC 1.8.4.9) (Adenosine 5'-phosphosulfate
5'-adenylylsulfate sulfotransferase 2) (APS
sulfotransferase 2) (Thioredoxin independent APS
reductase 2) (3'-phosphoadenosine-5'-phosphos
Length = 454
Score = 660 bits (1704), Expect = 0.0
Identities = 334/467 (71%), Positives = 388/467 (82%), Gaps = 19/467 (4%)
Query: 1 MALAVTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVNASQRRSFVKP---P 57
MALAVT S + S SS S + SS+ K QI SIR+S+ H SQRR +KP
Sbjct: 1 MALAVTSSSTAISGSSFSRSGASSESKALQICSIRLSDRTH-----LSQRRYSMKPLNAE 55
Query: 58 QRSR-DSIAPLAATIVASDVAET--EQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIA 114
SR +S A+T++A +V E E ++++QLA LE+ASPL+IMD ALE+FG+ IAIA
Sbjct: 56 SHSRSESWVTRASTLIAPEVEEKGGEVEDFEQLAKKLEDASPLEIMDKALERFGDQIAIA 115
Query: 115 FSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQ 174
FSGAEDVALIEYARLTG+PFRVFSLDTGRLNPETYR FD VEK YGI IEYMFPDAVEVQ
Sbjct: 116 FSGAEDVALIEYARLTGKPFRVFSLDTGRLNPETYRLFDAVEKQYGIRIEYMFPDAVEVQ 175
Query: 175 GLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVD 234
LVR+KGLFSFYEDGHQECCRVRKVRPLRRALKGL+AWITGQRKDQSPGTRSE+P+VQVD
Sbjct: 176 ALVRNKGLFSFYEDGHQECCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVD 235
Query: 235 PVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLP 294
PVFEG+DGGVGSLVKWNP+ANV+G D+W+FLRTM+VPVN+LH++GY+SIGCEPCTR VLP
Sbjct: 236 PVFEGLDGGVGSLVKWNPLANVEGADVWNFLRTMDVPVNALHAQGYVSIGCEPCTRPVLP 295
Query: 295 GQHEREGRWWWEDAKAKECGLHKGNVKQDAEAELNGNGVANTNGTATVADIFNTQNVVSL 354
GQHEREGRWWWEDAKAKECGLHKGN+K++ +G A++ A V +IF + NVV+L
Sbjct: 296 GQHEREGRWWWEDAKAKECGLHKGNIKEE-------DGAADSK-PAAVQEIFESNNVVAL 347
Query: 355 SRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQ 414
S+ G+ENL KLENRKE WLVVLYAPWCP+CQAME SY++LA+KLA GVKV KFRADGEQ
Sbjct: 348 SKGGVENLLKLENRKEAWLVVLYAPWCPFCQAMEASYIELAEKLAGKGVKVAKFRADGEQ 407
Query: 415 KEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNALR 461
KEFAK+EL LGSFPTI+ FPK + R IKYPSE+RDVDSLM+FVN LR
Sbjct: 408 KEFAKQELQLGSFPTILLFPKRAPRAIKYPSEHRDVDSLMSFVNLLR 454
>CYSH_PSEAE (O05927) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 267
Score = 290 bits (742), Expect = 5e-78
Identities = 133/234 (56%), Positives = 173/234 (73%), Gaps = 3/234 (1%)
Query: 87 LAVDLENASPLQIMDAALEKFGNHIAIAFSGAEDVALIEYARLTGRPFRVFSLDTGRLNP 146
LA L + SP I+ AA E FG+ + I+FSGAEDV L++ A R +VFSLDTGRL+P
Sbjct: 33 LASSLADKSPQDILKAAFEHFGDELWISFSGAEDVVLVDMAWKLNRNVKVFSLDTGRLHP 92
Query: 147 ETYRFFDEVEKHYGIHIEYMFPDAVEVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRAL 206
ETYRF D+V +HYGI I+ + PD ++ LV+ KGLFSFY DGH ECC +RK+ PL+R L
Sbjct: 93 ETYRFIDQVREHYGIAIDVLSPDPRLLEPLVKEKGLFSFYRDGHGECCGIRKIEPLKRKL 152
Query: 207 KGLRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLR 266
G+RAW TGQR+DQSPGTRS+V V+++D F + L K+NP++++ ++W ++R
Sbjct: 153 AGVRAWATGQRRDQSPGTRSQVAVLEIDGAFSTPE---KPLYKFNPLSSMTSEEVWGYIR 209
Query: 267 TMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLHKGNV 320
+ +P NSLH +GYISIGCEPCTR VLP QHEREGRWWWE+A KECGLH GN+
Sbjct: 210 MLELPYNSLHERGYISIGCEPCTRPVLPNQHEREGRWWWEEATHKECGLHAGNL 263
>CYSH_NEIMA (Q9JUD5) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 244
Score = 171 bits (433), Expect = 4e-42
Identities = 89/216 (41%), Positives = 128/216 (59%), Gaps = 15/216 (6%)
Query: 108 GNHIAIAFSG---AEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYG-IHI 163
G+H F+ AED+ + + +F+LDTG L+ ET D +E+ Y + I
Sbjct: 38 GSHRDARFASSLAAEDMVITDLIAGENLNIGIFTLDTGLLHAETLNLLDRIERVYPHMQI 97
Query: 164 EYMFPDAVEVQGLVRSKGLFSFYE--DGHQECCRVRKVRPLRRALKGLRAWITGQRKDQS 221
+ P + V SKG F+FY+ + +ECCR+RK PL RA+ G AW+TGQR++QS
Sbjct: 98 KRFQPIREDALHYVESKGRFAFYDSVEARRECCRIRKTEPLDRAIAGADAWLTGQRREQS 157
Query: 222 PGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYI 281
TR+E+P + D G+D K+NP+ + HD+W+++ NVP N L+ +G+
Sbjct: 158 -ATRTELPFAEYD-AGRGID-------KYNPIFDWSEHDVWAYILANNVPYNDLYRQGFP 208
Query: 282 SIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLHK 317
SIGC+PCTR V G+ R GRWWWED +KECGLHK
Sbjct: 209 SIGCDPCTRPVKAGEDIRAGRWWWEDKNSKECGLHK 244
>CYSH_NEIMB (Q9JRT1) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 246
Score = 167 bits (423), Expect = 5e-41
Identities = 87/216 (40%), Positives = 126/216 (58%), Gaps = 15/216 (6%)
Query: 108 GNHIAIAFSG---AEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYG-IHI 163
G+H F+ AED+ + + +F+LDTG L+ ET D + + Y + I
Sbjct: 40 GSHRDARFASSLAAEDMVITDLIAGENLNIGIFTLDTGLLHTETLNLLDRLGRAYPHLRI 99
Query: 164 EYMFPDAVEVQGLVRSKGLFSFYE--DGHQECCRVRKVRPLRRALKGLRAWITGQRKDQS 221
+ P + V SKG F+FY+ + +ECCR+RK PL RA+ G AW+TGQR++QS
Sbjct: 100 KRFRPVREDADRYVESKGRFAFYDSVEARRECCRIRKTEPLNRAIAGADAWLTGQRREQS 159
Query: 222 PGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYI 281
TR+E+P + D G+G K+NP+ + HD+W+++ NVP N L+ +G+
Sbjct: 160 -ATRTELPFAEYD-----AGRGIG---KYNPIFDWSEHDVWAYILANNVPYNDLYRQGFP 210
Query: 282 SIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLHK 317
SIGC+PCTR V G+ R GRWWWE +KECGLHK
Sbjct: 211 SIGCDPCTRPVKAGEDIRAGRWWWEGRNSKECGLHK 246
>CYSH_RHITR (O33579) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase) (Fragment)
Length = 180
Score = 159 bits (402), Expect = 1e-38
Identities = 80/180 (44%), Positives = 105/180 (57%), Gaps = 11/180 (6%)
Query: 139 LDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQGLVRSKGLFSFYE--DGHQECCRV 196
L TGRL PET + DE E Y I I P+ ++ GL FYE + CC V
Sbjct: 1 LQTGRLFPETLKLIDETENQYDIRISRYEPEQADIDAYAEKYGLNGFYESVEARHACCGV 60
Query: 197 RKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANV 256
RK++PL RAL G W+TG R+ QS R++ P + DP +L+K NP+A+
Sbjct: 61 RKLKPLARALSGATIWVTGLRRGQS-ANRADTPFAEYDPE--------RNLIKVNPLADW 111
Query: 257 KGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLH 316
I +++ VPVN LH +GY SIGCEPCTRA+ PG+ ER GRWWWE+ + +ECGLH
Sbjct: 112 DIDVIRAYVADNGVPVNPLHQRGYPSIGCEPCTRAIKPGEPERAGRWWWENDEKRECGLH 171
>CYSH_RHIME (P56891) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 265
Score = 159 bits (401), Expect = 2e-38
Identities = 100/252 (39%), Positives = 138/252 (54%), Gaps = 17/252 (6%)
Query: 67 LAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFSGAEDVALIEY 126
L A ++A D AE + N + ++DL A L ++ A LE G + G ED +
Sbjct: 13 LEADVMALD-AEAKALNDKLESLDL--AGRLALI-AGLE--GRAVFTTSLGIEDQVITAA 66
Query: 127 ARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQGLVRSKGLFSFY 186
V +L TGRL ET D+ E+ Y I I+ +P+ ++ V G+ FY
Sbjct: 67 IGSNRLDIEVATLKTGRLFNETVALIDQTEETYDILIKRYYPEKADIDAYVAQYGMNGFY 126
Query: 187 E--DGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGV 244
E + CC VRK++PL RAL G WITG R+ QS G R+ P + D V+ G
Sbjct: 127 ESVEARHACCGVRKLKPLARALDGASYWITGLRRGQS-GNRATTPFAEAD-----VERG- 179
Query: 245 GSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWW 304
L+K NP+A+ I + + +PVN LHS+GY SIGCEPCTRA+ PG+ ER GRWW
Sbjct: 180 --LIKINPLADWGIETIQAHVAAEGIPVNPLHSRGYPSIGCEPCTRAIKPGEPERAGRWW 237
Query: 305 WEDAKAKECGLH 316
WE+ + +ECGLH
Sbjct: 238 WENDEKRECGLH 249
>CYSH_AGRT5 (Q8UH67) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 260
Score = 158 bits (399), Expect = 3e-38
Identities = 84/208 (40%), Positives = 114/208 (54%), Gaps = 11/208 (5%)
Query: 117 GAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQGL 176
G ED + P V +L+TGRL ET DE E+ +GI I P+ ++
Sbjct: 52 GIEDQVITAAIGTHRLPIDVVTLETGRLFKETVDLIDETEERFGIEIRRFRPEQDDIDAY 111
Query: 177 VRSKGLFSFYE--DGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVD 234
GL FYE + CC VRK+ PL +AL+G WITG R+ QS G R+ P + D
Sbjct: 112 AAKYGLNGFYESVEARHACCHVRKLIPLGKALEGAAFWITGLRRGQS-GNRAATPFAEFD 170
Query: 235 PVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLP 294
+L+K N +A+ I +++ N+PVN LH +GY SIGCEPCTRA+ P
Sbjct: 171 --------AERNLIKINALADWDIEQIRAYVAEENIPVNPLHQRGYPSIGCEPCTRAIKP 222
Query: 295 GQHEREGRWWWEDAKAKECGLHKGNVKQ 322
G+ ER GRWWWE+ + +ECGLH +Q
Sbjct: 223 GEPERAGRWWWENDEKRECGLHVAGAEQ 250
>CYH2_BACSU (O06737) Probable phosphoadenosine phosphosulfate
reductase (EC 1.8.4.8) (PAPS reductase, thioredoxin
dependent) (PAdoPS reductase) (3'-phosphoadenylylsulfate
reductase) (PAPS sulfotransferase)
Length = 236
Score = 122 bits (307), Expect = 1e-27
Identities = 75/223 (33%), Positives = 114/223 (50%), Gaps = 15/223 (6%)
Query: 97 LQIMDAALEKFGNHIAIAFS-GAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEV 155
L ++ A +G I A S GAE + L++ + + LDTG ETY + V
Sbjct: 26 LDVLKWAYRTYGEKIVYACSFGAEGMVLLDLISKINKNAHIIFLDTGLHFQETYELIETV 85
Query: 156 EKHY-GIHIEYMFPDAVEVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWIT 214
++ Y G I+ + P+ + + G ++ CC++RK+ PL++ L G+ AWI+
Sbjct: 86 KERYPGFAIQMLEPELSLTEQGTKYGG--ELWKHNPNLCCQLRKIEPLKKHLSGMTAWIS 143
Query: 215 GQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNS 274
G R+DQSP TR + V +D FE L+K P+ + D+W+++R N+P N
Sbjct: 144 GLRRDQSP-TRKHIQYVNLDQKFE--------LIKICPLIHWTWDDVWTYIRLHNLPYNK 194
Query: 275 LHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLHK 317
LH + Y SIGCE CT ER GR W + ECGLH+
Sbjct: 195 LHDQHYPSIGCEMCTLPSPDPNDERAGR--WAGREKTECGLHQ 235
>CYSH_STAEP (Q8CMX3) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 243
Score = 114 bits (285), Expect = 5e-25
Identities = 78/244 (31%), Positives = 120/244 (48%), Gaps = 23/244 (9%)
Query: 82 DNYQ------QLAVDLENASPLQIMDAALEKFGNHIAIAFS-GAEDVALIEYARLTGRPF 134
DN+Q +L ++ E +I+ A + + N I + S GAE + LI+
Sbjct: 9 DNFQNDPFINELDINDETKGAYEILKWAYQTYENDIVYSCSFGAESMVLIDLISQIKPDA 68
Query: 135 RVFSLDTGRLNPETYRFFDEVEKHYG-IHIEYMFPD-AVEVQGLVRSKGLFSFYEDGHQE 192
++ LDT ETY D V+ Y + I+ P+ +E QG + L +++ +
Sbjct: 69 QIVFLDTDLHFQETYDLIDRVKDKYPQLRIKMKKPELTLEEQGEKYNPAL---WKNDPNQ 125
Query: 193 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNP 252
CC +RK++PL L G AWI+G R+ QSP TR+ + D F+ + V L+ W
Sbjct: 126 CCYIRKIKPLEDVLSGAVAWISGLRRAQSP-TRAHTNFINKDERFKSIK--VCPLIYWTE 182
Query: 253 VANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAKE 312
++WS++R ++P N LH + Y SIGC PCT V R GR W ++ E
Sbjct: 183 ------EEVWSYIRDKDLPYNELHDQNYPSIGCIPCTSPVFDSNDSRAGR--WSNSSKTE 234
Query: 313 CGLH 316
CGLH
Sbjct: 235 CGLH 238
>CYSH_BACHD (Q9KCT3) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 231
Score = 109 bits (273), Expect = 1e-23
Identities = 78/240 (32%), Positives = 114/240 (47%), Gaps = 17/240 (7%)
Query: 81 QDNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFS-GAEDVALIEYARLTGRPFRVFSL 139
Q N ++ LE L ++ A + +G+ + A S GAE + LI+ V L
Sbjct: 6 QVNINEVNQALERKDSLDVIKWAYKTYGDKLVYACSMGAEGMVLIDLISKVRPDAPVIFL 65
Query: 140 DTGRLNPETYRFFDEVEKHYG-IHIEYMFPDAV-EVQGLVRSKGLFSFYEDGHQECCRVR 197
DT ETY + V++ Y + ++ + P+ E Q L+ D CC++R
Sbjct: 66 DTDFHFSETYELIERVKERYPKLQLKLVKPELTPEEQAETYGDRLWERQPD---LCCKLR 122
Query: 198 KVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVK 257
K+ PL + L AW++G R+DQSP TR+ V D F + L+ W
Sbjct: 123 KLVPLEKELAQYDAWMSGLRRDQSP-TRTNTQFVNEDRRFGSTK--ICPLIHWT------ 173
Query: 258 GHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLHK 317
+IW ++ +P N LH K Y SIGCE CTR V G+ ER GR W + ECGLH+
Sbjct: 174 SEEIWMYIELHQLPYNDLHDKQYPSIGCEYCTRPVKEGEDERAGR--WSNTSKTECGLHQ 231
>CYH1_BACSU (P94498) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 233
Score = 109 bits (273), Expect = 1e-23
Identities = 80/244 (32%), Positives = 116/244 (46%), Gaps = 23/244 (9%)
Query: 82 DNYQQLAVDLENASP----LQIMDAALEKFGNHIAIAFS-GAEDVALIEYARLTGRPFRV 136
DN+++ + P L ++ A +G+ + A S G E + LI+ + +
Sbjct: 5 DNWEEPTITFPEDDPYKGALSVLKWAYGHYGDQLVYACSFGIEGIVLIDLIYKVKKDAEI 64
Query: 137 FSLDTGRLNPETYRFFDEVEKHY-GIHIEYMFPD-AVEVQGLVRSKGLFSFYEDGHQECC 194
LDTG ETY + V++ Y G++I PD +E Q L +E +CC
Sbjct: 65 VFLDTGLHFKETYETIERVKERYPGLNIILKKPDLTLEEQAEEHGDKL---WEREPNQCC 121
Query: 195 RVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVA 254
+RKV PLR AL G AW++G R+DQ P +R+ + D F+ V V L+ W
Sbjct: 122 YLRKVVPLREALSGHPAWLSGLRRDQGP-SRANTNFLNKDEKFKSVK--VCPLIHWT--- 175
Query: 255 NVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAK-EC 313
DIW + + N LH +GY SIGC PCT + R GRW + AK EC
Sbjct: 176 ---WKDIWRYTSRNELDYNPLHDQGYPSIGCAPCTSPAFTAEDLRSGRW---NGMAKTEC 229
Query: 314 GLHK 317
GLH+
Sbjct: 230 GLHE 233
>CYSH_MYCTU (P65668) Probable phosphoadenosine phosphosulfate
reductase (EC 1.8.4.8) (PAPS reductase, thioredoxin
dependent) (PAdoPS reductase) (3'-phosphoadenylylsulfate
reductase) (PAPS sulfotransferase)
Length = 254
Score = 109 bits (272), Expect = 2e-23
Identities = 73/205 (35%), Positives = 99/205 (47%), Gaps = 15/205 (7%)
Query: 113 IAFSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPD-AV 171
+ S D L++ A V LDTG ET D +E Y + + + P+ V
Sbjct: 62 VVASNMADAVLVDLAAKVRPGVPVIFLDTGYHFVETIGTRDAIESVYDVRVLNVTPEHTV 121
Query: 172 EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVV 231
Q + K LF+ ECCR+RKV PL + L+G AW+TG R+ +P TR+ P+V
Sbjct: 122 AEQDELLGKDLFA---RNPHECCRLRKVVPLGKTLRGYSAWVTGLRRVDAP-TRANAPLV 177
Query: 232 QVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRA 291
D F+ LVK NP+A D+ ++ +V VN L +GY SIGC PCT
Sbjct: 178 SFDETFK--------LVKVNPLAAWTDQDVQEYIADNDVLVNPLVREGYPSIGCAPCTAK 229
Query: 292 VLPGQHEREGRWWWEDAKAKECGLH 316
G R GR W+ ECGLH
Sbjct: 230 PAEGADPRSGR--WQGLAKTECGLH 252
>CYSH_MYCBO (P65669) Probable phosphoadenosine phosphosulfate
reductase (EC 1.8.4.8) (PAPS reductase, thioredoxin
dependent) (PAdoPS reductase) (3'-phosphoadenylylsulfate
reductase) (PAPS sulfotransferase)
Length = 254
Score = 109 bits (272), Expect = 2e-23
Identities = 73/205 (35%), Positives = 99/205 (47%), Gaps = 15/205 (7%)
Query: 113 IAFSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPD-AV 171
+ S D L++ A V LDTG ET D +E Y + + + P+ V
Sbjct: 62 VVASNMADAVLVDLAAKVRPGVPVIFLDTGYHFVETIGTRDAIESVYDVRVLNVTPEHTV 121
Query: 172 EVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVV 231
Q + K LF+ ECCR+RKV PL + L+G AW+TG R+ +P TR+ P+V
Sbjct: 122 AEQDELLGKDLFA---RNPHECCRLRKVVPLGKTLRGYSAWVTGLRRVDAP-TRANAPLV 177
Query: 232 QVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRA 291
D F+ LVK NP+A D+ ++ +V VN L +GY SIGC PCT
Sbjct: 178 SFDETFK--------LVKVNPLAAWTDQDVQEYIADNDVLVNPLVREGYPSIGCAPCTAK 229
Query: 292 VLPGQHEREGRWWWEDAKAKECGLH 316
G R GR W+ ECGLH
Sbjct: 230 PAEGADPRSGR--WQGLAKTECGLH 252
>CYSH_RHILO (Q98GP9) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 254
Score = 108 bits (270), Expect = 3e-23
Identities = 75/224 (33%), Positives = 114/224 (50%), Gaps = 19/224 (8%)
Query: 96 PLQIMD-AALEKFGNHIAIAFS-GAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFD 153
PL+I++ +A E F + IA S GA+ L+ R V LDTG+ ET + D
Sbjct: 34 PLEIIERSARELFHDEIAAVSSFGADSAVLLHMIAEIDRTLPVIFLDTGKHFEETLGYRD 93
Query: 154 EVEKHYGI-HIEYMFPDAVEVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAW 212
+ +G+ +I+ + P E L R + ++ CC VRKV PL R + RAW
Sbjct: 94 ALVADFGLTNIQVIKP---EEAALARIDPTGNLHQSNTDACCDVRKVEPLARGVAPFRAW 150
Query: 213 ITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPV 272
TG+++ Q+ TR+ +PV + VG+ ++ NP+A+ D ++R +
Sbjct: 151 FTGRKRFQA-STRAALPVFEA----------VGARIRINPLAHWTTSDQADYMRAHALRE 199
Query: 273 NSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLH 316
N L + GY+SIGC PCT+ V PG+ R GR W ECG+H
Sbjct: 200 NPLVAYGYLSIGCFPCTQPVQPGEDARSGR--WAGHAKTECGIH 241
>CYSH_BACCR (Q81FZ1) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 234
Score = 101 bits (252), Expect = 4e-21
Identities = 72/243 (29%), Positives = 118/243 (47%), Gaps = 19/243 (7%)
Query: 78 ETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFS-GAEDVALIEYARLTGRPFRV 136
ET ++N + + E L +++ A +++ + + A S G E + L+ +V
Sbjct: 5 ETWEENVVSFSEEDETKGALSVLNWAYKEYKDEVVYACSFGVEGMVLLHIINQVNPSAKV 64
Query: 137 FSLDTGRLNPETYRFFDEVEKHY-GIHIEYMFPD-AVEVQGLVRSKGLFSFYEDGHQECC 194
LDT ETY +V + + ++I P+ ++ Q + + L +E CC
Sbjct: 65 VFLDTNVHFQETYELIRKVRERFPSLNIIEKQPELTLDEQAKLHGEKL---WESNPNLCC 121
Query: 195 RVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVA 254
++RK+ PL ++L +AWI+G R++QS TR + D F+ + V L+ W
Sbjct: 122 KIRKILPLEKSLVVEKAWISGLRREQSE-TRKHTKFINQDHRFQSIK--VCPLIHWT--- 175
Query: 255 NVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAK-EC 313
++W ++ ++P N LH GY SIGCE CT V G R+GRW K K EC
Sbjct: 176 ---WKEVWRYVYKHSLPYNPLHDVGYPSIGCEKCTLPVGDGGDSRDGRW---AGKVKTEC 229
Query: 314 GLH 316
GLH
Sbjct: 230 GLH 232
>CYSH_BACAN (Q81T49) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 226
Score = 99.0 bits (245), Expect = 2e-20
Identities = 71/229 (31%), Positives = 109/229 (47%), Gaps = 19/229 (8%)
Query: 92 ENASPLQIMDAALEKFGNHIAIAFS-GAEDVALIEYARLTGRPFRVFSLDTGRLNPETYR 150
E L ++ A +++ + I A S G E + L++ +V LDT ETY
Sbjct: 11 ETKGALSVLSWAYKEYKSEIVYACSFGVEGMVLLDLINQVNPSAKVVFLDTNVHFQETYE 70
Query: 151 FFDEVEKHY-GIHIEYMFPD-AVEVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKG 208
+V + + ++I P ++ Q + L +E CC++RK+ PL +L
Sbjct: 71 LIQKVRERFPSLNIIEKQPKLTLDEQDKLHGDKL---WESNPNLCCKIRKILPLEESLAN 127
Query: 209 LRAWITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTM 268
+AWI+G R++QS TR + D F+ + V L+ W ++W ++
Sbjct: 128 EKAWISGLRREQSE-TRKHTKFINQDHRFQSIK--VCPLIHWT------WKEVWRYVYKH 178
Query: 269 NVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAK-ECGLH 316
++P NSLH GY SIGCE CT V G R+GRW K K ECGLH
Sbjct: 179 SLPYNSLHDIGYPSIGCEKCTLPVGEGGDSRDGRW---AGKVKTECGLH 224
>CYSH_XYLFA (Q9PD82) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 237
Score = 96.7 bits (239), Expect = 1e-19
Identities = 72/239 (30%), Positives = 117/239 (48%), Gaps = 17/239 (7%)
Query: 82 DNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFS-GAEDVALIEYARLTGRPFRVFSLD 140
D+ + L V LE S + ALE+ +H A++ S GA+ ++ V +D
Sbjct: 11 DDLETLNVHLETLSAENRVCWALERGPDHPALSSSFGAQSAVMLHLLTRFAPDIPVILVD 70
Query: 141 TGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQGLVRSKGLFSFYEDGHQECCRVRKVR 200
TG L PETYRF D + + ++++ P R L+ DG + + KV
Sbjct: 71 TGYLFPETYRFADTLTERLKLNLKVYQPLRSGAWTEARHGRLWEQGIDGINQYNTLHKVE 130
Query: 201 PLRRALKGLRA--WITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKG 258
P+RRAL+ L+ W TG R+ QS TR++ +VQ K +P+A+
Sbjct: 131 PMRRALEELQVGTWFTGLRRGQS-STRTQTSIVQQRD----------ERYKISPIADWTD 179
Query: 259 HDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLHK 317
DIW +++ ++P + L +GY+SIG TR + PG E + R++ +ECG+H+
Sbjct: 180 RDIWEYMKHHDLPYHPLWEQGYVSIGDIHTTRPLEPGMREEDTRFF---GFKRECGIHE 235
>CYSH_XANCP (Q8P607) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 241
Score = 94.4 bits (233), Expect = 6e-19
Identities = 73/248 (29%), Positives = 114/248 (45%), Gaps = 17/248 (6%)
Query: 73 ASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIAFSGAEDVALIEYARLTGR 132
A+ +A D+ L LE + + AL+ A++ S A+ + R
Sbjct: 6 ATSIAAPSLDDLDALNAHLETLRADERVAWALQHGPQQAALSSSFGAQSAVTLHLLTQQR 65
Query: 133 P-FRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQGLVRSKGLFSFYEDGHQ 191
P V +DTG L PETYRF D + + ++++ P R L+ G
Sbjct: 66 PDIPVILIDTGYLFPETYRFADALTERLSLNLQVYRPLVSRAWMEARHGRLWEQGMVGID 125
Query: 192 ECCRVRKVRPLRRALKGLRA--WITGQRKDQSPGTRSEVPVVQVDPVFEGVDGGVGSLVK 249
+ +RKV P+RRAL L W TG R+ QS G R++ P+VQ G K
Sbjct: 126 QYNNLRKVEPMRRALDELNVGTWFTGLRRSQSGG-RAQTPIVQKR----------GDRYK 174
Query: 250 WNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAK 309
+P+A+ D+W +L+ +P + L +GY+SIG TR PG E + R++
Sbjct: 175 ISPIADWTDRDVWQYLQAHALPYHPLWEQGYVSIGDFHTTRRWEPGMREEDTRFF---GL 231
Query: 310 AKECGLHK 317
+ECG+H+
Sbjct: 232 KRECGIHE 239
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,238,131
Number of Sequences: 164201
Number of extensions: 2281624
Number of successful extensions: 7181
Number of sequences better than 10.0: 181
Number of HSP's better than 10.0 without gapping: 94
Number of HSP's successfully gapped in prelim test: 87
Number of HSP's that attempted gapping in prelim test: 6832
Number of HSP's gapped (non-prelim): 259
length of query: 461
length of database: 59,974,054
effective HSP length: 114
effective length of query: 347
effective length of database: 41,255,140
effective search space: 14315533580
effective search space used: 14315533580
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0026.11


