

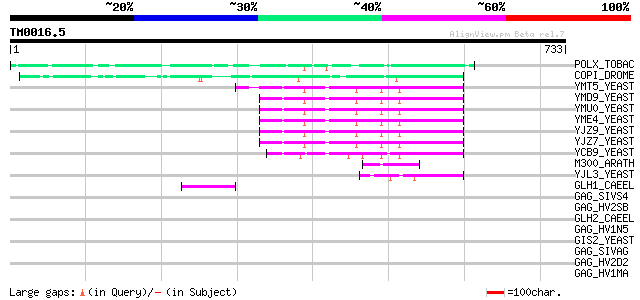
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0016.5
(733 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 137 1e-31
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 108 5e-23
YMT5_YEAST (Q04214) Transposon Ty1 protein B 83 3e-15
YMD9_YEAST (Q03434) Transposon Ty1 protein B 83 3e-15
YMU0_YEAST (Q04670) Transposon Ty1 protein B 82 5e-15
YME4_YEAST (Q04711) Transposon Ty1 protein B 82 5e-15
YJZ9_YEAST (P47100) Transposon Ty1 protein B 82 5e-15
YJZ7_YEAST (P47098) Transposon Ty1 protein B 80 1e-14
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 76 4e-13
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 48 8e-05
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 45 9e-04
GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-... 45 9e-04
GAG_SIVS4 (P12496) Gag polyprotein [Contains: Core protein p17; ... 42 0.004
GAG_HV2SB (P12450) Gag polyprotein [Contains: Core protein p16; ... 42 0.006
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 42 0.008
GAG_HV1N5 (P12493) Gag polyprotein [Contains: Core protein p17 (... 41 0.010
GIS2_YEAST (P53849) Zinc-finger protein GIS2 41 0.013
GAG_SIVAG (P27978) Gag polyprotein [Contains: Core protein p17; ... 41 0.013
GAG_HV2D2 (P15832) Gag polyprotein [Contains: Core protein p16; ... 41 0.013
GAG_HV1MA (P04594) Gag polyprotein [Contains: Core protein p17 (... 41 0.013
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 137 bits (345), Expect = 1e-31
Identities = 164/633 (25%), Positives = 251/633 (38%), Gaps = 92/633 (14%)
Query: 1 MSTEKYEVLLVRLNGKN-YSAWAFEFQIFVKGKSLWGHVDGSTSALDKEKQETEYADWEV 59
MS KYEV + NG N +S W + + + L +D + D K E DW
Sbjct: 1 MSGVKYEV--AKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAE----DWAD 54
Query: 60 KDNQVLAWIIRFVDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIAIFQQEI 119
D + + I + ++V N+ TA +W L+ +Y + L+ Q
Sbjct: 55 LDERAASAIRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKK-----QLYA 109
Query: 120 LSISEFYSQFMNLWADYTDIVYGSVSTEGLTSVQTVHETTK*DQFLMKLRSDFEGIRTNL 179
L +SE + +L V+ + T+ + E K L L S ++ + T +
Sbjct: 110 LHMSEGTNFLSHL------NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTI 163
Query: 180 MN-RATVPS*DACLNELLREERRLLTLATMEQHKSASLPVAYVVQEKPRGRDLSAVQC-- 236
++ + T+ D LL E+ R K + A + + + R S+
Sbjct: 164 LHGKTTIELKDVTSALLLNEKMR---------KKPENQGQALITEGRGRSYQRSSNNYGR 214
Query: 237 FCCKGFGHYASNCPRKSCNYCKKDGHVIKECPIRPPKKNATTFTTSVHSPIAPSFVDIAN 296
+G S ++C C + GH ++CP P K T A A
Sbjct: 215 SGARGKSKNRSKSRVRNCYNCNQPGHFKRDCP-NPRKGKGETSGQKNDDNTA------AM 267
Query: 297 VQHNAPTPVQALTPEIVEHMIILAFSALGISGKHSPNSSPWYFDYGASNHMANNAEALTN 356
VQ+N + E H+ S S W D AS+H +
Sbjct: 268 VQNNDNVVLFINEEEECMHL--------------SGPESEWVVDTAASHHATPVRDLFCR 313
Query: 357 -ITQNFGNLKIQVANGNHLPITAIGDISTSLN--------DVYVSPGLTSNLIFVGQLVD 407
+ +FG +K + N ++ I IGDI N DV P L NLI G +D
Sbjct: 314 YVAGDFGTVK--MGNTSYSKIAGIGDICIKTNVGCTLVLKDVRHVPDLRMNLI-SGIALD 370
Query: 408 NDCRVAFSKS-------GCLVQDQHSGKLIARGPKVGRLFPLCFSMSPCSSLPFVSCHSA 460
D ++ + G LV K +ARG +C +
Sbjct: 371 RDGYESYFANQKWRLTKGSLV----IAKGVARGTLYRTNAEICQGELNAAQ--------- 417
Query: 461 TVVNFELWHKRLGHPNSNVLYELLQSGVLGNKETPSLSTIKFDCTYCKLGKSKILPFPNH 520
++ +LWHKR+GH + L L + ++ + +T+K C YC GK + F
Sbjct: 418 DEISVDLWHKRMGHMSEKGLQILAKKSLISYAKG---TTVK-PCDYCLFGKQHRVSFQTS 473
Query: 521 QSNISQTFDMIHSDLWGITPVISHANYKYFVTFIDDFSRFTWVYFLRSKDEVFSAFKFFH 580
D+++SD+ G + S KYFVTFIDD SR WVY L++KD+VF F+ FH
Sbjct: 474 SERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFH 533
Query: 581 AYVETQFSSKIKILRSDNGGGKVHI*FDSRFFE 613
A VE + K+K LRSDNGG + SR FE
Sbjct: 534 ALVERETGRKLKRLRSDNGGE-----YTSREFE 561
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 108 bits (270), Expect = 5e-23
Identities = 131/615 (21%), Positives = 243/615 (39%), Gaps = 103/615 (16%)
Query: 14 NGKNYSAWAFEFQIFVKGKSLWGHVDGSTSALDKEKQETEYADWEVKDNQVLAWIIRFVD 73
+G+ Y+ W F + + + + VDG + E ++ W+ + + II ++
Sbjct: 12 DGEKYAIWKFRIRALLAEQDVLKVVDG---LMPNEVDDS----WKKAERCAKSTIIEYLS 64
Query: 74 PNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIAIFQQEILSISEFYSQFMNLW 133
+ + TA ++ L +Y + + A + L ++ +LS+ S M+L
Sbjct: 65 DSFLNFATSDITARQILENLDAVYERKSLASQLAL-------RKRLLSLK--LSSEMSLL 115
Query: 134 ADYTDIVYGSVSTEGLTSVQTVHETTK*DQFLMKLRSDFEGIRTNLMNRATVPS*DACLN 193
+ + ++ + +E L + + E K L+ L S ++GI T +
Sbjct: 116 SHFH--IFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAI-------------- 159
Query: 194 ELLREERRLLTLATMEQHKSASLPVAYVVQEKPRGRDLSAVQCFCCKGFGHYASNC---- 249
E L EE LTLA + K+ L ++ + Y +N
Sbjct: 160 ETLSEEN--LTLAFV---KNRLLDQEIKIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNR 214
Query: 250 ---PRK----------SCNYCKKDGHVIKECPIRPPKKNATTFTTSVHSPIAPSFVDIAN 296
P+K C++C ++GH+ K+C
Sbjct: 215 VTKPKKIFKGNSKYKVKCHHCGREGHIKKDC------------------------FHYKR 250
Query: 297 VQHNAPTPVQALTPEIVEHMIILAFSALGISGKHSPNSSPWYFDYGASNHMANNAEALTN 356
+ +N + H I AF ++ ++ + D GAS+H+ N+ T+
Sbjct: 251 ILNNKNKENEKQVQTATSHGI--AFMVKEVNNTSVMDNCGFVLDSGASDHLINDESLYTD 308
Query: 357 ITQNFGNLKIQVAN-GNHLPITAIG------DISTSLNDVYVSPGLTSNLIFVGQLVDND 409
+ LKI VA G + T G D +L DV NL+ V +L +
Sbjct: 309 SVEVVPPLKIAVAKQGEFIYATKRGIVRLRNDHEITLEDVLFCKEAAGNLMSVKRLQEAG 368
Query: 410 CRVAFSKSGCLVQDQHSGKLIARGPKVGRLFPLCFSMSPCSSLPFVSCHSATVVNFELWH 469
+ F KSG + +G ++ + + + P + S ++ NF LWH
Sbjct: 369 MSIEFDKSGVTISK--NGLMVVKNSGM-------LNNVPVINFQAYSINAKHKNNFRLWH 419
Query: 470 KRLGHPNSNVLYELLQSGVLGNKETPSLSTIKFDCTYCKL---GKSKILPFPN--HQSNI 524
+R GH + L E+ + + ++ L+ ++ C C+ GK LPF +++I
Sbjct: 420 ERFGHISDGKLLEIKRKNMFSDQSL--LNNLELSCEICEPCLNGKQARLPFKQLKDKTHI 477
Query: 525 SQTFDMIHSDLWGITPVISHANYKYFVTFIDDFSRFTWVYFLRSKDEVFSAFKFFHAYVE 584
+ ++HSD+ G ++ + YFV F+D F+ + Y ++ K +VFS F+ F A E
Sbjct: 478 KRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDFVAKSE 537
Query: 585 TQFSSKIKILRSDNG 599
F+ K+ L DNG
Sbjct: 538 AHFNLKVVYLYIDNG 552
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 82.8 bits (203), Expect = 3e-15
Identities = 74/325 (22%), Positives = 138/325 (41%), Gaps = 40/325 (12%)
Query: 299 HNAPTPVQALTPEIVEHMIILAFSALGISGKHSPNSSPWYF--DYGASNHMANNAEALTN 356
H T Q LT V H HS + P + D GAS + +A + +
Sbjct: 3 HTTFTLGQELTESTVNHT------------NHSDDKLPGHLLLDSGASRTLIRSAHHIHS 50
Query: 357 ITQNFGNLKIQVANGNHLPITAIGDISTSLND-------VYVSPGLTSNLIFVGQLVDND 409
+ N ++ + A ++PI AIGD+ D V +P + +L+ + +L D
Sbjct: 51 ASSN-PDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAVD 109
Query: 410 CRVAFSKSGCLVQDQHSGKLIARGPKVGRLFPLC--FSMSPCSSLPFVS----CHSATVV 463
F+K+ V ++ G ++A K G + + + + S+P ++ S
Sbjct: 110 ITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLPSNISVPTINNVHTSESTRKY 166
Query: 464 NFELWHKRLGHPNSNVLYELLQSGVL---GNKETPSLSTIKFDCTYCKLGKSK----ILP 516
+ H+ L H N+ + L++ + + S I + C C +GKS I
Sbjct: 167 PYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKG 226
Query: 517 FPNHQSNISQTFDMIHSDLWGITPVISHANYKYFVTFIDDFSRFTWVYFL--RSKDEVFS 574
N + F +H+D++G + + YF++F D+ ++F WVY L R +D +
Sbjct: 227 SRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILD 286
Query: 575 AFKFFHAYVETQFSSKIKILRSDNG 599
F A+++ QF + + +++ D G
Sbjct: 287 VFTTILAFIKNQFQASVLVIQMDRG 311
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 82.8 bits (203), Expect = 3e-15
Identities = 67/294 (22%), Positives = 131/294 (43%), Gaps = 28/294 (9%)
Query: 330 HSPNSSPWYF--DYGASNHMANNAEALTNITQNFGNLKIQVANGNHLPITAIGDISTSLN 387
HS + P + D GAS + +A + + + N ++ + A ++PI AIGD+
Sbjct: 22 HSDDELPGHLLLDSGASRTLIRSAHHIHSASSN-PDINVVDAQKRNIPINAIGDLQFHFQ 80
Query: 388 D-------VYVSPGLTSNLIFVGQLVDNDCRVAFSKSGCLVQDQHSGKLIARGPKVGRLF 440
D V +P + +L+ + +L D F+K+ V ++ G ++A K G +
Sbjct: 81 DNTKTSIKVLHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFY 137
Query: 441 PLC--FSMSPCSSLPFVS----CHSATVVNFELWHKRLGHPNSNVLYELLQSGVL---GN 491
+ + + S+P ++ S + H+ L H N+ + L++ +
Sbjct: 138 WVSKKYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNE 197
Query: 492 KETPSLSTIKFDCTYCKLGKSK----ILPFPNHQSNISQTFDMIHSDLWGITPVISHANY 547
+ S I + C C +GKS I N + F +H+D++G + +
Sbjct: 198 SDVDRSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAP 257
Query: 548 KYFVTFIDDFSRFTWVYFL--RSKDEVFSAFKFFHAYVETQFSSKIKILRSDNG 599
YF++F D+ ++F WVY L R +D + F A+++ QF + + +++ D G
Sbjct: 258 SYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRG 311
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 82.0 bits (201), Expect = 5e-15
Identities = 67/294 (22%), Positives = 131/294 (43%), Gaps = 28/294 (9%)
Query: 330 HSPNSSPWYF--DYGASNHMANNAEALTNITQNFGNLKIQVANGNHLPITAIGDISTSLN 387
HS + P + D GAS + +A + + + N ++ + A ++PI AIGD+
Sbjct: 22 HSDDELPGHLLLDSGASRTLIRSAHHIHSASSN-PDINVVDAQKRNIPINAIGDLQFHFQ 80
Query: 388 D-------VYVSPGLTSNLIFVGQLVDNDCRVAFSKSGCLVQDQHSGKLIARGPKVGRLF 440
D V +P + +L+ + +L D F+K+ V ++ G ++A K G +
Sbjct: 81 DNTKTSIKVLHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFY 137
Query: 441 PLC--FSMSPCSSLPFVS----CHSATVVNFELWHKRLGHPNSNVLYELLQSGVL---GN 491
+ + + S+P ++ S + H+ L H N+ + L++ +
Sbjct: 138 WVSKKYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNE 197
Query: 492 KETPSLSTIKFDCTYCKLGKSK----ILPFPNHQSNISQTFDMIHSDLWGITPVISHANY 547
+ S I + C C +GKS I N + F +H+D++G + +
Sbjct: 198 SDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAP 257
Query: 548 KYFVTFIDDFSRFTWVYFL--RSKDEVFSAFKFFHAYVETQFSSKIKILRSDNG 599
YF++F D+ ++F WVY L R +D + F A+++ QF + + +++ D G
Sbjct: 258 SYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRG 311
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 82.0 bits (201), Expect = 5e-15
Identities = 67/294 (22%), Positives = 131/294 (43%), Gaps = 28/294 (9%)
Query: 330 HSPNSSPWYF--DYGASNHMANNAEALTNITQNFGNLKIQVANGNHLPITAIGDISTSLN 387
HS + P + D GAS + +A + + + N ++ + A ++PI AIGD+
Sbjct: 22 HSDDELPGHLLLDSGASRTLIRSAHHIHSASSN-PDINVVDAQKRNIPINAIGDLQFHFQ 80
Query: 388 D-------VYVSPGLTSNLIFVGQLVDNDCRVAFSKSGCLVQDQHSGKLIARGPKVGRLF 440
D V +P + +L+ + +L D F+K+ V ++ G ++A K G +
Sbjct: 81 DNTKTSIKVLHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFY 137
Query: 441 PLC--FSMSPCSSLPFVS----CHSATVVNFELWHKRLGHPNSNVLYELLQSGVL---GN 491
+ + + S+P ++ S + H+ L H N+ + L++ +
Sbjct: 138 WVSKKYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNE 197
Query: 492 KETPSLSTIKFDCTYCKLGKSK----ILPFPNHQSNISQTFDMIHSDLWGITPVISHANY 547
+ S I + C C +GKS I N + F +H+D++G + +
Sbjct: 198 SDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAP 257
Query: 548 KYFVTFIDDFSRFTWVYFL--RSKDEVFSAFKFFHAYVETQFSSKIKILRSDNG 599
YF++F D+ ++F WVY L R +D + F A+++ QF + + +++ D G
Sbjct: 258 SYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRG 311
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 82.0 bits (201), Expect = 5e-15
Identities = 67/294 (22%), Positives = 131/294 (43%), Gaps = 28/294 (9%)
Query: 330 HSPNSSPWYF--DYGASNHMANNAEALTNITQNFGNLKIQVANGNHLPITAIGDISTSLN 387
HS + P + D GAS + +A + + + N ++ + A ++PI AIGD+
Sbjct: 449 HSDDELPGHLLLDSGASRTLIRSAHHIHSASSN-PDINVVDAQKRNIPINAIGDLQFHFQ 507
Query: 388 D-------VYVSPGLTSNLIFVGQLVDNDCRVAFSKSGCLVQDQHSGKLIARGPKVGRLF 440
D V +P + +L+ + +L D F+K+ V ++ G ++A K G +
Sbjct: 508 DNTKTSIKVLHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFY 564
Query: 441 PLC--FSMSPCSSLPFVS----CHSATVVNFELWHKRLGHPNSNVLYELLQSGVL---GN 491
+ + + S+P ++ S + H+ L H N+ + L++ +
Sbjct: 565 WVSKKYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNE 624
Query: 492 KETPSLSTIKFDCTYCKLGKSK----ILPFPNHQSNISQTFDMIHSDLWGITPVISHANY 547
+ S I + C C +GKS I N + F +H+D++G + +
Sbjct: 625 SDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAP 684
Query: 548 KYFVTFIDDFSRFTWVYFL--RSKDEVFSAFKFFHAYVETQFSSKIKILRSDNG 599
YF++F D+ ++F WVY L R +D + F A+++ QF + + +++ D G
Sbjct: 685 SYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRG 738
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 80.5 bits (197), Expect = 1e-14
Identities = 66/294 (22%), Positives = 131/294 (44%), Gaps = 28/294 (9%)
Query: 330 HSPNSSPWYF--DYGASNHMANNAEALTNITQNFGNLKIQVANGNHLPITAIGDISTSLN 387
HS + P + D GAS + +A + + + N ++ + A ++PI AIGD+
Sbjct: 449 HSDDELPGHLLLDSGASRTLIRSAHHIHSASSN-PDINVVDAQKRNIPINAIGDLQFHFQ 507
Query: 388 D-------VYVSPGLTSNLIFVGQLVDNDCRVAFSKSGCLVQDQHSGKLIARGPKVGRLF 440
D V +P + +L+ + +L D F+K+ V ++ G ++A + G +
Sbjct: 508 DNTKTSIKVLHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVQYGDFY 564
Query: 441 PLC--FSMSPCSSLPFVS----CHSATVVNFELWHKRLGHPNSNVLYELLQSGVL---GN 491
+ + + S+P ++ S + H+ L H N+ + L++ +
Sbjct: 565 WVSKRYLLPSNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNE 624
Query: 492 KETPSLSTIKFDCTYCKLGKSK----ILPFPNHQSNISQTFDMIHSDLWGITPVISHANY 547
+ S I + C C +GKS I N + F +H+D++G + +
Sbjct: 625 SDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAP 684
Query: 548 KYFVTFIDDFSRFTWVYFL--RSKDEVFSAFKFFHAYVETQFSSKIKILRSDNG 599
YF++F D+ ++F WVY L R +D + F A+++ QF + + +++ D G
Sbjct: 685 SYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRG 738
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 75.9 bits (185), Expect = 4e-13
Identities = 67/285 (23%), Positives = 132/285 (45%), Gaps = 32/285 (11%)
Query: 340 DYGASNHMANNAEALTNITQNFGNLKIQVANGNHLPITAIGDI--------STSLNDVYV 391
D GAS + +A L + T N + I A +PI AIG++ TS+ ++
Sbjct: 457 DSGASQTLVRSAHYLHHATPN-SEINIVDAQKQDIPINAIGNLHFNFQNGTKTSIKALH- 514
Query: 392 SPGLTSNLIFVGQLVDNDCRVAFSKSGCLVQDQHSGKLIARGPKVGRLFPLCFSM---SP 448
+P + +L+ + +L + + F+++ ++ G ++A K G + L S
Sbjct: 515 TPNIAYDLLSLSELANQNITACFTRN---TLERSDGTVLAPIVKHGDFYWLSKKYLIPSH 571
Query: 449 CSSLPFVSCHSATVVN---FELWHKRLGHPNSNVLYELLQSGVL-----GNKETPSLSTI 500
S L + + + VN + L H+ LGH N + + L+ + + E + ST
Sbjct: 572 ISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNAST- 630
Query: 501 KFDCTYCKLGKSK----ILPFPNHQSNISQTFDMIHSDLWGITPVISHANYKYFVTFIDD 556
+ C C +GKS + + F +H+D++G + + YF++F D+
Sbjct: 631 -YQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDE 689
Query: 557 FSRFTWVYFL--RSKDEVFSAFKFFHAYVETQFSSKIKILRSDNG 599
+RF WVY L R ++ + + F A+++ QF++++ +++ D G
Sbjct: 690 KTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRG 734
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 48.1 bits (113), Expect = 8e-05
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query: 467 LWHKRLGHPNSNVLYELLQSGVLGNKETPSLSTIKFDCTYCKLGKSKILPFPNHQSNISQ 526
LWH RL H + + L++ G L ++ +S++KF C C GK+ + F Q
Sbjct: 71 LWHSRLAHMSQRGMELLVKKGFL---DSSKVSSLKF-CEDCIYGKTHRVNFSTGQHTTKN 126
Query: 527 TFDMIHSDLWGITPV 541
D +HSDLWG V
Sbjct: 127 PLDYVHSDLWGAPSV 141
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 44.7 bits (104), Expect = 9e-04
Identities = 36/150 (24%), Positives = 65/150 (43%), Gaps = 20/150 (13%)
Query: 463 VNFELWHKRLGHPNSNVLYELLQSGVLGNKETPSLSTIK----FDCTYCKLGKSKILPFP 518
+ E HKR+GH + +++ + N SL IK F C CK+ K+
Sbjct: 556 ITLEDAHKRMGHTG----IQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKR--- 608
Query: 519 NHQSNISQTFDMIHS-------DLWGITPVISHANYKYFVTFIDDFSRF--TWVYFLRSK 569
NH + H D++G + +Y + +D+ +R+ T +F ++
Sbjct: 609 NHYTGSMNNHSTDHEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNA 668
Query: 570 DEVFSAFKFFHAYVETQFSSKIKILRSDNG 599
+ + + + YVETQF K++ + SD G
Sbjct: 669 ETILAQVRKNIQYVETQFDRKVREINSDRG 698
>GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-)
(Germline helicase-1)
Length = 763
Score = 44.7 bits (104), Expect = 9e-04
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 4/75 (5%)
Query: 228 GRDLSAVQCFCCKGFGHYASNCPR--KSCNYCKKDGHVIKEC--PIRPPKKNATTFTTSV 283
G + ++CF CKG GH ++ CP + C C + GH EC P +P + +
Sbjct: 236 GEERGPMKCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAKPREGVEGEGPKAT 295
Query: 284 HSPIAPSFVDIANVQ 298
+ P+ + D+ N+Q
Sbjct: 296 YVPVEDNMEDVFNMQ 310
Score = 34.3 bits (77), Expect = 1.2
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 8/50 (16%)
Query: 236 CFCCKGFGHYASNCPR-------KSCNYCKKDGHVIKEC-PIRPPKKNAT 277
CF C+ GH +S+CP + C C++ GH +EC R P++ T
Sbjct: 160 CFNCQQPGHRSSDCPEPRKEREPRVCYNCQQPGHTSRECTEERKPREGRT 209
>GAG_SIVS4 (P12496) Gag polyprotein [Contains: Core protein p17;
Core protein p24]
Length = 507
Score = 42.4 bits (98), Expect = 0.004
Identities = 26/76 (34%), Positives = 37/76 (48%), Gaps = 7/76 (9%)
Query: 198 EERRLLTLATMEQHKSASLPVAYVVQEKPRGRDLSAVQCFCCKGFGHYASNCP---RKSC 254
++ RL+ A E + LP A V Q+ R ++C+ C GH A C R+ C
Sbjct: 360 QKARLMAEALKEALRPDQLPFAAVQQKGQR----KTIKCWNCGKEGHSAKQCRAPRRQGC 415
Query: 255 NYCKKDGHVIKECPIR 270
C K GHV+ +CP R
Sbjct: 416 WKCGKTGHVMAKCPER 431
>GAG_HV2SB (P12450) Gag polyprotein [Contains: Core protein p16;
Core protein p26]
Length = 520
Score = 42.0 bits (97), Expect = 0.006
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 10/76 (13%)
Query: 198 EERRLLTLATMEQHKSASLPVAYVVQEKPRGRDLSAVQCFCCKGFGHYASNCP---RKSC 254
++ RL+ A E + A +P A Q++ A++C+ C GH A C R+ C
Sbjct: 359 QKARLMAEALKEAMRPAPIPFAAAQQKR-------AIKCWNCGKEGHSARQCRAPRRQGC 411
Query: 255 NYCKKDGHVIKECPIR 270
C K GH++ CP R
Sbjct: 412 WKCGKSGHIMANCPDR 427
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 41.6 bits (96), Expect = 0.008
Identities = 20/69 (28%), Positives = 35/69 (49%), Gaps = 4/69 (5%)
Query: 234 VQCFCCKGFGHYASNCPR--KSCNYCKKDGHVIKEC--PIRPPKKNATTFTTSVHSPIAP 289
++CF CKG GH ++ CP + C C + GH EC P +P + + + P+
Sbjct: 453 MKCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAKPREGAEGEGPKATYVPVED 512
Query: 290 SFVDIANVQ 298
+ ++ N+Q
Sbjct: 513 NMEEVFNMQ 521
Score = 35.8 bits (81), Expect = 0.41
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 8/54 (14%)
Query: 236 CFCCKGFGHYASNCPR-------KSCNYCKKDGHVIKECP-IRPPKKNATTFTT 281
CF C+ GH +++CP + C C++ GH ++CP R P++ FT+
Sbjct: 373 CFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPREGRNGFTS 426
Score = 35.4 bits (80), Expect = 0.54
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 8/53 (15%)
Query: 236 CFCCKGFGHYASNCPR-------KSCNYCKKDGHVIKECP-IRPPKKNATTFT 280
CF C+ GH +++CP + C C++ GH ++CP R P++ FT
Sbjct: 259 CFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPREGRNGFT 311
>GAG_HV1N5 (P12493) Gag polyprotein [Contains: Core protein p17
(Matrix protein); Core protein p24 (Core antigen); Core
protein p2; Core protein p7 (Nucleocapsid protein); Core
protein p1; Core protein p6]
Length = 499
Score = 41.2 bits (95), Expect = 0.010
Identities = 22/57 (38%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Query: 217 PVAYVVQEKPRGRDLSAVQCFCCKGFGHYASNC--PR-KSCNYCKKDGHVIKECPIR 270
P ++Q+ V+CF C GH A NC PR K C C K+GH +K+C R
Sbjct: 372 PATIMIQKGNFRNQRKTVKCFNCGKEGHIAKNCRAPRKKGCWKCGKEGHQMKDCTER 428
>GIS2_YEAST (P53849) Zinc-finger protein GIS2
Length = 153
Score = 40.8 bits (94), Expect = 0.013
Identities = 18/48 (37%), Positives = 25/48 (51%), Gaps = 2/48 (4%)
Query: 229 RDLSAVQCFCCKGFGHYASNCPRKSCNYCKKDGHVIKECPIRPPKKNA 276
R + QC+ C GH S C + C C + GH+ +ECP PKK +
Sbjct: 42 RTVEFKQCYNCGETGHVRSECTVQRCFNCNQTGHISRECP--EPKKTS 87
Score = 37.0 bits (84), Expect = 0.19
Identities = 13/39 (33%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 231 LSAVQCFCCKGFGHYASNCPR-KSCNYCKKDGHVIKECP 268
+S ++C+ C GH + +C + C C + GH+ K+CP
Sbjct: 113 ISGLKCYTCGQAGHMSRDCQNDRLCYNCNETGHISKDCP 151
Score = 33.5 bits (75), Expect = 2.1
Identities = 14/38 (36%), Positives = 19/38 (49%), Gaps = 1/38 (2%)
Query: 231 LSAVQCFCCKGFGHYASNC-PRKSCNYCKKDGHVIKEC 267
+S C+ C GH A +C + C C K GHV +C
Sbjct: 1 MSQKACYVCGKIGHLAEDCDSERLCYNCNKPGHVQTDC 38
>GAG_SIVAG (P27978) Gag polyprotein [Contains: Core protein p17;
Core protein p24; Core protein p15]
Length = 521
Score = 40.8 bits (94), Expect = 0.013
Identities = 18/44 (40%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Query: 227 RGRDLSAVQCFCCKGFGHYASNCP---RKSCNYCKKDGHVIKEC 267
RGR V+C+ C FGH CP + C C K GH+ K+C
Sbjct: 395 RGRPRPPVKCYNCGKFGHMQRQCPEPRKMRCLKCGKPGHLAKDC 438
>GAG_HV2D2 (P15832) Gag polyprotein [Contains: Core protein p16;
Core protein p26]
Length = 521
Score = 40.8 bits (94), Expect = 0.013
Identities = 27/76 (35%), Positives = 37/76 (48%), Gaps = 6/76 (7%)
Query: 198 EERRLLTLATMEQHKSASLPVAYVVQEKPRGRDLSAVQCFCCKGFGHYASNCP---RKSC 254
++ RL+ A E A +P A VQ+K R V C+ C GH A C R+ C
Sbjct: 355 QKARLMAEALKEALTPAPIPFA-AVQQKAGKR--GTVTCWNCGKQGHTARQCRAPRRQGC 411
Query: 255 NYCKKDGHVIKECPIR 270
C K GH++ +CP R
Sbjct: 412 WKCGKTGHIMSKCPER 427
>GAG_HV1MA (P04594) Gag polyprotein [Contains: Core protein p17
(Matrix protein); Core protein p24 (Core antigen); Core
protein p2; Core protein p7 (Nucleocapsid protein); Core
protein p1; Core protein p6]
Length = 504
Score = 40.8 bits (94), Expect = 0.013
Identities = 22/65 (33%), Positives = 33/65 (49%), Gaps = 3/65 (4%)
Query: 209 EQHKSASLPVAYVVQEKPRGRDLSAVQCFCCKGFGHYASNC--PR-KSCNYCKKDGHVIK 265
E A+ A ++ ++ + ++CF C GH A NC PR K C C K+GH +K
Sbjct: 370 EAMSQATNSTAAIMMQRGNFKGQKRIKCFNCGKEGHLARNCRAPRKKGCWKCGKEGHQMK 429
Query: 266 ECPIR 270
+C R
Sbjct: 430 DCTER 434
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.331 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 83,653,339
Number of Sequences: 164201
Number of extensions: 3471060
Number of successful extensions: 9420
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 71
Number of HSP's that attempted gapping in prelim test: 9155
Number of HSP's gapped (non-prelim): 202
length of query: 733
length of database: 59,974,054
effective HSP length: 118
effective length of query: 615
effective length of database: 40,598,336
effective search space: 24967976640
effective search space used: 24967976640
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0016.5


