

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.9
(388 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
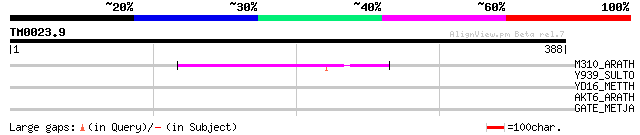
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 88 3e-17
Y939_SULTO (Q973G3) Hypothetical protein ST0939 precursor 32 2.1
YD16_METTH (O27371) Hypothetical UPF0215 protein MTH1316 32 2.8
AKT6_ARATH (Q8GXE6) Potassium channel AKT6 (Shaker pollen inward... 31 4.7
GATE_METJA (Q60325) Glutamyl-tRNA(Gln) amidotransferase subunit ... 30 8.0
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 88.2 bits (217), Expect = 3e-17
Identities = 52/152 (34%), Positives = 75/152 (49%), Gaps = 7/152 (4%)
Query: 118 AIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKR-DGGLGFRF 176
A+P Y MSCF L +C + + F+W +R + W W LCK+K DGGLGFR
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 177 FRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKG---YITWSSILKAGGK 233
F +AL+AK RI P +L+ R+ + Y P SS++ G W SI+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 234 IEGGGGGLRWRVGDGFQINLLQDKWLPDGSSL 265
+ GL +GDG + D+W+ D + L
Sbjct: 122 L---SRGLLRTIGDGIHTKVWLDRWIMDETPL 150
>Y939_SULTO (Q973G3) Hypothetical protein ST0939 precursor
Length = 709
Score = 32.3 bits (72), Expect = 2.1
Identities = 16/48 (33%), Positives = 25/48 (51%), Gaps = 2/48 (4%)
Query: 292 ELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFI 339
+++ SS P I+ + + N T L WYDTFNG Y + Y ++
Sbjct: 393 DIISATPSSVPNPPIIKVKIGNLNAT--LTWYDTFNGGYPIEGYYLYV 438
>YD16_METTH (O27371) Hypothetical UPF0215 protein MTH1316
Length = 204
Score = 32.0 bits (71), Expect = 2.8
Identities = 25/83 (30%), Positives = 37/83 (44%), Gaps = 6/83 (7%)
Query: 60 VKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKE--KSLSRAGREILIKSIAQ 117
V ++ + GLP I+ K +K + R WK +S+ RAGR I +
Sbjct: 96 VVDIVELSERTGLPVIVVVRKHPDMERIKRALKHRFSDWKARWRSIERAGR---IHRVES 152
Query: 118 AIPSYIMSCFILPDSICADIDRM 140
P YI + I PD A+I R+
Sbjct: 153 REPLYIQTAGIEPDD-AAEIVRI 174
>AKT6_ARATH (Q8GXE6) Potassium channel AKT6 (Shaker pollen inward
rectifier K(+) channel) (Potassium channel SPIK)
Length = 888
Score = 31.2 bits (69), Expect = 4.7
Identities = 15/32 (46%), Positives = 19/32 (58%)
Query: 256 DKWLPDGSSLVYMHEAIAELRLEKVSDLLDNG 287
D LPDG+ +H A++E LE V LLD G
Sbjct: 666 DVTLPDGNGTTALHRAVSEGHLEIVKFLLDQG 697
>GATE_METJA (Q60325) Glutamyl-tRNA(Gln) amidotransferase subunit E
(EC 6.3.5.-) (Glu-ADT subunit E)
Length = 630
Score = 30.4 bits (67), Expect = 8.0
Identities = 30/96 (31%), Positives = 48/96 (49%), Gaps = 11/96 (11%)
Query: 216 MAKKGYITWSSILKAGGKIEGGGGGLRWRVG----DGFQINL--LQDKWLPDGSSLVYMH 269
MAK+ IL+A GK++ G G +R + DG +I + +QD D V +
Sbjct: 198 MAKEAARRIGEILRATGKVKRGLGTIRQDINISIKDGARIEVKGVQDL---DLIEKVVEN 254
Query: 270 EAIAELRLEKVSDLLDNGRWNLELLEEVFSSSPMQK 305
E I +L L K+ D L N E++E++F + + K
Sbjct: 255 EVIRQLNLLKIRDELRER--NAEVVEKIFDVTEIFK 288
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.334 0.147 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,819,011
Number of Sequences: 164201
Number of extensions: 1906570
Number of successful extensions: 6383
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 6379
Number of HSP's gapped (non-prelim): 5
length of query: 388
length of database: 59,974,054
effective HSP length: 112
effective length of query: 276
effective length of database: 41,583,542
effective search space: 11477057592
effective search space used: 11477057592
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0023.9


