

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
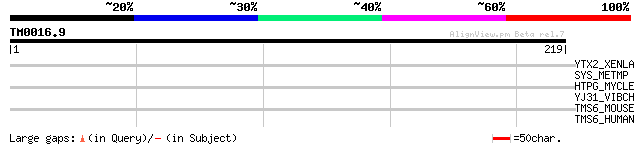
Query= TM0016.9
(219 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 39 0.009
SYS_METMP (O30520) Seryl-tRNA synthetase (EC 6.1.1.11) (Serine--... 29 7.5
HTPG_MYCLE (O33012) Chaperone protein htpG (Heat shock protein h... 29 7.5
YJ31_VIBCH (Q9KQR7) Hypothetical UPF0061 protein VC1931 29 9.8
TMS6_MOUSE (Q9DBI0) Transmembrane protease, serine 6 (EC 3.4.21.... 29 9.8
TMS6_HUMAN (Q8IU80) Transmembrane protease, serine 6 (EC 3.4.21.... 29 9.8
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 38.9 bits (89), Expect = 0.009
Identities = 39/141 (27%), Positives = 61/141 (42%), Gaps = 16/141 (11%)
Query: 1 IGGDFNAILNDGERKGLNSNTYGDVHFKHFVETYALIDLPLGND----EFTWGSTR-GDG 55
IGGDFN L+ +R + + + ++L+D+ + FT+ R G
Sbjct: 140 IGGDFNYTLDARDRNVPKKRDSSESVLRELIAHFSLVDVWREQNPETVAFTYVRVRDGHV 199
Query: 56 LWSKIDRWLVNDEAILQFDGAT----QSADHRPVSLALGSHDFGPK-PFCFFNYWLMED- 109
S+IDR ++ + + +T +DH VSL + PK + FN L+ED
Sbjct: 200 SQSRIDRIYISSHLMSRAQSSTIRLAPFSDHNCVSLRMSIAPSLPKAAYWHFNNSLLEDE 259
Query: 110 GFKKMVEEWWSSAVVEGWSNF 130
GF K V + W GW F
Sbjct: 260 GFAKSVRDTW-----RGWRAF 275
>SYS_METMP (O30520) Seryl-tRNA synthetase (EC 6.1.1.11)
(Serine--tRNA ligase) (SerRS)
Length = 514
Score = 29.3 bits (64), Expect = 7.5
Identities = 19/53 (35%), Positives = 26/53 (48%), Gaps = 8/53 (15%)
Query: 13 ERKGLNSNTYGDVHFKHFVETYALIDLPLGNDEFTWGSTRGDGLWSKIDRWLV 65
ERKG+ + T ++H HFVE + + D D W G GL RWL+
Sbjct: 436 ERKGV-AVTSANIHGTHFVEGFGIKDY---KDRKVWTGCTGYGL----SRWLI 480
>HTPG_MYCLE (O33012) Chaperone protein htpG (Heat shock protein
htpG) (High temperature protein G)
Length = 656
Score = 29.3 bits (64), Expect = 7.5
Identities = 24/120 (20%), Positives = 45/120 (37%), Gaps = 13/120 (10%)
Query: 62 RWLVNDEAILQFDGATQSADHRPVSLALGSHDFGPKPFCFFNYWLMEDGFKKMVEEWWSS 121
RW + EA + ++ V+L L DF ED EW
Sbjct: 160 RWSSDGEATYTIESVDEAPQGTSVTLHLKPEDF-------------EDELHDYTSEWKIR 206
Query: 122 AVVEGWSNFALMQKLKGLKVKIREWKENRGIRGSEKIKGLEDNLHEGECLVTKIKDSLAQ 181
+V+ +S+F ++ + + G G E++ ++ + L TK KD +++
Sbjct: 207 ELVKKYSDFIAWPIRMEVERRAPATSDGEGADGEEQVTIETQTINSMKALWTKSKDEVSE 266
>YJ31_VIBCH (Q9KQR7) Hypothetical UPF0061 protein VC1931
Length = 489
Score = 28.9 bits (63), Expect = 9.8
Identities = 12/30 (40%), Positives = 15/30 (50%)
Query: 66 NDEAILQFDGATQSADHRPVSLALGSHDFG 95
NDE +L G AD PV++ H FG
Sbjct: 52 NDELLLSLSGQQLPADFSPVAMKYAGHQFG 81
>TMS6_MOUSE (Q9DBI0) Transmembrane protease, serine 6 (EC 3.4.21.-)
(Matriptase-2)
Length = 811
Score = 28.9 bits (63), Expect = 9.8
Identities = 17/40 (42%), Positives = 19/40 (47%), Gaps = 1/40 (2%)
Query: 67 DEAILQFDG-ATQSADHRPVSLALGSHDFGPKPFCFFNYW 105
D A+LQ D SA RPV L SH F P C+ W
Sbjct: 668 DVALLQLDHPVVYSATVRPVCLPARSHFFEPGQHCWITGW 707
>TMS6_HUMAN (Q8IU80) Transmembrane protease, serine 6 (EC 3.4.21.-)
(Matriptase-2)
Length = 811
Score = 28.9 bits (63), Expect = 9.8
Identities = 24/70 (34%), Positives = 32/70 (45%), Gaps = 12/70 (17%)
Query: 48 WGSTRGDGLWS-KIDRWLVN----------DEAILQFDG-ATQSADHRPVSLALGSHDFG 95
W ++R G S K+ R L++ D A+LQ D +SA RPV L SH F
Sbjct: 638 WQNSRWPGEVSFKVSRLLLHPYHEEDSHDYDVALLQLDHPVVRSAAVRPVCLPARSHFFE 697
Query: 96 PKPFCFFNYW 105
P C+ W
Sbjct: 698 PGLHCWITGW 707
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.329 0.145 0.479
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,143,742
Number of Sequences: 164201
Number of extensions: 1125336
Number of successful extensions: 2588
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 2585
Number of HSP's gapped (non-prelim): 7
length of query: 219
length of database: 59,974,054
effective HSP length: 106
effective length of query: 113
effective length of database: 42,568,748
effective search space: 4810268524
effective search space used: 4810268524
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0016.9


