

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
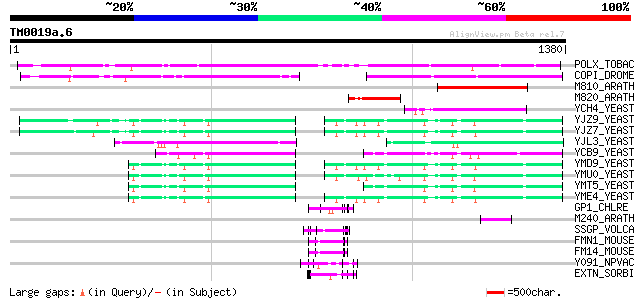
Query= TM0019a.6
(1380 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 442 e-123
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 280 1e-74
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 217 2e-55
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 104 1e-21
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 103 3e-21
YJZ9_YEAST (P47100) Transposon Ty1 protein B 96 7e-19
YJZ7_YEAST (P47098) Transposon Ty1 protein B 96 9e-19
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 95 2e-18
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 88 2e-16
YMD9_YEAST (Q03434) Transposon Ty1 protein B 86 7e-16
YMU0_YEAST (Q04670) Transposon Ty1 protein B 85 1e-15
YMT5_YEAST (Q04214) Transposon Ty1 protein B 85 1e-15
YME4_YEAST (Q04711) Transposon Ty1 protein B 85 1e-15
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 79 9e-14
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 70 5e-11
SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor ... 62 1e-08
FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity p... 62 1e-08
FM14_MOUSE (Q05859) Formin 1 isoform IV (Limb deformity protein) 62 1e-08
Y091_NPVAC (P41479) Hypothetical 24.1 kDa protein in LEF4-P33 in... 61 2e-08
EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein) 60 3e-08
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 442 bits (1138), Expect = e-123
Identities = 390/1406 (27%), Positives = 595/1406 (41%), Gaps = 152/1406 (10%)
Query: 20 KLNSGN-FLLWSKQVTAILQNQNLFDIVDPIVAPPSEFMLQPVSSALHHVVPAPNPLFVQ 78
K N N F W +++ +L Q L ++D P +
Sbjct: 10 KFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAE------------------D 51
Query: 79 WQARDRNVNSLLLSSLTEEALSETLSATTARDVWTALHSAYASRNKAREMRLRDELQL-- 136
W D S + L+++ ++ + TAR +WT L S Y S+ ++ L+ +L
Sbjct: 52 WADLDERAASAIRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALH 111
Query: 137 MRRGSLSVSEYG---------RKIR*SVANDDKVHWFLRGLGPSYANFSTGQLDQVPLPL 187
M G+ +S + + +DK L L SY N +T L
Sbjct: 112 MSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKTTIE 171
Query: 188 FTDILWKVESHAIFQASLEDYVTPPSAAFHARNPTRSSGSQSSGGGHRGNSSSGSRPRRD 247
D+ + + + E+ R+ RSS + G RG S + S+ R
Sbjct: 172 LKDVTSALLLNEKMRKKPENQGQALITEGRGRSYQRSSNNYGRSGA-RGKSKNRSKSRVR 230
Query: 248 NGGSHCRGSYTPRCQLCRKQGHYAAKCP-VRWDRPSESANLTHSFAAGCSLNNSN----- 301
N C C + GH+ CP R + S A NN N
Sbjct: 231 N------------CYNCNQPGHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLFI 278
Query: 302 ------------RSDKYMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGNGAGLHITHIGS 349
S+ +DT A+ H T + L + V +GN + I IG
Sbjct: 279 NEEEECMHLSGPESEWVVDTAASHHAT-PVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGD 337
Query: 350 ----RSASHSVPLSNVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLGK 405
+ ++ L +V VP L NL+S L RD + F + + + V+ K
Sbjct: 338 ICIKTNVGCTLVLKDVRHVPDLRMNLISGIALDRD-GYESYFANQKWRLT--KGSLVIAK 394
Query: 406 GPCDKGFYVLDQGSQALLATSSSLPRASFELWHSRLDHVN---FDIIKKLHKLGCFNVSS 462
G +G +++ S +LWH R+ H++ I+ K + ++
Sbjct: 395 GVA-RGTLYRTNAEICQGELNAAQDEISVDLWHKRMGHMSEKGLQILAKKSLISYAKGTT 453
Query: 463 ILPKPICCTSCQMAKSKRLVCYDNHKRASAVLDLVHCDLWGPSPVASVDGFSYFVIFVDD 522
+ P C C K R+ + +R +LDLV+ D+ GP + S+ G YFV F+DD
Sbjct: 454 VKP----CDYCLFGKQHRVSFQTSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDD 509
Query: 523 FSQFTWFYPLKRKSDFYDVLVRFKAFVENQFSRSLKVFQSDNGTEFTNNKVQALFSSSGV 582
S+ W Y LK K + V +F A VE + R LK +SDNG E+T+ + + SS G+
Sbjct: 510 ASRKLWVYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGI 569
Query: 583 LHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHSHVSTSYWVHAFSTAVYIINRVPSKVL 642
H P T NG ER +R ++E +ML + + S+W A TA Y+INR PS L
Sbjct: 570 RHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPL 629
Query: 643 SDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYMKNKFSPRGTPCIFLGYNSHHKGFKCF 702
+ +IP ++ +Y++ FGCR F + + K + PCIF+GY G++ +
Sbjct: 630 AFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLW 689
Query: 703 DPTTSRTYVSRHAQF--DEFCFPLTGSKSSPNDLVV-FTFYEPAAGSPPSLQPVILVVPE 759
DP + SR F E S+ N ++ F + +P S + V E
Sbjct: 690 DPVKKKVIRSRDVVFRESEVRTAADMSEKVKNGIIPNFVTIPSTSNNPTSAESTTDEVSE 749
Query: 760 SSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPT 819
G + ++ D V H PT PL ++ P
Sbjct: 750 QGEQPGEV---------IEQGEQLDEGVEEVEH--------PTQGEEQHQPLRRSERPRV 792
Query: 820 SPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNPRYALVHAQPTGLLTALHTVT*PKGFK 879
P T+ L D P + + L H + L+ A
Sbjct: 793 ESRRYPS--TEYVLISDDREPESL----------KEVLSHPEKNQLMKA----------- 829
Query: 880 SAMKHPHWLPAMEDELSALHKKFTWTLVPRPYTTNVVGSK*VFRTKFHSDGTVERLKTCL 939
M++E+ +L K T+ LV P + K VF+ K D + R K L
Sbjct: 830 -----------MQEEMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARL 878
Query: 940 VAQGLTQIPGFDYSLTFSPVVKATTVRLILSLVVLND*QLHQLDVKNAFLHGHLTETVYM 999
V +G Q G D+ FSPVVK T++R ILSL D ++ QLDVK AFLHG L E +YM
Sbjct: 879 VVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYM 938
Query: 1000 EQPHGF-VDLVSRLMCVG*TRL--STASNRRLVLGFSA*ALFSCVLVFRVVGLIHPCSFF 1056
EQP GF V ++C L + R+ + F + L PC +F
Sbjct: 939 EQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYL----KTYSDPCVYF 994
Query: 1057 --YKGHITLYLLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYLGKLGYFLGLEIT--YT 1112
+ + + LL+YVDD+++ G D L+ L+ F +K LG LG++I T
Sbjct: 995 KRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERT 1054
Query: 1113 ADGLFLGHAKYAHDLLSHALMLEASHVLTPLAAGSHL-------VSSGEGYSDPTHYRSL 1165
+ L+L KY +L M A V TPLA L +G Y S
Sbjct: 1055 SRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSA 1114
Query: 1166 VGALQY-LTITRPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSSP 1224
VG+L Y + TRPD+++AV VS+FL+ P +H++AVK I+RY+ L F S P
Sbjct: 1115 VGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCF-GGSDP 1173
Query: 1225 AVLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPSVAHSSCESEYRAMANTASE 1284
+ GY+DAD A D+R+S+ GY +SW +K Q VA S+ E+EY A T E
Sbjct: 1174 ILKGYTDADMAGDIDNRKSSTGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKE 1233
Query: 1285 LVWLLNLLHELRVRLSAPPLFLSDNQSALFMAQNPVAHKHAKHIDLDCHFVRELVSSGRL 1344
++WL L EL + ++ D+QSA+ +++N + H KHID+ H++RE+V L
Sbjct: 1234 MIWLKRFLQELGLHQKEYVVY-CDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESL 1292
Query: 1345 AVRHVPTSLQLADIFTKVLPRPLFDL 1370
V + T+ AD+ TKV+PR F+L
Sbjct: 1293 KVLKISTNENPADMLTKVVPRNKFEL 1318
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 280 bits (717), Expect = 1e-74
Identities = 178/498 (35%), Positives = 264/498 (52%), Gaps = 14/498 (2%)
Query: 887 WLPAMEDELSALHKKFTWTLVPRPYTTNVVGSK*VFRTKFHSDGTVERLKTCLVAQGLTQ 946
W A+ EL+A TWT+ RP N+V S+ VF K++ G R K LVA+G TQ
Sbjct: 906 WEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQ 965
Query: 947 IPGFDYSLTFSPVVKATTVRLILSLVVLND*QLHQLDVKNAFLHGHLTETVYMEQPHGFV 1006
DY TF+PV + ++ R ILSLV+ + ++HQ+DVK AFL+G L E +YM P G
Sbjct: 966 KYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGIS 1025
Query: 1007 DLVSRLMCVG*TRLSTASNRRLVLGFSA*ALFSCVLVFRVVGLIHPCSFFY-KGHIT--L 1063
+ + R AL C V V C + KG+I +
Sbjct: 1026 CNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDR---CIYILDKGNINENI 1082
Query: 1064 YLLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYLGKLGYFLGLEITYTADGLFLGHAKY 1123
Y+L+YVDD+++ D + + F L ++F + L ++ +F+G+ I D ++L + Y
Sbjct: 1083 YVLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAY 1142
Query: 1124 AHDLLSHALMLEASHVLTPLAA--GSHLVSSGEGYSDPTHYRSLVGALQYLTI-TRPDLS 1180
+LS M + V TPL + L++S E + P RSL+G L Y+ + TRPDL+
Sbjct: 1143 VKKILSKFNMENCNAVSTPLPSKINYELLNSDEDCNTPC--RSLIGCLMYIMLCTRPDLT 1200
Query: 1181 YAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSS--PAVLGYSDADWARCT 1238
AVN +S++ + +Q +KR++RY+ L F++ + ++GY D+DWA
Sbjct: 1201 TAVNILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSE 1260
Query: 1239 DHRRSTYGYAI-FLGYNLLSWSAKKQPSVAHSSCESEYRAMANTASELVWLLNLLHELRV 1297
R+ST GY +NL+ W+ K+Q SVA SS E+EY A+ E +WL LL + +
Sbjct: 1261 IDRKSTTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINI 1320
Query: 1298 RLSAPPLFLSDNQSALFMAQNPVAHKHAKHIDLDCHFVRELVSSGRLAVRHVPTSLQLAD 1357
+L P DNQ + +A NP HK AKHID+ HF RE V + + + ++PT QLAD
Sbjct: 1321 KLENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLAD 1380
Query: 1358 IFTKVLPRPLFDLFRSKL 1375
IFTK LP F R KL
Sbjct: 1381 IFTKPLPAARFVELRDKL 1398
Score = 153 bits (387), Expect = 3e-36
Identities = 172/739 (23%), Positives = 298/739 (40%), Gaps = 94/739 (12%)
Query: 26 FLLWSKQVTAILQNQNLFDIVDPIVAPPSEFMLQPVSSALHHVVPAPNPLFVQWQARDRN 85
+ +W ++ A+L Q++ +VD ++ PN + W+ +R
Sbjct: 16 YAIWKFRIRALLAEQDVLKVVDGLM---------------------PNEVDDSWKKAERC 54
Query: 86 VNSLLLSSLTEEALSETLSATTARDVWTALHSAYASRNKAREMRLRDE-LQLMRRGSLSV 144
S ++ L++ L+ S TAR + L + Y ++ A ++ LR L L +S+
Sbjct: 55 AKSTIIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSL 114
Query: 145 SEY--------------GRKIR*SVANDDKVHWFLRGLGPSYANFSTG--QLDQVPLPL- 187
+ G KI DK+ L L Y T L + L L
Sbjct: 115 LSHFHIFDELISELLAAGAKIE----EMDKISHLLITLPSCYDGIITAIETLSEENLTLA 170
Query: 188 FTDILWKVESHAIFQASLEDYVTPPSAAFHARNPTRSSGSQSSGGGHRGN--SSSGSRPR 245
F + I + +A H N T ++ N + ++P+
Sbjct: 171 FVKNRLLDQEIKIKNDHNDTSKKVMNAIVHNNNNT-----------YKNNLFKNRVTKPK 219
Query: 246 RDNGGSHCRGSYTPRCQLCRKQGHYAAKCPVRWDRPSESAN----------LTHSFAAGC 295
+ G+ Y +C C ++GH C + R + N +H A
Sbjct: 220 KIFKGN---SKYKVKCHHCGREGHIKKDC-FHYKRILNNKNKENEKQVQTATSHGIAFMV 275
Query: 296 S-LNNSNRSDK---YMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGN-GAGLHITHIG-- 348
+NN++ D +D+GA+ H+ + S T S ++ V G ++ T G
Sbjct: 276 KEVNNTSVMDNCGFVLDSGASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIV 335
Query: 349 SRSASHSVPLSNVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLGKGPC 408
H + L +VL NL+SV +L ++ + F+ I V G
Sbjct: 336 RLRNDHEITLEDVLFCKEAAGNLMSVKRL-QEAGMSIEFDKSGVTISKNGLMVVKNSGML 394
Query: 409 DKGFYVLDQGSQALLATSSSLPRASFELWHSRLDHVNFDIIKKLHKLGCFNVSSILP--K 466
+ V++ + ++ A + +F LWH R H++ + ++ + F+ S+L +
Sbjct: 395 NN-VPVINFQAYSINAKHKN----NFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLE 449
Query: 467 PIC--CTSCQMAKSKRLVCYD--NHKRASAVLDLVHCDLWGPSPVASVDGFSYFVIFVDD 522
C C C K RL + L +VH D+ GP ++D +YFVIFVD
Sbjct: 450 LSCEICEPCLNGKQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQ 509
Query: 523 FSQFTWFYPLKRKSDFYDVLVRFKAFVENQFSRSLKVFQSDNGTEFTNNKVQALFSSSGV 582
F+ + Y +K KSD + + F A E F+ + DNG E+ +N+++ G+
Sbjct: 510 FTHYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGI 569
Query: 583 LHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHSHVSTSYWVHAFSTAVYIINRVPSKVL 642
+ PHT NG ER R + E M+ + + S+W A TA Y+INR+PS+ L
Sbjct: 570 SYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRAL 629
Query: 643 --SDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYMKNKFSPRGTPCIFLGYNSHHKGFK 700
S + P+++ P + FG V+ ++ + KF + IF+GY + GFK
Sbjct: 630 VDSSKTPYEMWHNKKPYLKHLRVFGATVYVHIK-NKQGKFDDKSFKSIFVGYEPN--GFK 686
Query: 701 CFDPTTSRTYVSRHAQFDE 719
+D + V+R DE
Sbjct: 687 LWDAVNEKFIVARDVVVDE 705
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 217 bits (553), Expect = 2e-55
Identities = 112/225 (49%), Positives = 150/225 (65%)
Query: 1063 LYLLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYLGKLGYFLGLEITYTADGLFLGHAK 1122
+YLL+YVDDI+LTGS +LL I L+ F++K LG + YFLG++I GLFL K
Sbjct: 1 MYLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTK 60
Query: 1123 YAHDLLSHALMLEASHVLTPLAAGSHLVSSGEGYSDPTHYRSLVGALQYLTITRPDLSYA 1182
YA +L++A ML+ + TPL + S Y DP+ +RS+VGALQYLT+TRPD+SYA
Sbjct: 61 YAEQILNNAGMLDCKPMSTPLPLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYA 120
Query: 1183 VNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSSPAVLGYSDADWARCTDHRR 1242
VN V Q + PT+ F +KR++RYV GL + S V + D+DWA CT RR
Sbjct: 121 VNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRR 180
Query: 1243 STYGYAIFLGYNLLSWSAKKQPSVAHSSCESEYRAMANTASELVW 1287
ST G+ FLG N++SWSAK+QP+V+ SS E+EYRA+A TA+EL W
Sbjct: 181 STTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 104 bits (260), Expect = 1e-21
Identities = 59/130 (45%), Positives = 79/130 (60%), Gaps = 8/130 (6%)
Query: 842 TRS*NGIFKPNPRYALVHAQPTGLLTALHTVT*PKGFKSAMKHPHWLPAMEDELSALHKK 901
TRS GI K NP+Y+L + T + PK A+K P W AM++EL AL +
Sbjct: 3 TRSKAGINKLNPKYSLT------ITTTIKKE--PKSVIFALKDPGWCQAMQEELDALSRN 54
Query: 902 FTWTLVPRPYTTNVVGSK*VFRTKFHSDGTVERLKTCLVAQGLTQIPGFDYSLTFSPVVK 961
TW LVP P N++G K VF+TK HSDGT++RLK LVA+G Q G + T+SPVV+
Sbjct: 55 KTWILVPPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVR 114
Query: 962 ATTVRLILSL 971
T+R IL++
Sbjct: 115 TATIRTILNV 124
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 103 bits (257), Expect = 3e-21
Identities = 96/321 (29%), Positives = 144/321 (43%), Gaps = 30/321 (9%)
Query: 982 LDVKNAFLHGHLTETVYMEQPHGFV-----DLVSRLMCVG*TRLSTA--------SNRRL 1028
+DV AFL+ + E +Y++QP GFV D V L G L A +N
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYG-GMYGLKQAPLLWNEHINNTLK 59
Query: 1029 VLGFSA*ALFSCVLVFRVVGLIHPCSFFYKGHITLYLLVYVDDIILTGSDPSLLT*FIAC 1088
+GF R G H F +Y+ VYVDD+++ P +
Sbjct: 60 KIGFC-----------RHEGE-HGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQE 107
Query: 1089 LNDEFAIKYLGKLGYFLGLEITYTADG-LFLGHAKYAHDLLSHALMLEASHVLTPLAAGS 1147
L +++K LGK+ FLGL I + +G + L Y S + + TPL
Sbjct: 108 LTKLYSMKDLGKVDKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSK 167
Query: 1148 HLVSSGEGY-SDPTHYRSLVGALQYLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRII 1205
L + + D T Y+S+VG L + T RPD+SY V+ +S+FL+ P H ++ +R++
Sbjct: 168 PLFETTSPHLKDITPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVL 227
Query: 1206 RYVCAMQHFGLTFRRTSSPAVLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPS 1265
RY+ + L +R S A+ Y DA D ST GY L ++WS+KK
Sbjct: 228 RYLYTTRSMCLKYRSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKG 287
Query: 1266 VAH-SSCESEYRAMANTASEL 1285
V S E+EY + T E+
Sbjct: 288 VIPVPSTEAEYITASETVMEI 308
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 95.9 bits (237), Expect = 7e-19
Identities = 163/747 (21%), Positives = 280/747 (36%), Gaps = 108/747 (14%)
Query: 25 NFLLWSKQVTAILQNQNLFDIVDPIVAPPSEFMLQPVSSALHHV--VPAPNPLFVQWQAR 82
+F W K LQN NL I+ + P + + L++ + AP+ W
Sbjct: 180 DFPNWVKTYIKFLQNSNLGGIIPTVNGKPVRQITDDELTFLYNTFQIFAPSQFLPTWVKD 239
Query: 83 DRNVNSLLLSSLTEEALSETLSATT-ARDVWTALHSAYASRNKAR--EMRLRDELQLMRR 139
+V+ + + +++ + S T A D+ T + Y A E ++ + + +
Sbjct: 240 ILSVDYTDIMKILSKSIEKMQSDTQEANDIVTLANLQYNGSTPADAFETKVTNIIDRLNN 299
Query: 140 GSLSVSEYGRKIR*SVANDDKVHWFLRGLGPSYANFSTGQ---LDQVPLPLFTDILWKVE 196
+ ++ N +RGL Y + L+ LF DI E
Sbjct: 300 NGIHIN-----------NKVACQLIMRGLSGEYKFLRYTRHRHLNMTVAELFLDIHAIYE 348
Query: 197 SHAIFQASLEDYVTPPSAAFH--------------ARNPTRSSGSQSSGGGHRGNSSSGS 242
+ S +Y PS + ARNP +++ S+S S+S +
Sbjct: 349 EQQGSRNSKPNYRRNPSDEKNDSRSYTNTTKPKVIARNPQKTNNSKSKTARAHNVSTSNN 408
Query: 243 RPRRDNGGSHCRGSYTPRCQLCRKQGHYAAKCPVRWDRPSESANLTHSFAAGCSLNNSNR 302
P DN S T QL K + LT S ++N++N
Sbjct: 409 SPSTDNDS--ISKSTTEPIQLNNKHDLILGQ------------KLTES-----TVNHTNH 449
Query: 303 SDK------YMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGNGA--GLHITHIGSRS--- 351
SD +D+GA+ + S + H+ S N + V + + I IG
Sbjct: 450 SDDELPGHLLLDSGASRTLIRSAHHI---HSASSNPDINVVDAQKRNIPINAIGDLQFHF 506
Query: 352 ASHSVPLSNVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLGKGPCDKG 411
++ VL P + +L+S+++L ++ A F + + R G VL
Sbjct: 507 QDNTKTSIKVLHTPNIAYDLLSLNELAAV-DITACFTKN---VLERSDGTVLAPIVKYGD 562
Query: 412 FYVLDQGSQALLATSSSLPRAS------------FELWHSRLDHVNFDIIK---KLHKLG 456
FY + + + LL ++ S+P + + H L H N I+ K + +
Sbjct: 563 FYWVSK--KYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTIT 620
Query: 457 CFNVSSILPKPIC---CTSCQMAKSKRLVCYDNHKRASAV--------LDLVHCDLWGPS 505
FN S + C C + KS + H + S + +H D++GP
Sbjct: 621 YFNESDVDWSSAIDYQCPDCLIGKSTK----HRHIKGSRLKYQNSYEPFQYLHTDIFGPV 676
Query: 506 PVASVDGFSYFVIFVDDFSQFTWFYPL--KRKSDFYDVLVRFKAFVENQFSRSLKVFQSD 563
SYF+ F D+ ++F W YPL +R+ DV AF++NQF S+ V Q D
Sbjct: 677 HNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMD 736
Query: 564 NGTEFTNNKVQALFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHSHVSTSYW 623
G+E+TN + +G+ + +G ER +R +L+ L S + W
Sbjct: 737 RGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLW 796
Query: 624 VHAFSTAVYIINRVPSKVLSDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYMKN-KFSP 682
A + + N + S S + Q + PFG V + + N K P
Sbjct: 797 FSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI--VNDHNPNSKIHP 853
Query: 683 RGTPCIFLGYNSHHKGFKCFDPTTSRT 709
RG P L + + G+ + P+ +T
Sbjct: 854 RGIPGYALHPSRNSYGYIIYLPSLKKT 880
Score = 65.9 bits (159), Expect = 7e-10
Identities = 150/693 (21%), Positives = 257/693 (36%), Gaps = 136/693 (19%)
Query: 782 VDDAPPSPVPHNDAPPLPAPTSPTPPATP------LVVTQAPPTSPPTTPPAVTQVPLAP 835
+D +PP ++ P+ PT+ + T L + PP SP P ++P P
Sbjct: 1096 IDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELP--P 1153
Query: 836 VDARPRTRS*NGIFKPNPRYALVHA---------------------------QPTGLLTA 868
+++R S GI N Y +++ +P
Sbjct: 1154 INSRQTNSSLGGIGDSNA-YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKR 1212
Query: 869 LHTVT*PKGFKS------------AMKHPHWLPAMEDELSALHKKFTWTLVPRPYTTN-- 914
+H + K KS A+ + + E + A HK+ L + + T+
Sbjct: 1213 IHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEY 1272
Query: 915 ----------VVGSK*VFRTKFHSDGTVERLKTCLVAQGLTQIPGFDYSLTFSPVVKATT 964
V+ S +F K DGT K VA+G Q P S S V
Sbjct: 1273 YDRKEIDPKRVINSMFIFNKK--RDGTH---KARFVARGDIQHPDTYDSGMQSNTVHHYA 1327
Query: 965 VRLILSLVVLND*QLHQLDVKNAFLHGHLTETVYMEQPH--GFVDLVSRLMCVG*TRLST 1022
+ LSL + N+ + QLD+ +A+L+ + E +Y+ P G D + RL +
Sbjct: 1328 LMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLIRLKKSLYGLKQS 1387
Query: 1023 ASNRRLVL--------GFSA*ALFSCVLVFRVVGLIHPCSFFYKGHITLYLLVYVDDIIL 1074
+N + G +SCV F +T+ L +VDD++L
Sbjct: 1388 GANWYETIKSYLIQQCGMEEVRGWSCV--------------FKNSQVTICL--FVDDMVL 1431
Query: 1075 TGSDPSLLT*FIACLNDEFAIKYLG------KLGY-FLGLEITYTADGLFLGHAKYAHDL 1127
+ + I L ++ K + ++ Y LGLEI Y KY
Sbjct: 1432 FSKNLNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQ-------RGKYMKLG 1484
Query: 1128 LSHALMLEASHVLTPL-AAGSHLVSSGE---------------GYSDPTH-YRSLVGALQ 1170
+ ++L + + PL G L + G+ Y H + L+G
Sbjct: 1485 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLAS 1544
Query: 1171 YLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTS----SPA 1225
Y+ R DL Y +NT++Q + P+ +I+++ + L + ++ +
Sbjct: 1545 YVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNK 1604
Query: 1226 VLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPSVAHSSCESEYRAMANTASEL 1285
++ SDA + + +S G L ++ + K S+ E+E A+ SE
Sbjct: 1605 LVVISDASYGN-QPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SES 1659
Query: 1286 VWLLNLLHELRVRLSAPPL---FLSDNQSAL-FMAQNPVAHKHAKHIDLDCHFVRELVSS 1341
V LLN L L L P+ L+D++S + + N + +R+ VS
Sbjct: 1660 VPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSG 1719
Query: 1342 GRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSK 1374
L V ++ T +AD+ TK LP F L +K
Sbjct: 1720 NHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1752
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 95.5 bits (236), Expect = 9e-19
Identities = 159/739 (21%), Positives = 283/739 (37%), Gaps = 92/739 (12%)
Query: 25 NFLLWSKQVTAILQNQNLFDIVDPIVAPPSEFMLQPVSSALHHV--VPAPNPLFVQWQAR 82
+F W K LQN NL I+ + P + + L++ + AP+ W
Sbjct: 180 DFPNWVKTYIKFLQNSNLGGIIPTVNGKPVRQITDDELTFLYNTFQIFAPSQFLPTWVKD 239
Query: 83 DRNVNSLLLSSLTEEALSETLSATT-ARDVWTALHSAYASRNKAR--EMRLRDELQLMRR 139
+V+ + + +++ + S T A D+ T + Y A E ++ + + +
Sbjct: 240 ILSVDYTDIMKILSKSIEKMQSDTQEANDIVTLANLQYNGSTPADAFETKVTNIIDRLNN 299
Query: 140 GSLSVSEYG--RKIR*SVANDDKVHWFLRGLGPSYANFSTGQLDQVPLPLFTDILWKVES 197
+ ++ + I ++ + K FLR + N + +L ++ + S
Sbjct: 300 NGIHINNKVACQLIMRGLSGEYK---FLRYTRHRHLNMTVAELFLDIHAIYEEQQGSRNS 356
Query: 198 HAIFQASLED-------YVTPPSAAFHARNPTRSSGSQSSGGGHRGNSSSGSRPRRDNGG 250
++ +L D Y ARNP +++ S+S S+S + P DN
Sbjct: 357 KPNYRRNLSDEKNDSRSYTNTTKPKVIARNPQKTNNSKSKTARAHNVSTSNNSPSTDNDS 416
Query: 251 SHCRGSYTPRCQLCRKQGHYAAKCPVRWDRPSESANLTHSFAAGCSLNNSNRSDK----- 305
S T QL K + LT S ++N++N SD
Sbjct: 417 --ISKSTTEPIQLNNKHDLTLGQ------------ELTES-----TVNHTNHSDDELPGH 457
Query: 306 -YMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGNGA--GLHITHIGSRS---ASHSVPLS 359
+D+GA+ + S + H+ S N + V + + I IG ++
Sbjct: 458 LLLDSGASRTLIRSAHHI---HSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI 514
Query: 360 NVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLGKGPCDKGFYVLDQGS 419
VL P + +L+S+++L ++ A F + + R G VL FY + +
Sbjct: 515 KVLHTPNIAYDLLSLNELAAV-DITACFTKN---VLERSDGTVLAPIVQYGDFYWVSK-- 568
Query: 420 QALLATSSSLPRAS------------FELWHSRLDHVNFDIIK---KLHKLGCFNVSSIL 464
+ LL ++ S+P + + H L H N I+ K + + FN S +
Sbjct: 569 RYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVD 628
Query: 465 PKPIC---CTSCQMAKSKRLVCYDNHKRASAV--------LDLVHCDLWGPSPVASVDGF 513
C C + KS + H + S + +H D++GP
Sbjct: 629 WSSAIDYQCPDCLIGKSTK----HRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAP 684
Query: 514 SYFVIFVDDFSQFTWFYPL--KRKSDFYDVLVRFKAFVENQFSRSLKVFQSDNGTEFTNN 571
SYF+ F D+ ++F W YPL +R+ DV AF++NQF S+ V Q D G+E+TN
Sbjct: 685 SYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNR 744
Query: 572 KVQALFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHSHVSTSYWVHAFSTAV 631
+ +G+ + +G ER +R +L+ L S + W A +
Sbjct: 745 TLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFST 804
Query: 632 YIINRVPSKVLSDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYMKN-KFSPRGTPCIFL 690
+ N + S S + Q + PFG V + + N K PRG P L
Sbjct: 805 IVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI--VNDHNPNSKIHPRGIPGYAL 861
Query: 691 GYNSHHKGFKCFDPTTSRT 709
+ + G+ + P+ +T
Sbjct: 862 HPSRNSYGYIIYLPSLKKT 880
Score = 71.2 bits (173), Expect = 2e-11
Identities = 150/693 (21%), Positives = 258/693 (36%), Gaps = 136/693 (19%)
Query: 782 VDDAPPSPVPHNDAPPLPAPTSPTPPATP------LVVTQAPPTSPPTTPPAVTQVPLAP 835
+D +PP ++ P+ PT+ + T L + PP SP P ++P P
Sbjct: 1096 IDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELP--P 1153
Query: 836 VDARPRTRS*NGIFKPNPRYALVHA---------------------------QPTGLLTA 868
+++ S GI N Y +++ +P
Sbjct: 1154 INSHQTNSSLGGIGDSNA-YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKR 1212
Query: 869 LHTVT*PKGFKS------------AMKHPHWLPAMEDELSALHKKFTWTLVPRPYTTN-- 914
+H + K KS A+ + + E + A HK+ L + + T+
Sbjct: 1213 IHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEY 1272
Query: 915 ----------VVGSK*VFRTKFHSDGTVERLKTCLVAQGLTQIPGFDYSLTFSPVVKATT 964
V+ S +F K DGT K VA+G Q P + S V
Sbjct: 1273 YDRKEIDPKRVINSMFIFNKK--RDGTH---KARFVARGDIQHPDTYDTGMQSNTVHHYA 1327
Query: 965 VRLILSLVVLND*QLHQLDVKNAFLHGHLTETVYMEQPH--GFVDLVSRLMCVG*TRLST 1022
+ LSL + N+ + QLD+ +A+L+ + E +Y+ P G D + RL +
Sbjct: 1328 LMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLIRLKKSHYGLKQS 1387
Query: 1023 ASNRRLVL--------GFSA*ALFSCVLVFRVVGLIHPCSFFYKGHITLYLLVYVDDIIL 1074
+N + G +SCV F +T+ L +VDD+IL
Sbjct: 1388 GANWYETIKSYLIKQCGMEEVRGWSCV--------------FKNSQVTICL--FVDDMIL 1431
Query: 1075 TGSDPSLLT*FIACLNDEFAIKYLG------KLGY-FLGLEITYTADGLFLGHAKYAHDL 1127
D + I L ++ K + ++ Y LGLEI Y KY
Sbjct: 1432 FSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQ-------RGKYMKLG 1484
Query: 1128 LSHALMLEASHVLTPL-AAGSHLVSSGEG---------------YSDPTH-YRSLVGALQ 1170
+ ++L + + PL G L + G+ Y + H + L+G
Sbjct: 1485 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLAS 1544
Query: 1171 YLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSSPA---- 1225
Y+ R DL Y +NT++Q + P+ +I+++ + L + +
Sbjct: 1545 YVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNK 1604
Query: 1226 VLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPSVAHSSCESEYRAMANTASEL 1285
++ SDA + + +S G L ++ + K S+ E+E A+ SE
Sbjct: 1605 LVAISDASYGN-QPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SES 1659
Query: 1286 VWLLNLLHELRVRLSAPPL---FLSDNQSALFMAQNPVAHK-HAKHIDLDCHFVRELVSS 1341
V LLN L L L+ P+ L+D++S + + ++ K + +R+ VS
Sbjct: 1660 VPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSG 1719
Query: 1342 GRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSK 1374
L V ++ T +AD+ TK LP F L +K
Sbjct: 1720 NNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1752
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 94.7 bits (234), Expect = 2e-18
Identities = 115/516 (22%), Positives = 208/516 (40%), Gaps = 77/516 (14%)
Query: 261 CQLCRKQGHYAAKCPVRWDRPSESANLTHSFAAGCSLNNSNRSDKYMDTGATSHMTHSLS 320
C C+ H + C +P+ + LT + + +R + ++ + + +
Sbjct: 342 CMYCKSVFHCSINCK---KKPNRNLGLTRPISQKPIIYKVHRDNNHLSPVQNEQKSWNKT 398
Query: 321 QLTYSHAYSGNDRVLVGNGAGLHITHIGSRSASHSVPLSNVLVVPRLT-------NNLVS 373
Q + Y+ V++ G+G++IT+ ++ H+ SN R T N+ VS
Sbjct: 399 QKRSNKVYNSKKLVIIDTGSGVNITN--DKTLLHNYEDSN-----RSTRFFGIGKNSSVS 451
Query: 374 VS-----KLTRDHN--------VRAIFEADSFVIQ----NRQTGAVLGKGPCDKGFYVL- 415
V K+ HN + E +S +I ++T VL + G ++
Sbjct: 452 VKGYGYIKIKNGHNNTDNKCLLTYYVPEEESTIISCYDLAKKTKMVLSRKYTRLGNKIIK 511
Query: 416 -----------------------DQGSQALLATSS---SLPRASFELW--HSRLDHVNFD 447
D A+ TSS L + S L H R+ H
Sbjct: 512 IKTKIVNGVIHVKMNELIERPSDDSKINAIKPTSSPGFKLNKRSITLEDAHKRMGHTGIQ 571
Query: 448 IIKKLHKLGCFNVSSIL---PKPICCTSCQMAKSKRLVCY----DNHKRASAVLDLVHCD 500
I+ K + S L P C +C+++K+ + Y +NH D
Sbjct: 572 QIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTDHEPGSSWCMD 631
Query: 501 LWGPSPVASVDGFSYFVIFVDDFSQF--TWFYPLKRKSDFYDVLVRFKAFVENQFSRSLK 558
++GP ++ D Y +I VD+ +++ T + K + + +VE QF R ++
Sbjct: 632 IFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQYVETQFDRKVR 691
Query: 559 VFQSDNGTEFTNNKVQALFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHSHV 618
SD GTEFTN++++ F S G+ H A NGR ER R ++ +L S++
Sbjct: 692 EINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITDATTLLRQSNL 751
Query: 619 STSYWVHAFSTAVYIINRVPSKVLSDQIPFQLLFQ--VAPTYANFHPFGCRVFPCLRPYM 676
+W +A ++A I N + K + ++P + + + V +F PFG + + +
Sbjct: 752 RVKFWEYAVTSATNIRNYLEHK-STGKLPLKAISRQPVTVRLMSFLPFGEK--GIIWNHN 808
Query: 677 KNKFSPRGTPCIFLGYNSHHKGFKCFDPTTSRTYVS 712
K P G P I L + + G+K F P+ ++ S
Sbjct: 809 HKKLKPSGLPSIILCKDPNSYGYKFFIPSKNKIVTS 844
Score = 58.9 bits (141), Expect = 9e-08
Identities = 97/468 (20%), Positives = 176/468 (36%), Gaps = 43/468 (9%)
Query: 936 KTCLVAQGLTQIPGFDYSLTFSPVVKATTVRLILSLVVLND*QLHQLDVKNAFLHGHLTE 995
K +V +G TQ P YS+ + + +++ L + + + LD+ +AFL+ L E
Sbjct: 1338 KARIVCRGDTQSPD-TYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLEE 1396
Query: 996 TVYMEQPHG---FVDLVSRLMCVG*TRLSTASNRRLVLGFSA*ALFSCVLVFRVVGL--- 1049
+Y+ PH V L L + + + R L +GL
Sbjct: 1397 EIYIPHPHDRRCVVKLNKALYGLKQSPKEWNDHLRQYLNG--------------IGLKDN 1442
Query: 1050 IHPCSFFYKGHITLYLLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYLGKL------GY 1103
+ + L + VYVDD ++ S+ L FI L F +K G L
Sbjct: 1443 SYTPGLYQTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTD 1502
Query: 1104 FLGLEITY---------TADGLFLGHAKYAHDLLSHALMLEASHVLTPLAAGSHLV---S 1151
LG+++ Y T K ++ L H+ T V S
Sbjct: 1503 ILGMDLVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMS 1562
Query: 1152 SGEGYSDPTHYRSLVGALQYLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCA 1210
E + L+G L Y+ R D+ +AV V++ + P F + +II+Y+
Sbjct: 1563 EEEFRQGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVR 1622
Query: 1211 MQHFGLTFRR--TSSPAVLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPSVAH 1268
+ G+ + R V+ +DA D +S G ++ G N+ + + K +
Sbjct: 1623 YKDIGIHYDRDCNKDKKVIAITDASVGSEYD-AQSRIGVILWYGMNIFNVYSNKSTNRCV 1681
Query: 1269 SSCESEYRAMANTASELVWLLNLLHELRVRLSAPPLFLSDNQSALFMAQNPVAHKHAKHI 1328
SS E+E A+ ++ L L EL + + ++D++ A+ K
Sbjct: 1682 SSTEAELHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFT 1741
Query: 1329 DLDCHFVRELVSSGRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSKLR 1376
+ ++E + + + + +AD+ TK + F F L+
Sbjct: 1742 WIKTEIIKEKIKEKSIKLLKITGKGNIADLLTKPVSASDFKRFIQVLK 1789
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 87.8 bits (216), Expect = 2e-16
Identities = 91/374 (24%), Positives = 151/374 (40%), Gaps = 36/374 (9%)
Query: 362 LVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLGKGPCDKGFYVLDQG--- 418
L P + +L+S+S+L + N+ A F ++ R G VL FY L +
Sbjct: 513 LHTPNIAYDLLSLSELA-NQNITACFTRNTL---ERSDGTVLAPIVKHGDFYWLSKKYLI 568
Query: 419 ----SQALLAT---SSSLPRASFELWHSRLDHVNFDIIKKLHKLGCF------NVSSILP 465
S+ + S S+ + + L H L H NF I+K K ++
Sbjct: 569 PSHISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNA 628
Query: 466 KPICCTSCQMAKSKRLVCYDNHKRASAV--------LDLVHCDLWGPSPVASVDGFSYFV 517
C C + KS + H + S + +H D++GP SYF+
Sbjct: 629 STYQCPDCLIGKSTK----HRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFI 684
Query: 518 IFVDDFSQFTWFYPL--KRKSDFYDVLVRFKAFVENQFSRSLKVFQSDNGTEFTNNKVQA 575
F D+ ++F W YPL +R+ +V AF++NQF+ + V Q D G+E+TN +
Sbjct: 685 SFTDEKTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHK 744
Query: 576 LFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHSHVSTSYWVHAFSTAVYIIN 635
F++ G+ + +G ER +R +L +L+ S + W A + I N
Sbjct: 745 FFTNRGITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAVEFSTIIRN 804
Query: 636 RVPSKVLSDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYMKNKFSPRGTPCIFLGYNSH 695
+ S +D+ Q PFG V +K PRG P L + +
Sbjct: 805 SLVSP-KNDKSARQHAGLAGLDITTILPFGQPVI-VNNHNPDSKIHPRGIPGYALHPSRN 862
Query: 696 HKGFKCFDPTTSRT 709
G+ + P+ +T
Sbjct: 863 SYGYIIYLPSLKKT 876
Score = 78.6 bits (192), Expect = 1e-13
Identities = 129/536 (24%), Positives = 215/536 (40%), Gaps = 65/536 (12%)
Query: 879 KSAMKHPHWLPAMEDELSALHKKFTWTLVPRPYTTNVVGSK*VFRTKF----HSDGTVER 934
K + ++ A E+S L K TW + Y N + K V + F DGT
Sbjct: 1257 KDNKEKDRYVEAYHKEISQLLKMNTWD-TNKYYDRNDIDPKKVINSMFIFNKKRDGTH-- 1313
Query: 935 LKTCLVAQGLTQIPGFDYSLTFSPVVKATTVRLILSLVVLND*QLHQLDVKNAFLHGHLT 994
K VA+G Q P S S V + LS+ + ND + QLD+ +A+L+ +
Sbjct: 1314 -KARFVARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIK 1372
Query: 995 ETVYMEQPH--GFVDLVSRL-MCVG*TRLSTASNRRLVLGFSA*ALFSCVLVFRVVGLIH 1051
E +Y+ P G D + RL + + S A+ + + L +C + V G
Sbjct: 1373 EELYIRPPPHLGLNDKLLRLRKSLYGLKQSGANWYETIKSY----LINCCDMQEVRGW-- 1426
Query: 1052 PCSFFYKGHITLYLLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYL------GKLGY-F 1104
F +T+ L +VDD+IL D + I L ++ K + ++ Y
Sbjct: 1427 -SCVFKNSQVTICL--FVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDI 1483
Query: 1105 LGLEITYTADGLFLGHAKYAHDLLSHALMLEASHVLTPL--------AAG--SHLVSSGE 1154
LGLEI Y +KY + +L + + PL A G H + E
Sbjct: 1484 LGLEIKYQ-------RSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDE 1536
Query: 1155 GYSDPTHYR-------SLVGALQYLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRIIR 1206
D Y+ L+G Y+ R DL Y +NT++Q + P+ +I+
Sbjct: 1537 LEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQ 1596
Query: 1207 YVCAMQHFGLTFRRTS----SPAVLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKK 1262
++ + L + + ++ SDA + + +S G L ++ + K
Sbjct: 1597 FMWDTRDKQLIWHKNKPTKPDNKLVAISDASYGN-QPYYKSQIGNIFLLNGKVIGGKSTK 1655
Query: 1263 QPSVAHSSCESEYRAMANTASELVWLLNLLHELRVRLSAPPL---FLSDNQSALFMAQNP 1319
A +C S A + SE + LLN L L L+ P+ L+D++S + + ++
Sbjct: 1656 ----ASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPIIKGLLTDSRSTISIIKST 1711
Query: 1320 VAHK-HAKHIDLDCHFVRELVSSGRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSK 1374
K + +R+ VS L V ++ T +AD+ TK LP F L +K
Sbjct: 1712 NEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1767
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 85.9 bits (211), Expect = 7e-16
Identities = 106/454 (23%), Positives = 183/454 (39%), Gaps = 56/454 (12%)
Query: 296 SLNNSNRSDK------YMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGNGA--GLHITHI 347
++N++N SD +D+GA+ + S + H+ S N + V + + I I
Sbjct: 16 TVNHTNHSDDELPGHLLLDSGASRTLIRSAHHI---HSASSNPDINVVDAQKRNIPINAI 72
Query: 348 GSRS---ASHSVPLSNVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLG 404
G ++ VL P + +L+S+++L ++ A F + + R G VL
Sbjct: 73 GDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAV-DITACFTKN---VLERSDGTVLA 128
Query: 405 KGPCDKGFYVLDQGSQALLATSSSLPRAS------------FELWHSRLDHVNFDIIK-- 450
FY + + + LL ++ S+P + + H L H N I+
Sbjct: 129 PIVKYGDFYWVSK--KYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYS 186
Query: 451 -KLHKLGCFNVSSILPKPIC---CTSCQMAKSKRLVCYDNHKRASAV--------LDLVH 498
K + + FN S + C C + KS + H + S + +H
Sbjct: 187 LKNNTITYFNESDVDRSSAIDYQCPDCLIGKSTK----HRHIKGSRLKYQNSYEPFQYLH 242
Query: 499 CDLWGPSPVASVDGFSYFVIFVDDFSQFTWFYPL--KRKSDFYDVLVRFKAFVENQFSRS 556
D++GP SYF+ F D+ ++F W YPL +R+ DV AF++NQF S
Sbjct: 243 TDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQAS 302
Query: 557 LKVFQSDNGTEFTNNKVQALFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHS 616
+ V Q D G+E+TN + +G+ + +G ER +R +L+ L S
Sbjct: 303 VLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCS 362
Query: 617 HVSTSYWVHAFSTAVYIINRVPSKVLSDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYM 676
+ W A + + N + S S + Q + PFG V + +
Sbjct: 363 GLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI--VNDHN 419
Query: 677 KN-KFSPRGTPCIFLGYNSHHKGFKCFDPTTSRT 709
N K PRG P L + + G+ + P+ +T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
Score = 71.2 bits (173), Expect = 2e-11
Identities = 151/693 (21%), Positives = 258/693 (36%), Gaps = 136/693 (19%)
Query: 782 VDDAPPSPVPHNDAPPLPAPTSPTPPATP------LVVTQAPPTSPPTTPPAVTQVPLAP 835
+D +PP ++ P+ PT+ + T L + PP SP P ++P P
Sbjct: 669 IDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELP--P 726
Query: 836 VDARPRTRS*NGIFKPNPRYALVHA---------------------------QPTGLLTA 868
+++ S GI N Y +++ +P
Sbjct: 727 INSHQTNSSLGGIGDSNA-YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKR 785
Query: 869 LHTVT*PKGFKS------------AMKHPHWLPAMEDELSALHKKFTWTLVPRPYTTN-- 914
+H + K KS A+ + + E + A HK+ L + + T+
Sbjct: 786 IHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEY 845
Query: 915 ----------VVGSK*VFRTKFHSDGTVERLKTCLVAQGLTQIPGFDYSLTFSPVVKATT 964
V+ S +F K DGT K VA+G Q P S S V
Sbjct: 846 YDRKEIDPKRVINSMFIFNKK--RDGTH---KARFVARGDIQHPDTYDSGMQSNTVHHYA 900
Query: 965 VRLILSLVVLND*QLHQLDVKNAFLHGHLTETVYMEQPH--GFVDLVSRLMCVG*TRLST 1022
+ LSL + N+ + QLD+ +A+L+ + E +Y+ P G D + RL +
Sbjct: 901 LMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLIRLKKSLYGLKQS 960
Query: 1023 ASNRRLVL--------GFSA*ALFSCVLVFRVVGLIHPCSFFYKGHITLYLLVYVDDIIL 1074
+N + G +SCV F +T+ L +VDD+IL
Sbjct: 961 GANWYETIKSYLIKQCGMEEVRGWSCV--------------FKNSQVTICL--FVDDMIL 1004
Query: 1075 TGSDPSLLT*FIACLNDEFAIKYLG------KLGY-FLGLEITYTADGLFLGHAKYAHDL 1127
D + I L ++ K + ++ Y LGLEI Y KY
Sbjct: 1005 FSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQ-------RGKYMKLG 1057
Query: 1128 LSHALMLEASHVLTPL-AAGSHLVSSGEG---------------YSDPTH-YRSLVGALQ 1170
+ ++L + + PL G L + G+ Y + H + L+G
Sbjct: 1058 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLAS 1117
Query: 1171 YLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSSPA---- 1225
Y+ R DL Y +NT++Q + P+ +I+++ + L + +
Sbjct: 1118 YVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNK 1177
Query: 1226 VLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPSVAHSSCESEYRAMANTASEL 1285
++ SDA + + +S G L ++ + K S+ E+E A+ SE
Sbjct: 1178 LVAISDASYGN-QPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SES 1232
Query: 1286 VWLLNLLHELRVRLSAPPL---FLSDNQSALFMAQNPVAHK-HAKHIDLDCHFVRELVSS 1341
V LLN L L L+ P+ L+D++S + + ++ K + +R+ VS
Sbjct: 1233 VPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSG 1292
Query: 1342 GRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSK 1374
L V ++ T +AD+ TK LP F L +K
Sbjct: 1293 NNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 85.1 bits (209), Expect = 1e-15
Identities = 106/454 (23%), Positives = 183/454 (39%), Gaps = 56/454 (12%)
Query: 296 SLNNSNRSDK------YMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGNGA--GLHITHI 347
++N++N SD +D+GA+ + S + H+ S N + V + + I I
Sbjct: 16 TVNHTNHSDDELPGHLLLDSGASRTLIRSAHHI---HSASSNPDINVVDAQKRNIPINAI 72
Query: 348 GSRS---ASHSVPLSNVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLG 404
G ++ VL P + +L+S+++L ++ A F + + R G VL
Sbjct: 73 GDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAV-DITACFTKN---VLERSDGTVLA 128
Query: 405 KGPCDKGFYVLDQGSQALLATSSSLPRAS------------FELWHSRLDHVNFDIIK-- 450
FY + + + LL ++ S+P + + H L H N I+
Sbjct: 129 PIVKYGDFYWVSK--KYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYS 186
Query: 451 -KLHKLGCFNVSSILPKPIC---CTSCQMAKSKRLVCYDNHKRASAV--------LDLVH 498
K + + FN S + C C + KS + H + S + +H
Sbjct: 187 LKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTK----HRHIKGSRLKYQNSYEPFQYLH 242
Query: 499 CDLWGPSPVASVDGFSYFVIFVDDFSQFTWFYPL--KRKSDFYDVLVRFKAFVENQFSRS 556
D++GP SYF+ F D+ ++F W YPL +R+ DV AF++NQF S
Sbjct: 243 TDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQAS 302
Query: 557 LKVFQSDNGTEFTNNKVQALFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHS 616
+ V Q D G+E+TN + +G+ + +G ER +R +L+ L S
Sbjct: 303 VLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCS 362
Query: 617 HVSTSYWVHAFSTAVYIINRVPSKVLSDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYM 676
+ W A + + N + S S + Q + PFG V + +
Sbjct: 363 GLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI--VNDHN 419
Query: 677 KN-KFSPRGTPCIFLGYNSHHKGFKCFDPTTSRT 709
N K PRG P L + + G+ + P+ +T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
Score = 72.8 bits (177), Expect = 6e-12
Identities = 152/697 (21%), Positives = 262/697 (36%), Gaps = 144/697 (20%)
Query: 782 VDDAPPSPVPHNDAPPLPAPTSPTPPATP------LVVTQAPPTSPPTTPPAVTQVPLAP 835
+D +PP ++ P+ PT+ + T L + PP SP P ++P P
Sbjct: 669 IDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELP--P 726
Query: 836 VDARPRTRS*NGIFKPNPRYALVHAQPTGL------------------------------ 865
+++R S GI N Y ++++ L
Sbjct: 727 INSRQTNSSLGGIGDSNA-YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKR 785
Query: 866 ---LTALHTVT*PKGFKSAMKHPH-------------WLPAMEDELSALHKKFTWTLVPR 909
+ A+ V K ++ +++ ++ A E++ L K TW R
Sbjct: 786 IHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIQAYHKEVNQLLKMKTWD-TDR 844
Query: 910 PYTTNVVGSK*VFRTKF----HSDGTVERLKTCLVAQGLTQIPGFDYSLTFSPVVKATTV 965
Y + K V + F DGT K VA+G Q P T+ P +++ TV
Sbjct: 845 YYDRKEIDPKRVINSMFIFNRKRDGTH---KARFVARGDIQHPD-----TYDPGMQSNTV 896
Query: 966 R-----LILSLVVLND*QLHQLDVKNAFLHGHLTETVYMEQPH--GFVDLVSRLMCVG*T 1018
LSL + N+ + QLD+ +A+L+ + E +Y+ P G D + RL
Sbjct: 897 HHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLIRLKKSLYG 956
Query: 1019 RLSTASNRRLVL--------GFSA*ALFSCVLVFRVVGLIHPCSFFYKGHITLYLLVYVD 1070
+ +N + G +SCV F +T+ L +VD
Sbjct: 957 LKQSGANWYETIKSYLIKQCGMEEVRGWSCV--------------FKNSQVTICL--FVD 1000
Query: 1071 DIILTGSDPSLLT*FIACLNDEFAIKYLG------KLGY-FLGLEITYTADGLFLGHAKY 1123
D+IL D + I L ++ K + ++ Y LGLEI Y KY
Sbjct: 1001 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQ-------RGKY 1053
Query: 1124 AHDLLSHALMLEASHVLTPL-AAGSHLVSSGE---------------GYSDPTH-YRSLV 1166
+ ++L + + PL G L + G+ Y H + L+
Sbjct: 1054 MKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLI 1113
Query: 1167 GALQYLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTS--- 1222
G Y+ R DL Y +NT++Q + P+ +I+++ + L + ++
Sbjct: 1114 GLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVK 1173
Query: 1223 -SPAVLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPSVAHSSCESEYRAMANT 1281
+ ++ SDA + + +S G L ++ + K S+ E+E A+
Sbjct: 1174 PTNKLVVISDASYGN-QPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAI--- 1229
Query: 1282 ASELVWLLNLLHELRVRLSAPPL---FLSDNQSAL-FMAQNPVAHKHAKHIDLDCHFVRE 1337
SE V LLN L L L+ P+ L+D++S + + N + +R+
Sbjct: 1230 -SESVPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRD 1288
Query: 1338 LVSSGRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSK 1374
VS L V ++ T +AD+ TK LP F L +K
Sbjct: 1289 EVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 85.1 bits (209), Expect = 1e-15
Identities = 106/454 (23%), Positives = 183/454 (39%), Gaps = 56/454 (12%)
Query: 296 SLNNSNRSDK------YMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGNGA--GLHITHI 347
++N++N SD +D+GA+ + S + H+ S N + V + + I I
Sbjct: 16 TVNHTNHSDDKLPGHLLLDSGASRTLIRSAHHI---HSASSNPDINVVDAQKRNIPINAI 72
Query: 348 GSRS---ASHSVPLSNVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLG 404
G ++ VL P + +L+S+++L ++ A F + + R G VL
Sbjct: 73 GDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAV-DITACFTKN---VLERSDGTVLA 128
Query: 405 KGPCDKGFYVLDQGSQALLATSSSLPRAS------------FELWHSRLDHVNFDIIK-- 450
FY + + + LL ++ S+P + + H L H N I+
Sbjct: 129 PIVKYGDFYWVSK--KYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYS 186
Query: 451 -KLHKLGCFNVSSILPKPIC---CTSCQMAKSKRLVCYDNHKRASAV--------LDLVH 498
K + + FN S + C C + KS + H + S + +H
Sbjct: 187 LKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTK----HRHIKGSRLKYQNSYEPFQYLH 242
Query: 499 CDLWGPSPVASVDGFSYFVIFVDDFSQFTWFYPL--KRKSDFYDVLVRFKAFVENQFSRS 556
D++GP SYF+ F D+ ++F W YPL +R+ DV AF++NQF S
Sbjct: 243 TDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQAS 302
Query: 557 LKVFQSDNGTEFTNNKVQALFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHS 616
+ V Q D G+E+TN + +G+ + +G ER +R +L+ L S
Sbjct: 303 VLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCS 362
Query: 617 HVSTSYWVHAFSTAVYIINRVPSKVLSDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYM 676
+ W A + + N + S S + Q + PFG V + +
Sbjct: 363 GLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI--VNDHN 419
Query: 677 KN-KFSPRGTPCIFLGYNSHHKGFKCFDPTTSRT 709
N K PRG P L + + G+ + P+ +T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
Score = 61.2 bits (147), Expect = 2e-08
Identities = 124/543 (22%), Positives = 210/543 (37%), Gaps = 79/543 (14%)
Query: 879 KSAMKHPHWLPAMEDELSALHKKFTWTLVPRPYTTNVVGSK*VFRTKF----HSDGTVER 934
K + ++ A E++ L K TW + Y + K V + F DGT
Sbjct: 815 KDIKEKEKYIEAYHKEVNQLLKMKTWD-TDKYYDRKEIDPKRVINSMFIFNRKRDGTH-- 871
Query: 935 LKTCLVAQGLTQIPGFDYSLTFSPVVKATTVRLILSLVVLND*QLHQLDVKNAFLHGHLT 994
K VA+G Q P S S V + LSL + N+ + QLD+ +A+L+ +
Sbjct: 872 -KARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIK 930
Query: 995 ETVYMEQPH--GFVDLVSRLMCVG*TRLSTASNRRLVL--------GFSA*ALFSCVLVF 1044
E +Y+ P G D + RL + +N + G +SCV
Sbjct: 931 EELYIRPPPHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCV--- 987
Query: 1045 RVVGLIHPCSFFYKGHITLYLLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYLG----- 1099
F +T+ L +VDD++L + + I L ++ K +
Sbjct: 988 -----------FENSQVTICL--FVDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESD 1034
Query: 1100 -KLGY-FLGLEITYTADGLFLGHAKYAHDLLSHALMLEASHVLTPL-AAGSHLVSSGE-- 1154
++ Y LGLEI Y KY + ++L + + PL G L + G+
Sbjct: 1035 EEIQYDILGLEIKYQ-------RGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPG 1087
Query: 1155 -------------GYSDPTH-YRSLVGALQYLTIT-RPDLSYAVNTVSQFLQTPTVDHFQ 1199
Y H + L+G Y+ R DL Y +NT++Q + P+
Sbjct: 1088 LYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLD 1147
Query: 1200 AVKRIIRYVCAMQHFGLTFRRTS----SPAVLGYSDADWARCTDHRRSTYGYAIFLGYNL 1255
+I+++ + L + ++ + ++ SDA + + +S G L +
Sbjct: 1148 MTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGN-QPYYKSQIGNIYLLNGKV 1206
Query: 1256 LSWSAKKQPSVAHSSCESEYRAMANTASELVWLLNLLHELRVRLSAPPL---FLSDNQSA 1312
+ + K S+ E+E A+ SE V LLN L L L P+ L+D++S
Sbjct: 1207 IGGKSTKASLTCTSTTEAEIHAI----SESVPLLNNLSYLIQELDKKPITKGLLTDSKST 1262
Query: 1313 L-FMAQNPVAHKHAKHIDLDCHFVRELVSSGRLAVRHVPTSLQLADIFTKVLPRPLFDLF 1371
+ + N + +R+ VS L V ++ T +AD+ TK LP F L
Sbjct: 1263 ISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLL 1322
Query: 1372 RSK 1374
+K
Sbjct: 1323 TNK 1325
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 85.1 bits (209), Expect = 1e-15
Identities = 106/454 (23%), Positives = 183/454 (39%), Gaps = 56/454 (12%)
Query: 296 SLNNSNRSDK------YMDTGATSHMTHSLSQLTYSHAYSGNDRVLVGNGA--GLHITHI 347
++N++N SD +D+GA+ + S + H+ S N + V + + I I
Sbjct: 16 TVNHTNHSDDELPGHLLLDSGASRTLIRSAHHI---HSASSNPDINVVDAQKRNIPINAI 72
Query: 348 GSRS---ASHSVPLSNVLVVPRLTNNLVSVSKLTRDHNVRAIFEADSFVIQNRQTGAVLG 404
G ++ VL P + +L+S+++L ++ A F + + R G VL
Sbjct: 73 GDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAV-DITACFTKN---VLERSDGTVLA 128
Query: 405 KGPCDKGFYVLDQGSQALLATSSSLPRAS------------FELWHSRLDHVNFDIIK-- 450
FY + + + LL ++ S+P + + H L H N I+
Sbjct: 129 PIVKYGDFYWVSK--KYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYS 186
Query: 451 -KLHKLGCFNVSSILPKPIC---CTSCQMAKSKRLVCYDNHKRASAV--------LDLVH 498
K + + FN S + C C + KS + H + S + +H
Sbjct: 187 LKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTK----HRHIKGSRLKYQNSYEPFQYLH 242
Query: 499 CDLWGPSPVASVDGFSYFVIFVDDFSQFTWFYPL--KRKSDFYDVLVRFKAFVENQFSRS 556
D++GP SYF+ F D+ ++F W YPL +R+ DV AF++NQF S
Sbjct: 243 TDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQAS 302
Query: 557 LKVFQSDNGTEFTNNKVQALFSSSGVLHRFLCPHTQAQNGRVERKHRHVLELGLAMLYHS 616
+ V Q D G+E+TN + +G+ + +G ER +R +L+ L S
Sbjct: 303 VLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCS 362
Query: 617 HVSTSYWVHAFSTAVYIINRVPSKVLSDQIPFQLLFQVAPTYANFHPFGCRVFPCLRPYM 676
+ W A + + N + S S + Q + PFG V + +
Sbjct: 363 GLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI--VNDHN 419
Query: 677 KN-KFSPRGTPCIFLGYNSHHKGFKCFDPTTSRT 709
N K PRG P L + + G+ + P+ +T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
Score = 69.7 bits (169), Expect = 5e-11
Identities = 151/693 (21%), Positives = 255/693 (36%), Gaps = 136/693 (19%)
Query: 782 VDDAPPSPVPHNDAPPLPAPTSPTPPATP------LVVTQAPPTSPPTTPPAVTQVPLAP 835
+D +PP ++ P+ PT+ + T L + PP SP P ++P P
Sbjct: 669 IDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELP--P 726
Query: 836 VDARPRTRS*NGIFKPNPRYALVHA---------------------------QPTGLLTA 868
+++ S GI N Y +++ +P
Sbjct: 727 INSHQTNSSLGGIGDSNA-YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKR 785
Query: 869 LHTVT*PKGFKS------------AMKHPHWLPAMEDELSALHKKFTWTLVPRPYTTN-- 914
+H + K KS A+ + + E + A HK+ L + T+
Sbjct: 786 IHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMNTWDTDKY 845
Query: 915 ----------VVGSK*VFRTKFHSDGTVERLKTCLVAQGLTQIPGFDYSLTFSPVVKATT 964
V+ S +F K DGT K VA+G Q P S S V
Sbjct: 846 YDRKEIDPKRVINSMFIFNRK--RDGTH---KARFVARGDIQHPDTYDSGMQSNTVHHYA 900
Query: 965 VRLILSLVVLND*QLHQLDVKNAFLHGHLTETVYMEQPH--GFVDLVSRLMCVG*TRLST 1022
+ LSL + N+ + QLD+ +A+L+ + E +Y+ P G D + RL +
Sbjct: 901 LMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLIRLKKSLYGLKQS 960
Query: 1023 ASNRRLVL--------GFSA*ALFSCVLVFRVVGLIHPCSFFYKGHITLYLLVYVDDIIL 1074
+N + G +SCV F +T+ L +VDD+IL
Sbjct: 961 GANWYETIKSYLIKQCGMEEVRGWSCV--------------FKNSQVTICL--FVDDMIL 1004
Query: 1075 TGSDPSLLT*FIACLNDEFAIKYLG------KLGY-FLGLEITYTADGLFLGHAKYAHDL 1127
D + I L ++ K + ++ Y LGLEI Y KY
Sbjct: 1005 FSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQ-------RGKYMKLG 1057
Query: 1128 LSHALMLEASHVLTPL-AAGSHLVSSGEG---------------YSDPTH-YRSLVGALQ 1170
+ ++L + + PL G L + G+ Y + H + L+G
Sbjct: 1058 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLAS 1117
Query: 1171 YLTIT-RPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSSPA---- 1225
Y+ R DL Y +NT++Q + P+ +I+++ + L + +
Sbjct: 1118 YVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNK 1177
Query: 1226 VLGYSDADWARCTDHRRSTYGYAIFLGYNLLSWSAKKQPSVAHSSCESEYRAMANTASEL 1285
++ SDA + + +S G L ++ + K S+ E+E A+ SE
Sbjct: 1178 LVAISDASYGN-QPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SES 1232
Query: 1286 VWLLNLLHELRVRLSAPPL---FLSDNQSAL-FMAQNPVAHKHAKHIDLDCHFVRELVSS 1341
V LLN L L L+ P+ L+D++S + + N + +R+ VS
Sbjct: 1233 VPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSG 1292
Query: 1342 GRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSK 1374
L V ++ T +AD+ TK LP F L +K
Sbjct: 1293 NHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 79.0 bits (193), Expect = 9e-14
Identities = 42/99 (42%), Positives = 49/99 (49%), Gaps = 4/99 (4%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P + +PPS P V P +PP+ P P P P P APPSPVP + APP PAP
Sbjct: 199 PPSPAPPSPAPP--VPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAP 256
Query: 802 TSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDARP 840
SP PPA P +PP PP PP P+ P P
Sbjct: 257 PSPKPPAPP--PPPSPPPPPPPRPPFPANTPMPPSPPSP 293
Score = 71.6 bits (174), Expect = 1e-11
Identities = 36/100 (36%), Positives = 50/100 (50%), Gaps = 1/100 (1%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P + +PPS P P +PP+ P P P P P P +P P+P + APP P+P
Sbjct: 98 PPSPAPPSPAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSP 157
Query: 802 TSPTPPATPLVVTQAPPT-SPPTTPPAVTQVPLAPVDARP 840
P P+ P+ + APP+ +PP+ P V P P A P
Sbjct: 158 PVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPP 197
Score = 71.6 bits (174), Expect = 1e-11
Identities = 45/118 (38%), Positives = 56/118 (47%), Gaps = 11/118 (9%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPL--- 798
PA SPPS P P SPP P P P P + P P +P PVP + +PP+
Sbjct: 116 PAPPSPPSPAP-----PSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPS 170
Query: 799 PAPTSPTP--PATPLVVTQAPPT-SPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNP 853
PAP SPTP P+ P+ + APP+ +PP P P PV P S P+P
Sbjct: 171 PAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSP 228
Score = 71.6 bits (174), Expect = 1e-11
Identities = 43/117 (36%), Positives = 54/117 (45%), Gaps = 5/117 (4%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDP----DVQPVPVDDAPPSPVPHNDAPP 797
PA SPPS P P +PP+ P P+ P P P +PPSP P + +PP
Sbjct: 73 PAPPSPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPPSPAPPSPSPP 132
Query: 798 LPAPTSPTPPATPLVVTQAPPT-SPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNP 853
P SP PA PL + APP+ SPP P VP +P P S + P+P
Sbjct: 133 APPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSP 189
Score = 70.9 bits (172), Expect = 2e-11
Identities = 40/113 (35%), Positives = 49/113 (42%), Gaps = 6/113 (5%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P+ +PPS P P +PP P P P P V P P PPSP P + PP P+P
Sbjct: 129 PSPPAPPSPSP-----PSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSP 183
Query: 802 TSPTPPATPLVVTQAPPT-SPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNP 853
P PA P PP+ +PP+ P V P P P S P+P
Sbjct: 184 PVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSP 236
Score = 69.7 bits (169), Expect = 5e-11
Identities = 42/113 (37%), Positives = 53/113 (46%), Gaps = 3/113 (2%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P + +PPS P P +PP+ P P P P P APPSP P + APP PAP
Sbjct: 40 PPSPAPPSPAPPSPAPPSPAPPSPAPPSPGPPSP-APPSPPSPAPPSPAPPSPAPPSPAP 98
Query: 802 TSPTPPATPLVVTQAPPT-SPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNP 853
SP PP +P + APP+ +PP+ P P P P S P+P
Sbjct: 99 PSPAPP-SPAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSP 150
Score = 69.7 bits (169), Expect = 5e-11
Identities = 44/118 (37%), Positives = 55/118 (46%), Gaps = 9/118 (7%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQ--PVPVDDAPPSPVPHNDAPPLP 799
P + +PPS P V P SPP P P P P PVP APPSP P PP P
Sbjct: 147 PPSPAPPSPSPP--VPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAP--PVPPSP 202
Query: 800 APTSPTPPATPLVVTQAPPTSPPTTPPA---VTQVPLAPVDARPRTRS*NGIFKPNPR 854
AP SP PP P +PP+ P +PP+ + P AP P + + P+P+
Sbjct: 203 APPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPK 260
Score = 68.2 bits (165), Expect = 2e-10
Identities = 41/100 (41%), Positives = 48/100 (48%), Gaps = 12/100 (12%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
PA PPS P P +PP+ P P P P P PV PPSP P + APP P P
Sbjct: 207 PAPPVPPSPAPPS--PPSPAPPSPPSPAPPSPSPPAPPSPV---PPSPAPPSPAPPSPKP 261
Query: 802 TSPTPPATPLVVTQAPPTSPPTTP-PAVTQVPLAPVDARP 840
+P PP +P PP PP P PA T +P +P P
Sbjct: 262 PAPPPPPSP------PPPPPPRPPFPANTPMPPSPPSPPP 295
Score = 65.1 bits (157), Expect = 1e-09
Identities = 43/115 (37%), Positives = 48/115 (41%), Gaps = 15/115 (13%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHND------- 794
P+ PPS P P SPP P P P P V P P +P PVP +
Sbjct: 163 PSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPS 222
Query: 795 -A---PPLPAPTSPTPPATPLVVTQAP----PTSPPTTPPAVTQVPLAPVDARPR 841
A PP PAP SP+PPA P V +P P P PPA P P PR
Sbjct: 223 PAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPR 277
Score = 60.8 bits (146), Expect = 2e-08
Identities = 36/92 (39%), Positives = 45/92 (48%), Gaps = 6/92 (6%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCP-SCVDPDVQPVPVDDAPPSPVPHNDAPPLPA 800
P SPPS P +PPT P P P S V P PVP APPSP P PP PA
Sbjct: 285 PMPPSPPSPPPS---PAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAP--SPPPSPA 339
Query: 801 PTSPTPPATPLVVTQAPPTSPPTTPPAVTQVP 832
P +P+P +P P+ P+ P+ + +P
Sbjct: 340 PPTPSPSPSPSPSPSPSPSPSPSPSPSPSPIP 371
Score = 58.5 bits (140), Expect = 1e-07
Identities = 39/105 (37%), Positives = 44/105 (41%), Gaps = 8/105 (7%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPS-PVPHNDAPPLPA 800
PA SPPS P P P P P P P P PPS P P PP PA
Sbjct: 223 PAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPA 282
Query: 801 -----PTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDARP 840
P+ P+PP +P T PPT P +PP+ APV P
Sbjct: 283 NTPMPPSPPSPPPSPAPPT--PPTPPSPSPPSPVPPSPAPVPPSP 325
Score = 58.2 bits (139), Expect = 2e-07
Identities = 33/92 (35%), Positives = 44/92 (46%), Gaps = 6/92 (6%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHN-----DAP 796
P + +PPS P P PP P P P P P+ +PPSP P P
Sbjct: 246 PPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAPPTPPTP 305
Query: 797 PLPAPTSPTPPA-TPLVVTQAPPTSPPTTPPA 827
P P+P SP PP+ P+ + APP+ P+ PP+
Sbjct: 306 PSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPS 337
Score = 56.2 bits (134), Expect = 6e-07
Identities = 31/82 (37%), Positives = 40/82 (47%), Gaps = 1/82 (1%)
Query: 772 CVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQV 831
CV + P APPSP P + APP PAP SP PP +P + APP+ P PP+
Sbjct: 31 CVPGGIFNCPPSPAPPSPAPPSPAPPSPAPPSPAPP-SPGPPSPAPPSPPSPAPPSPAPP 89
Query: 832 PLAPVDARPRTRS*NGIFKPNP 853
AP P + + P+P
Sbjct: 90 SPAPPSPAPPSPAPPSPAPPSP 111
Score = 55.1 bits (131), Expect = 1e-06
Identities = 33/99 (33%), Positives = 44/99 (44%), Gaps = 8/99 (8%)
Query: 742 PAAGSPPSLQPVILVVPESSP----PTGPLPCPSCVDPDVQPVPVDDAP----PSPVPHN 793
P SPP P P ++P P P P P+ P P P +P P+PVP +
Sbjct: 265 PPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAPPTPPTPPSPSPPSPVPPSPAPVPPS 324
Query: 794 DAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVP 832
APP PAP+ P PA P P+ P+ P+ + P
Sbjct: 325 PAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSPSPSP 363
Score = 54.3 bits (129), Expect = 2e-06
Identities = 33/103 (32%), Positives = 44/103 (42%), Gaps = 12/103 (11%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPP----SPVPHNDAPP 797
P+ +PPS P P +PP+ P P P P P PP +P+P + P
Sbjct: 236 PSPPAPPSPVPPSPAPPSPAPPSPKPPAPP--PPPSPPPPPPPRPPFPANTPMPPSPPSP 293
Query: 798 LPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDARP 840
P+P PTPP P P+ P PP+ VP +P P
Sbjct: 294 PPSPAPPTPPTPP------SPSPPSPVPPSPAPVPPSPAPPSP 330
Score = 51.6 bits (122), Expect = 1e-05
Identities = 38/122 (31%), Positives = 51/122 (41%), Gaps = 16/122 (13%)
Query: 742 PAAGSPPSLQPVILVVPESSP---PTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPL 798
P G+ L V+ V ++ P G CP P P APPSP P + APP
Sbjct: 9 PLVGAVNVLMVVLAFVASANAQCVPGGIFNCPP------SPAPPSPAPPSPAPPSPAPPS 62
Query: 799 PAP---TSPTP----PATPLVVTQAPPTSPPTTPPAVTQVPLAPVDARPRTRS*NGIFKP 851
PAP P+P P +P + APP+ P +P + P +P P S P
Sbjct: 63 PAPPSPGPPSPAPPSPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPP 122
Query: 852 NP 853
+P
Sbjct: 123 SP 124
Score = 47.0 bits (110), Expect = 4e-04
Identities = 32/92 (34%), Positives = 41/92 (43%), Gaps = 6/92 (6%)
Query: 742 PAAGSPPSLQPVILVVPESSP-PTGPLPCPSCVDPDVQPVPVDDAP-PSPVPHNDAPPLP 799
P +PPS P V P +P P P P P P P +P PSP P P P
Sbjct: 300 PTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSP 359
Query: 800 APTSPTPPATPLVVTQAPPTSPPTTPPAVTQV 831
+P SP+P +P+ +P P +P AV V
Sbjct: 360 SP-SPSPSPSPI---PSPSPKPSPSPVAVKLV 387
Score = 45.4 bits (106), Expect = 0.001
Identities = 32/97 (32%), Positives = 40/97 (40%), Gaps = 12/97 (12%)
Query: 742 PAAGSPPS-LQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPA 800
P + SPPS + P VP S P P P P P APP+P P P P+
Sbjct: 305 PPSPSPPSPVPPSPAPVPPSPAPPSPAPSP----------PPSPAPPTPSPSPSPSPSPS 354
Query: 801 PTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVD 837
P SP+P +P P+ P P+ V L D
Sbjct: 355 P-SPSPSPSPSPSPSPIPSPSPKPSPSPVAVKLVWAD 390
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 69.7 bits (169), Expect = 5e-11
Identities = 34/77 (44%), Positives = 46/77 (59%)
Query: 1171 YLTITRPDLSYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSSPAVLGYS 1230
YLTITRPDL++AVN +SQF QAV +++ YV GL + TS + ++
Sbjct: 2 YLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAFA 61
Query: 1231 DADWARCTDHRRSTYGY 1247
D+DWA C D RRS G+
Sbjct: 62 DSDWASCPDTRRSVTGF 78
>SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor
(SSG 185)
Length = 485
Score = 62.0 bits (149), Expect = 1e-08
Identities = 35/99 (35%), Positives = 45/99 (45%), Gaps = 2/99 (2%)
Query: 730 SPNDLVVFTFYEPAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSP 789
+PN+ + +P A S P P P PP+ P P P P P PP P
Sbjct: 223 APNNSPLPPSPQPTASSRPPSPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPP 282
Query: 790 VPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAV 828
P PP P P SP+PP P + +PP PP +PP+V
Sbjct: 283 PPPPPPPPPPPPPSPSPPRKP--PSPSPPVPPPPSPPSV 319
Score = 62.0 bits (149), Expect = 1e-08
Identities = 34/89 (38%), Positives = 38/89 (42%), Gaps = 1/89 (1%)
Query: 747 PPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTP 806
PPS QP P S PP+ P P P P P PP P P PP P P P P
Sbjct: 230 PPSPQPTASSRPPSPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPPPP 289
Query: 807 PATPLVVTQAPPTSPPTTPPAVTQVPLAP 835
P P + +PP PP+ P V P P
Sbjct: 290 PPPP-PPSPSPPRKPPSPSPPVPPPPSPP 317
Score = 55.8 bits (133), Expect = 8e-07
Identities = 37/99 (37%), Positives = 43/99 (43%), Gaps = 7/99 (7%)
Query: 742 PAAGSPPSLQPVILVVPESSP-PTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPA 800
P S P++ P I P +SP P P P S P P P PPSP P + +PP P
Sbjct: 209 PTGLSGPNVNP-IGPAPNNSPLPPSPQPTASSRPPSPPPSP---RPPSPPPPSPSPPPPP 264
Query: 801 PTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDAR 839
P P PP P PP PP PP P +P R
Sbjct: 265 PPPPPPPPPP--PPSPPPPPPPPPPPPPPPPPPSPSPPR 301
Score = 55.5 bits (132), Expect = 1e-06
Identities = 32/82 (39%), Positives = 37/82 (45%), Gaps = 2/82 (2%)
Query: 763 PTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPP 822
P GP P S + P QP PPSP P + PP P P SP+PP P PP PP
Sbjct: 219 PIGPAPNNSPLPPSPQPT-ASSRPPSPPP-SPRPPSPPPPSPSPPPPPPPPPPPPPPPPP 276
Query: 823 TTPPAVTQVPLAPVDARPRTRS 844
+ PP P P P + S
Sbjct: 277 SPPPPPPPPPPPPPPPPPPSPS 298
Score = 53.9 bits (128), Expect = 3e-06
Identities = 36/98 (36%), Positives = 41/98 (41%), Gaps = 11/98 (11%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P SPP P P PP P P PS P P P PP P P + +PP P
Sbjct: 248 PRPPSPPPPSPSPPPPPPPPPPPPPPPPPS---PPPPPPPPPPPPPPPPPPSPSPP-RKP 303
Query: 802 TSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDAR 839
SP+PP PP SPP+ PA T P +R
Sbjct: 304 PSPSPPV-------PPPPSPPSVLPAATGFPFCECVSR 334
Score = 48.1 bits (113), Expect = 2e-04
Identities = 32/97 (32%), Positives = 40/97 (40%), Gaps = 2/97 (2%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPS--PVPHNDAPPLP 799
P + SPP P P PP+ P P P P P P +PP P P PP P
Sbjct: 255 PPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPSPSPPRKPPSPSPPVPPPP 314
Query: 800 APTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPV 836
+P S P AT + SP + P VT ++ V
Sbjct: 315 SPPSVLPAATGFPFCECVSRSPSSYPWRVTVANVSAV 351
>FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity
protein)
Length = 1468
Score = 61.6 bits (148), Expect = 1e-08
Identities = 40/99 (40%), Positives = 47/99 (47%), Gaps = 9/99 (9%)
Query: 742 PAAGSPPSLQPVILVVPESSPP-TGPLPCPSCVDPDVQPV----PVDDAPPSPVPHNDAP 796
PA +PP L P ++ P PP GPLP P+ P V PV P PP+PVP +D P
Sbjct: 871 PAPPTPPPLPPPLIPPPPPLPPGLGPLP-PAPPIPPVCPVSPPPPPPPPPPTPVPPSDGP 929
Query: 797 PLPAPTSPTPP---ATPLVVTQAPPTSPPTTPPAVTQVP 832
P P P P P A P PP PP PP + P
Sbjct: 930 PPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPP 968
Score = 48.1 bits (113), Expect = 2e-04
Identities = 31/88 (35%), Positives = 37/88 (41%), Gaps = 6/88 (6%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P G P P+ V P S PP P P P+ V P P P PP P+P+ A P
Sbjct: 892 PGLGPLPPAPPIPPVCPVSPPPPPPPPPPTPVPPSDGPPP-PPPPPPPLPNVLALPNSGG 950
Query: 802 TSPTPPATPLVVTQAPPTSPPTTPPAVT 829
P PP P PP P PP ++
Sbjct: 951 PPPPPPPPP-----PPPGLAPPPPPGLS 973
Score = 47.0 bits (110), Expect = 4e-04
Identities = 32/86 (37%), Positives = 37/86 (42%), Gaps = 9/86 (10%)
Query: 761 SPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLP--APTSPTPPATPLVVTQAPP 818
SPP P P P + P + P P PP P APP+P P SP PP P T PP
Sbjct: 869 SPPAPPTPPP--LPPPLIPPP-PPLPPGLGPLPPAPPIPPVCPVSPPPPPPPPPPTPVPP 925
Query: 819 TS----PPTTPPAVTQVPLAPVDARP 840
+ PP PP + V P P
Sbjct: 926 SDGPPPPPPPPPPLPNVLALPNSGGP 951
Score = 42.7 bits (99), Expect = 0.007
Identities = 23/68 (33%), Positives = 26/68 (37%), Gaps = 5/68 (7%)
Query: 773 VDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVP 832
+ P P P PP P PP P P PP P+ P SPP PP P
Sbjct: 868 ISPPAPPTPPPLPPPLIPPPPPLPPGLGPLPPAPPIPPVC-----PVSPPPPPPPPPPTP 922
Query: 833 LAPVDARP 840
+ P D P
Sbjct: 923 VPPSDGPP 930
Score = 40.8 bits (94), Expect = 0.026
Identities = 28/89 (31%), Positives = 34/89 (37%), Gaps = 6/89 (6%)
Query: 747 PPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTP 806
PP P V P PP P P P P+V +P PP P P PP P P P
Sbjct: 914 PPPPPPPTPVPPSDGPPPPPPPPPPL--PNVLALPNSGGPPPPPP--PPPPPPGLAPPPP 969
Query: 807 PATPLVVTQAPPTSP--PTTPPAVTQVPL 833
P ++ + P P P+ PL
Sbjct: 970 PGLSFGLSSSSSQYPRKPAIEPSCPMKPL 998
>FM14_MOUSE (Q05859) Formin 1 isoform IV (Limb deformity protein)
Length = 1206
Score = 61.6 bits (148), Expect = 1e-08
Identities = 40/99 (40%), Positives = 47/99 (47%), Gaps = 9/99 (9%)
Query: 742 PAAGSPPSLQPVILVVPESSPP-TGPLPCPSCVDPDVQPV----PVDDAPPSPVPHNDAP 796
PA +PP L P ++ P PP GPLP P+ P V PV P PP+PVP +D P
Sbjct: 645 PAPPTPPPLPPPLIPPPPPLPPGLGPLP-PAPPIPPVCPVSPPPPPPPPPPTPVPPSDGP 703
Query: 797 PLPAPTSPTPP---ATPLVVTQAPPTSPPTTPPAVTQVP 832
P P P P P A P PP PP PP + P
Sbjct: 704 PPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPP 742
Score = 48.1 bits (113), Expect = 2e-04
Identities = 31/88 (35%), Positives = 37/88 (41%), Gaps = 6/88 (6%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P G P P+ V P S PP P P P+ V P P P PP P+P+ A P
Sbjct: 666 PGLGPLPPAPPIPPVCPVSPPPPPPPPPPTPVPPSDGPPP-PPPPPPPLPNVLALPNSGG 724
Query: 802 TSPTPPATPLVVTQAPPTSPPTTPPAVT 829
P PP P PP P PP ++
Sbjct: 725 PPPPPPPPP-----PPPGLAPPPPPGLS 747
Score = 47.0 bits (110), Expect = 4e-04
Identities = 32/86 (37%), Positives = 37/86 (42%), Gaps = 9/86 (10%)
Query: 761 SPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLP--APTSPTPPATPLVVTQAPP 818
SPP P P P + P + P P PP P APP+P P SP PP P T PP
Sbjct: 643 SPPAPPTPPP--LPPPLIPPP-PPLPPGLGPLPPAPPIPPVCPVSPPPPPPPPPPTPVPP 699
Query: 819 TS----PPTTPPAVTQVPLAPVDARP 840
+ PP PP + V P P
Sbjct: 700 SDGPPPPPPPPPPLPNVLALPNSGGP 725
Score = 42.7 bits (99), Expect = 0.007
Identities = 23/68 (33%), Positives = 26/68 (37%), Gaps = 5/68 (7%)
Query: 773 VDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVP 832
+ P P P PP P PP P P PP P+ P SPP PP P
Sbjct: 642 ISPPAPPTPPPLPPPLIPPPPPLPPGLGPLPPAPPIPPVC-----PVSPPPPPPPPPPTP 696
Query: 833 LAPVDARP 840
+ P D P
Sbjct: 697 VPPSDGPP 704
Score = 40.8 bits (94), Expect = 0.026
Identities = 28/89 (31%), Positives = 34/89 (37%), Gaps = 6/89 (6%)
Query: 747 PPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTP 806
PP P V P PP P P P P+V +P PP P P PP P P P
Sbjct: 688 PPPPPPPTPVPPSDGPPPPPPPPPPL--PNVLALPNSGGPPPPPP--PPPPPPGLAPPPP 743
Query: 807 PATPLVVTQAPPTSP--PTTPPAVTQVPL 833
P ++ + P P P+ PL
Sbjct: 744 PGLSFGLSSSSSQYPRKPAIEPSCPMKPL 772
>Y091_NPVAC (P41479) Hypothetical 24.1 kDa protein in LEF4-P33
intergenic region
Length = 224
Score = 61.2 bits (147), Expect = 2e-08
Identities = 39/112 (34%), Positives = 50/112 (43%), Gaps = 10/112 (8%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P PP+ P+ VP +PP P P PS P P P+ PP+P P PP P P
Sbjct: 37 PPIVPPPTSPPI---VPLPTPP--PTPTPSPPSPTPPPTPI---PPTPTP--TPPPTPIP 86
Query: 802 TSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNP 853
+PTP P + P SPP +P T P P P T + + P+P
Sbjct: 87 PTPTPTPPPSPIPPTPTPSPPPSPIPPTPTPSPPPSPIPPTPTPSPPPSPSP 138
Score = 57.4 bits (137), Expect = 3e-07
Identities = 37/109 (33%), Positives = 48/109 (43%), Gaps = 18/109 (16%)
Query: 755 LVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVT 814
+V P +SPP PLP P P P +PPSP P PP P P +PTP P +
Sbjct: 39 IVPPPTSPPIVPLPTPP-------PTPTP-SPPSPTP----PPTPIPPTPTPTPPPTPIP 86
Query: 815 QAPPTSPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNPRYALVHAQPT 863
P +PP +P T P P P T + P+P + + PT
Sbjct: 87 PTPTPTPPPSPIPPTPTPSPPPSPIPPTPT------PSPPPSPIPPTPT 129
Score = 52.0 bits (123), Expect = 1e-05
Identities = 35/111 (31%), Positives = 49/111 (43%), Gaps = 19/111 (17%)
Query: 723 PLTGSKSSPNDLVVFTFYEPAAGSPPSLQPVILVVPESSPPTGP------LPCPSCVDPD 776
P+ +SP + + T SPPS P +P + PT P P P+
Sbjct: 38 PIVPPPTSPPIVPLPTPPPTPTPSPPSPTPPPTPIPPTPTPTPPPTPIPPTPTPTPPPSP 97
Query: 777 VQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPA 827
+ P P PPSP+P P PT P+PP +P+ PPT P+ PP+
Sbjct: 98 IPPTPTPSPPPSPIP-------PTPT-PSPPPSPI-----PPTPTPSPPPS 135
Score = 43.5 bits (101), Expect = 0.004
Identities = 42/140 (30%), Positives = 55/140 (39%), Gaps = 19/140 (13%)
Query: 723 PLTGSKSSPNDLVVFTFYEPAAGSPPSLQPVILVVPESSPPTGPLP---CPSCVDPDVQP 779
P T + S P+ T P P P I P +PP P+P PS + P
Sbjct: 55 PPTPTPSPPSPTPPPTPIPPTPTPTPPPTP-IPPTPTPTPPPSPIPPTPTPSPPPSPIPP 113
Query: 780 VPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDAR 839
P PPSP+P P P SP+P P+ P++ T + L P+ A
Sbjct: 114 TPTPSPPPSPIPPTPTPS--PPPSPSPLGEPMYY----PSNIDTNEALINF--LRPLCAT 165
Query: 840 PRTR-------S*NGIFKPN 852
RTR + NGIF N
Sbjct: 166 DRTRATYAVPWNCNGIFHCN 185
>EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein)
Length = 283
Score = 60.5 bits (145), Expect = 3e-08
Identities = 38/111 (34%), Positives = 47/111 (42%), Gaps = 9/111 (8%)
Query: 746 SPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPT 805
+PP+ P P PP P P P P P P +P P P + P SP
Sbjct: 70 TPPTYTPSPKPTP---PPATPKPTP----PTYTPSPKPKSPVYPPPPKASTPPTYTPSPK 122
Query: 806 PPAT--PLVVTQAPPTSPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNPR 854
PPAT P T PP + P TPP T P PV P + ++ PNP+
Sbjct: 123 PPATKPPTYPTPKPPATKPPTPPVYTPSPKPPVTKPPTPKPTPPVYTPNPK 173
Score = 57.4 bits (137), Expect = 3e-07
Identities = 41/107 (38%), Positives = 45/107 (41%), Gaps = 13/107 (12%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P A +PP+ P P + PPT P P P P PV PSP P PP P P
Sbjct: 109 PKASTPPTYTPSPKP-PATKPPTYPTPKPPATKPPTPPVYT----PSPKPPVTKPPTPKP 163
Query: 802 TSP--TPPATPLVV---TQAPPTSPPT---TPPAVTQVPLAPVDARP 840
T P TP P V T P PPT TPP T P P + P
Sbjct: 164 TPPVYTPNPKPPVTKPPTHTPSPKPPTSKPTPPVYTPSPKPPKPSPP 210
Score = 51.2 bits (121), Expect = 2e-05
Identities = 41/132 (31%), Positives = 54/132 (40%), Gaps = 13/132 (9%)
Query: 740 YEPAAGSPPSLQPVILVVPE---SSPPTGPLPCPSCVDPDVQPVPVDDAP----PSPVPH 792
Y P+ PP+ +P P+ + PPT P+ PS P +P P P+P P
Sbjct: 117 YTPSP-KPPATKPPTYPTPKPPATKPPTPPVYTPSPKPPVTKPPTPKPTPPVYTPNPKPP 175
Query: 793 NDAPP--LPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDARPRTRS*NGIFK 850
PP P+P PT TP V T + P P +PP T P P P + + K
Sbjct: 176 VTKPPTHTPSPKPPTSKPTPPVYTPS-PKPPKPSPPTYTPTPKPPATKPPTSTPTHP--K 232
Query: 851 PNPRYALVHAQP 862
P P A P
Sbjct: 233 PTPHTPYPQAHP 244
Score = 48.9 bits (115), Expect = 9e-05
Identities = 37/114 (32%), Positives = 49/114 (42%), Gaps = 12/114 (10%)
Query: 746 SPPSLQPVILVVPESS-PPT-GPLPCPSCVDPDVQPVPVDDA--PPSPVPHNDAPPLPAP 801
SP PV P++S PPT P P P P P P A PP+P + +P P
Sbjct: 97 SPKPKSPVYPPPPKASTPPTYTPSPKPPATKPPTYPTPKPPATKPPTPPVYTPSPKPPVT 156
Query: 802 TSPTPPATPLVVTQAPPTSPPTTPPAVTQVPL-APVDARPRTRS*NGIFKPNPR 854
PTP TP V T P P VT+ P P P ++ ++ P+P+
Sbjct: 157 KPPTPKPTPPVYT-------PNPKPPVTKPPTHTPSPKPPTSKPTPPVYTPSPK 203
Score = 47.8 bits (112), Expect = 2e-04
Identities = 29/93 (31%), Positives = 37/93 (39%), Gaps = 13/93 (13%)
Query: 747 PPSLQPVILVVPESSPPTGPLP---CPSCVDPDVQPVPVDDAPPSPVPHNDAP------- 796
PP+ +P V S P P P P+ P +P P P PH P
Sbjct: 188 PPTSKPTPPVYTPSPKPPKPSPPTYTPTPKPPATKPPTSTPTHPKPTPHTPYPQAHPPTY 247
Query: 797 ---PLPAPTSPTPPATPLVVTQAPPTSPPTTPP 826
P P+P +PTPP V+ P + PP PP
Sbjct: 248 KPAPKPSPPAPTPPTYTPPVSHTPSSPPPPPPP 280
Score = 45.4 bits (106), Expect = 0.001
Identities = 34/103 (33%), Positives = 38/103 (36%), Gaps = 8/103 (7%)
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P +P P P+ P P P + P P P A P PP P
Sbjct: 82 PPPATPKPTPPTYTPSPKPKSPVYPPPPKASTPPTYTPSPKPPATKPPTYPTPKPPATKP 141
Query: 802 TSPTPPA-TPLVVTQAPPTSPPT---TPPAVTQVPLAPVDARP 840
PTPP TP + P T PPT TPP T P PV P
Sbjct: 142 --PTPPVYTP--SPKPPVTKPPTPKPTPPVYTPNPKPPVTKPP 180
Score = 45.4 bits (106), Expect = 0.001
Identities = 35/109 (32%), Positives = 42/109 (38%), Gaps = 11/109 (10%)
Query: 741 EPAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPV--PHNDAP-- 796
+P PP+ P P +S PT P+ PS P P P P P P
Sbjct: 173 KPPVTKPPTHTPS--PKPPTSKPTPPVYTPSPKPPKPSPPTYTPTPKPPATKPPTSTPTH 230
Query: 797 PLPAPTSPTPPATPLVVTQAPPTSP-----PTTPPAVTQVPLAPVDARP 840
P P P +P P A P AP SP PT P V+ P +P P
Sbjct: 231 PKPTPHTPYPQAHPPTYKPAPKPSPPAPTPPTYTPPVSHTPSSPPPPPP 279
Score = 45.4 bits (106), Expect = 0.001
Identities = 38/109 (34%), Positives = 43/109 (38%), Gaps = 16/109 (14%)
Query: 741 EPAAGSPPSLQPVILV-VPESSPPTG--PLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPP 797
+P PP+ +P V P PP P PS P +P P PSP P +PP
Sbjct: 152 KPPVTKPPTPKPTPPVYTPNPKPPVTKPPTHTPSPKPPTSKPTP-PVYTPSPKPPKPSPP 210
Query: 798 LPAPTSPTPPAT-----------PLVVTQAPPTSPPTTPPAVTQVPLAP 835
PT P PPAT P T P PPT PA P AP
Sbjct: 211 TYTPT-PKPPATKPPTSTPTHPKPTPHTPYPQAHPPTYKPAPKPSPPAP 258
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.138 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 165,888,345
Number of Sequences: 164201
Number of extensions: 7556536
Number of successful extensions: 53807
Number of sequences better than 10.0: 927
Number of HSP's better than 10.0 without gapping: 292
Number of HSP's successfully gapped in prelim test: 684
Number of HSP's that attempted gapping in prelim test: 34882
Number of HSP's gapped (non-prelim): 6615
length of query: 1380
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1257
effective length of database: 39,777,331
effective search space: 50000105067
effective search space used: 50000105067
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0019a.6


