

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0016.18
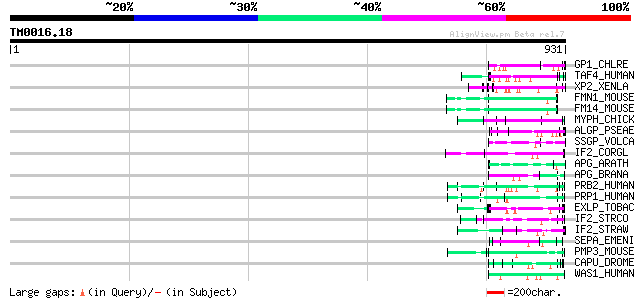
(931 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 76 5e-13
TAF4_HUMAN (O00268) Transcription initiation factor TFIID subuni... 72 7e-12
XP2_XENLA (P17437) Skin secretory protein xP2 precursor (APEG pr... 65 8e-10
FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity p... 65 1e-09
FM14_MOUSE (Q05859) Formin 1 isoform IV (Limb deformity protein) 65 1e-09
MYPH_CHICK (Q05623) Myosin-binding protein H (MyBP-H) (H-protein... 64 1e-09
ALGP_PSEAE (P15276) Transcriptional regulatory protein algP (Alg... 64 1e-09
SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor ... 64 2e-09
IF2_CORGL (Q8NP40) Translation initiation factor IF-2 64 2e-09
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 63 3e-09
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 62 7e-09
PRB2_HUMAN (P02812) Basic salivary proline-rich protein 2 (Saliv... 62 9e-09
PRP1_HUMAN (P04280) Basic salivary proline-rich protein 1 precur... 61 1e-08
EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precur... 61 1e-08
IF2_STRCO (Q8CJQ8) Translation initiation factor IF-2 61 2e-08
IF2_STRAW (Q82K53) Translation initiation factor IF-2 61 2e-08
SEPA_EMENI (P78621) Cytokinesis protein sepA (FH1/2 protein) (Fo... 60 4e-08
PMP3_MOUSE (P05143) Proline-rich protein MP-3 (Fragment) 59 5e-08
CAPU_DROME (Q24120) Cappuccino protein 59 5e-08
WAS1_HUMAN (Q92558) Wiskott-Aldrich syndrome protein family memb... 59 8e-08
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 75.9 bits (185), Expect = 5e-13
Identities = 45/132 (34%), Positives = 54/132 (40%), Gaps = 13/132 (9%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQ----APQPPPSPAAAARARHQAPPPPPASPAAAARAR 859
P PPA P P PA + A AP PPPSP PPPPP P A
Sbjct: 236 PSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPP---------PPPPPRPPFPANTPM 286
Query: 860 HQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA 919
+P PP SPA +P PP P P + A P + + S P PA P+
Sbjct: 287 PPSPPSPPPSPAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPS 346
Query: 920 AQPTKKPATKPS 931
P+ P+ PS
Sbjct: 347 PSPSPSPSPSPS 358
Score = 71.2 bits (173), Expect = 1e-11
Identities = 45/130 (34%), Positives = 56/130 (42%), Gaps = 11/130 (8%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
PLPP+ P+PS P + + +P PPSPA + PP P SPA + A P
Sbjct: 145 PLPPSPAPPSPSPP----VPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPP 200
Query: 864 QPPPASPAAAARARHQAPQPP---PPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAA 920
P P SPA P PP PPSP + A P+ + P P+ P PA
Sbjct: 201 SPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPP----SPSPPAPPSPVPPSPAPPSPAP 256
Query: 921 QPTKKPATKP 930
K PA P
Sbjct: 257 PSPKPPAPPP 266
Score = 70.1 bits (170), Expect = 3e-11
Identities = 44/131 (33%), Positives = 57/131 (42%), Gaps = 11/131 (8%)
Query: 804 PLPPAKKQPTPSQPAAAAIAT---NQAPQPPPSPAAAARARHQAPPPPPASPAAAARARH 860
P PP+ P+P+ P+ A + + AP PPSPA + A PP PA P+ A
Sbjct: 48 PAPPSPAPPSPAPPSPAPPSPGPPSPAPPSPPSPAPPSPAPPSPAPPSPAPPSPA----- 102
Query: 861 QAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAA 920
P P P SPA + A P P PPSP+ A P + + P PA P
Sbjct: 103 -PPSPAPPSPAPPSPAPPSPPSPAPPSPSPPAPP--SPSPPSPAPPLPPSPAPPSPSPPV 159
Query: 921 QPTKKPATKPS 931
P+ P PS
Sbjct: 160 PPSPSPPVPPS 170
Score = 68.9 bits (167), Expect = 6e-11
Identities = 45/129 (34%), Positives = 55/129 (41%), Gaps = 8/129 (6%)
Query: 804 PLPPAKKQPTPSQPAAAAIAT-NQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQA 862
P PP+ P+PS P + A + AP PPSPA + A P P P SP + A +
Sbjct: 171 PAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPS 230
Query: 863 PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
P PP SP A P P PPSPA + A +P A P P P
Sbjct: 231 PAPPSPSPPAP-------PSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPAN 283
Query: 923 TKKPATKPS 931
T P + PS
Sbjct: 284 TPMPPSPPS 292
Score = 68.2 bits (165), Expect = 1e-10
Identities = 42/130 (32%), Positives = 53/130 (40%), Gaps = 2/130 (1%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP+ P+P PA + P PP P A +PP PP SPA +P
Sbjct: 249 PAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAPPTPPTPPSP 308
Query: 864 QPP-PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPS-KPAAQ 921
PP P P+ A AP P PSP + + S S P+ PS P+
Sbjct: 309 SPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSPSPSPSPSPS 368
Query: 922 PTKKPATKPS 931
P P+ KPS
Sbjct: 369 PIPSPSPKPS 378
Score = 68.2 bits (165), Expect = 1e-10
Identities = 45/136 (33%), Positives = 54/136 (39%), Gaps = 19/136 (13%)
Query: 803 CPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQA 862
CP PA P P PA + A P P P + A +P PP +P + A
Sbjct: 39 CPPSPAPPSPAPPSPAPPSPAPPSPAPPSPGPPSPAPPSPPSPAPPSPAPPSPAPPSPAP 98
Query: 863 PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA--- 919
P P P SPA + A P P PPSP + PA S S P P+ P PA
Sbjct: 99 PSPAPPSPAPPSPA---PPSPAPPSPPS---------PAPPSPSPPAPPSPSPPSPAPPL 146
Query: 920 ----AQPTKKPATKPS 931
A P+ P PS
Sbjct: 147 PPSPAPPSPSPPVPPS 162
Score = 66.6 bits (161), Expect = 3e-10
Identities = 42/137 (30%), Positives = 56/137 (40%), Gaps = 9/137 (6%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQP----PPSPAAAARARHQAPPPPPASPAAAARAR 859
P PA PTP P+ + P P PPSPA + A P P P SP + A
Sbjct: 168 PPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPS 227
Query: 860 HQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP-----AVQ 914
+P PP SP A +P PP P+P + P+ P+ P +
Sbjct: 228 PPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPANTPMP 287
Query: 915 PSKPAAQPTKKPATKPS 931
PS P+ P+ P T P+
Sbjct: 288 PSPPSPPPSPAPPTPPT 304
Score = 66.2 bits (160), Expect = 4e-10
Identities = 41/128 (32%), Positives = 50/128 (39%), Gaps = 12/128 (9%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP+ P P PA + A P P+P + A P P P SP + A P
Sbjct: 73 PAPPSPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPPSPAPPSPSPP 132
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPT 923
PP SP + A +P PP PSP P S S P P+ P P P+
Sbjct: 133 APPSPSPPSPAPPLPPSPAPPSPSP-----------PVPPSPSPPVPPSPAPPSP-TPPS 180
Query: 924 KKPATKPS 931
P PS
Sbjct: 181 PSPPVPPS 188
Score = 65.9 bits (159), Expect = 5e-10
Identities = 43/132 (32%), Positives = 53/132 (39%), Gaps = 8/132 (6%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQP-PPSPAAAARARHQAPPPPPASPAAAARARHQA 862
P PP+ P P P+ A + P P PP P + A P PP SP +
Sbjct: 116 PAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPS 175
Query: 863 PQPPPASPAAAARARHQAPQPP-PPSPAAAARARHEGEPAARSASQPKKPAVQPSKP--A 919
P PP SP +P PP PPSPA + A P S + P P+ P P
Sbjct: 176 PTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPA----PPVPPSPAPPSPPSPAPPSPPSP 231
Query: 920 AQPTKKPATKPS 931
A P+ P PS
Sbjct: 232 APPSPSPPAPPS 243
Score = 65.5 bits (158), Expect = 6e-10
Identities = 45/141 (31%), Positives = 56/141 (38%), Gaps = 22/141 (15%)
Query: 804 PLPPAKKQPTPSQPAAA--------------AIATNQAPQPPPSPAAAARARHQAPPPPP 849
P PPA P+PS P+ A + + +P PPSPA + PP P
Sbjct: 129 PSPPAP--PSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVP 186
Query: 850 ASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPK 909
SPA + A P P P SPA P PP P+P + PA S S P
Sbjct: 187 PSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPP------SPAPPSPSPPA 240
Query: 910 KPAVQPSKPAAQPTKKPATKP 930
P+ P PA P+ KP
Sbjct: 241 PPSPVPPSPAPPSPAPPSPKP 261
Score = 65.1 bits (157), Expect = 8e-10
Identities = 46/129 (35%), Positives = 54/129 (41%), Gaps = 4/129 (3%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQP-PPSPAAAARARHQAPPPPPASPAAAARARHQA 862
P PA P P P + A P P PPSPA + A PP PA P+ A +
Sbjct: 55 PPSPAPPSPAPPSPGPPSPAPPSPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPS-PAP 113
Query: 863 PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
P P P SP + A P PP PSP + A PA S S P P+ P P + P
Sbjct: 114 PSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPL-PPSPAPPSPSPPVPPSPSPPVPPS-P 171
Query: 923 TKKPATKPS 931
T PS
Sbjct: 172 APPSPTPPS 180
Score = 65.1 bits (157), Expect = 8e-10
Identities = 46/130 (35%), Positives = 53/130 (40%), Gaps = 13/130 (10%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQP-PPSPAAAARARHQAPPPPPASPAAAARARHQA 862
P PP+ P P PA + A P P PPSP + A +P PP SP A +
Sbjct: 189 PAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPS 248
Query: 863 PQPP-PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQ 921
P PP PA P+ A P PPPP P P + P P P P A
Sbjct: 249 PAPPSPAPPSPKPPAPPPPPSPPPPPPPR--------PPFPANTPMPPSPPSPPPSP-AP 299
Query: 922 PTKKPATKPS 931
PT P T PS
Sbjct: 300 PT--PPTPPS 307
Score = 64.7 bits (156), Expect = 1e-09
Identities = 43/128 (33%), Positives = 56/128 (43%), Gaps = 11/128 (8%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPP--PSPAAAARARHQAPPPPPASPAAAARARHQ 861
P+PP+ P P PA + T +P PP PSPA + A PP P SPA + A
Sbjct: 158 PVPPSPSPPVPPSPAPPS-PTPPSPSPPVPPSPAPPSPA-----PPVPPSPAPPSPAPPV 211
Query: 862 APQPPPASPAAAARARHQAPQPP---PPSPAAAARARHEGEPAARSASQPKKPAVQPSKP 918
P P P SP + A +P PP PP+P + A + +P P PS P
Sbjct: 212 PPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPP 271
Query: 919 AAQPTKKP 926
P + P
Sbjct: 272 PPPPPRPP 279
Score = 64.7 bits (156), Expect = 1e-09
Identities = 41/130 (31%), Positives = 58/130 (44%), Gaps = 5/130 (3%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP+ P+P+ P+ A + AP PPSPA + + P P P SPA P
Sbjct: 96 PAPPSPAPPSPAPPSPA--PPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPP 153
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA--AQ 921
P P P + + +P PP P+P + + PA S + P P+ P PA
Sbjct: 154 SPSPPVPPSPSPPVPPSPAPPSPTPPSPS-PPVPPSPAPPSPAPPVPPSPAPPSPAPPVP 212
Query: 922 PTKKPATKPS 931
P+ P + PS
Sbjct: 213 PSPAPPSPPS 222
Score = 63.9 bits (154), Expect = 2e-09
Identities = 42/132 (31%), Positives = 58/132 (43%), Gaps = 9/132 (6%)
Query: 804 PLPPAKKQPTPSQPAAAAIA-TNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQA 862
P PP+ P+P+ P+ + A + +P PPSP+ + A P P P SP+
Sbjct: 106 PAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSP 165
Query: 863 PQPP---PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPA-VQPSKP 918
P PP P SP + + P P PPSPA PA S + P P+ PS P
Sbjct: 166 PVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPV----PPSPAPPSPAPPVPPSPAPPSPP 221
Query: 919 AAQPTKKPATKP 930
+ P P+ P
Sbjct: 222 SPAPPSPPSPAP 233
Score = 62.8 bits (151), Expect = 4e-09
Identities = 45/138 (32%), Positives = 52/138 (37%), Gaps = 11/138 (7%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQP---PPSPAAAARARHQAPPPPPASPAAAARARH 860
P PA P P PA + A P PPSPA + A P P P SP+ A
Sbjct: 78 PPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSP 137
Query: 861 QAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA- 919
P P P P + A P PP PSP + P S S P P+ P PA
Sbjct: 138 SPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPP-SPAPPSPTPPSPSPPVPPSPAPPSPAP 196
Query: 920 ------AQPTKKPATKPS 931
A P+ P PS
Sbjct: 197 PVPPSPAPPSPAPPVPPS 214
Score = 55.1 bits (131), Expect = 9e-07
Identities = 40/131 (30%), Positives = 53/131 (39%), Gaps = 24/131 (18%)
Query: 804 PLPPAKKQP----TPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARAR 859
P PP + P TP P+ + + AP PP+P P P P SP
Sbjct: 271 PPPPPPRPPFPANTPMPPSPPSPPPSPAPPTPPTP----------PSPSPPSPVP----- 315
Query: 860 HQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA 919
+P P P SPA + A P P PP+P+ + P S S P+ PS P
Sbjct: 316 -PSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSP---SPSPSPSPSPSPS-PI 370
Query: 920 AQPTKKPATKP 930
P+ KP+ P
Sbjct: 371 PSPSPKPSPSP 381
Score = 45.8 bits (107), Expect = 5e-04
Identities = 28/89 (31%), Positives = 44/89 (48%), Gaps = 7/89 (7%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPP-PSPAAAARARHQAPPPPPASPAAAARARHQA 862
P PP P+P P + + AP PP P+P + A + +P PP SP+ + + +
Sbjct: 300 PTPPTPPSPSPPSP----VPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSP-SPSPS 354
Query: 863 PQPPPASPAAAARARHQAPQP-PPPSPAA 890
P P P+ + + + +P P P PSP A
Sbjct: 355 PSPSPSPSPSPSPSPIPSPSPKPSPSPVA 383
>TAF4_HUMAN (O00268) Transcription initiation factor TFIID subunit 4
(Transcription initiation factor TFIID 135 kDa subunit)
(TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130)
(TAFII130)
Length = 1083
Score = 72.0 bits (175), Expect = 7e-12
Identities = 47/119 (39%), Positives = 56/119 (46%), Gaps = 6/119 (5%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P P TP AAA AP PP +PA AA A APPPPP +PA AR
Sbjct: 229 PAAPGTVIQTPPFVGAAAPPAPAAPSPPAAPAPAAPAA--APPPPPPAPATLARPPGHPA 286
Query: 864 QPPPASPA----AAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKP 918
PP A+PA AAA+ A P P+PAA A G+P +A+ P V+ P
Sbjct: 287 GPPTAAPAVPPPAAAQNGGSAGAAPAPAPAAGGPAGVSGQPGPGAAAAAPAPGVKAESP 345
Score = 56.6 bits (135), Expect = 3e-07
Identities = 42/116 (36%), Positives = 51/116 (43%), Gaps = 8/116 (6%)
Query: 807 PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPP 866
PA P P A + Q PP AAA A P PPA+PA AA A AP PP
Sbjct: 220 PAALLPLPKPAAPGTVI-----QTPPFVGAAAPPA-PAAPSPPAAPAPAAPAA--APPPP 271
Query: 867 PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
P +PA AR PP +PA A + +A +A P A P+ + QP
Sbjct: 272 PPAPATLARPPGHPAGPPTAAPAVPPPAAAQNGGSAGAAPAPAPAAGGPAGVSGQP 327
Score = 54.7 bits (130), Expect = 1e-06
Identities = 65/230 (28%), Positives = 77/230 (33%), Gaps = 70/230 (30%)
Query: 758 GPGRPKKLRRREPDEPDKKNPTKLK-RGGSSYTCGRCHQKGHNQRKCPLPPAKK-----Q 811
GP RP R P P P K R + G C P+P A +
Sbjct: 101 GPQRPGPPSPRRPLVPAGPAPPAAKLRPPPEGSAGAC---------APVPAAAAVAAGPE 151
Query: 812 PTPSQPAA----AAIATNQAPQPPPSP---------------------AAAARARHQAPP 846
P P+ PA AA+A P P P P AA + H A P
Sbjct: 152 PAPAGPAKPAGPAALAARAGPGPGPGPGPGPGPGKPAGPGAAQTLNGSAALLNSHHAAAP 211
Query: 847 ---------------PPPASPA----------AAARARHQAPQPPPASPAAAARARHQAP 881
P PA+P AAA AP PP A+PA AA A
Sbjct: 212 AVSLVNNGPAALLPLPKPAAPGTVIQTPPFVGAAAPPAPAAPSPP-AAPAPAA----PAA 266
Query: 882 QPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKPS 931
PPPP PA A AR G PA + P P ++ PA P+
Sbjct: 267 APPPPPPAPATLARPPGHPAGPPTAAPAVPPPAAAQNGGSAGAAPAPAPA 316
Score = 47.0 bits (110), Expect = 2e-04
Identities = 38/131 (29%), Positives = 49/131 (37%), Gaps = 8/131 (6%)
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQPP------PSPAAAARARHQAPPPPPASPAAAARAR 859
P A P P PA A PP P PAAA P PA A
Sbjct: 264 PAAAPPPPPPAPATLARPPGHPAGPPTAAPAVPPPAAAQNGGSAGAAPAPAPAAGGPAGV 323
Query: 860 HQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPS--K 917
P P A+ A A + ++P+ + AA+ PA+ +AS P +Q +
Sbjct: 324 SGQPGPGAAAAAPAPGVKAESPKRVVQAAPPAAQTLAASGPASTAASMVIGPTMQGALPS 383
Query: 918 PAAQPTKKPAT 928
PAA P P T
Sbjct: 384 PAAVPPPAPGT 394
Score = 43.1 bits (100), Expect = 0.003
Identities = 46/139 (33%), Positives = 54/139 (38%), Gaps = 24/139 (17%)
Query: 813 TPSQPAAAA-------IATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ- 864
TP AAAA ++ + A PAA A A P PP PA AR PQ
Sbjct: 47 TPEVRAAAAGALGNHVVSGSPAGAAGAGPAAPAEGAPGAAPEPP--PAGRARPGGGGPQR 104
Query: 865 PPPASPAAAARARHQAP-----QPPP--------PSPAAAARARHEGEPAARSASQPKKP 911
P P SP AP +PPP P PAAAA A EPA ++P P
Sbjct: 105 PGPPSPRRPLVPAGPAPPAAKLRPPPEGSAGACAPVPAAAAVAAGP-EPAPAGPAKPAGP 163
Query: 912 AVQPSKPAAQPTKKPATKP 930
A ++ P P P
Sbjct: 164 AALAARAGPGPGPGPGPGP 182
Score = 42.0 bits (97), Expect = 0.008
Identities = 40/120 (33%), Positives = 44/120 (36%), Gaps = 20/120 (16%)
Query: 814 PSQPAAAAIATNQAPQPPPSPAAAARAR-----HQAPPPP----PASPAAAARARHQAPQ 864
P+ PA A AP+PPP A RAR Q P PP P PA A +
Sbjct: 75 PAAPAEGA--PGAAPEPPP----AGRARPGGGGPQRPGPPSPRRPLVPAGPAPPAAKLRP 128
Query: 865 PPPAS-----PAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA 919
PP S P AA A P+P P PA A A P P KPA
Sbjct: 129 PPEGSAGACAPVPAAAAVAAGPEPAPAGPAKPAGPAALAARAGPGPGPGPGPGPGPGKPA 188
Score = 40.4 bits (93), Expect = 0.022
Identities = 36/112 (32%), Positives = 46/112 (40%), Gaps = 14/112 (12%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPA---SPAAAARARH 860
P P + +P P + + P P PA A A+ + PP A +P AA A
Sbjct: 89 PPPAGRARPGGGGPQRPGPPSPRRPLVPAGPAPPA-AKLRPPPEGSAGACAPVPAAAAVA 147
Query: 861 QAPQPPPASPA----AAARARHQAPQP-PPPSPAAAARARHEGEPAARSASQ 907
P+P PA PA AA A P P P P P G+PA A+Q
Sbjct: 148 AGPEPAPAGPAKPAGPAALAARAGPGPGPGPGPGPG-----PGKPAGPGAAQ 194
Score = 38.9 bits (89), Expect = 0.064
Identities = 36/101 (35%), Positives = 43/101 (41%), Gaps = 14/101 (13%)
Query: 835 AAAARARHQAPPPPPASPAAAARARHQAPQPPPAS-----PAAAAR-ARHQAPQPPPPSP 888
A+AA H AP P AAA + PA PAA A A AP+PPP
Sbjct: 35 ASAAHHHHLAPRTPEVRAAAAGALGNHVVSGSPAGAAGAGPAAPAEGAPGAAPEPPP--- 91
Query: 889 AAAARAR-HEGEPAARSASQPKKPAVQ--PSKPAAQPTKKP 926
A RAR G P P++P V P+ PAA+ P
Sbjct: 92 --AGRARPGGGGPQRPGPPSPRRPLVPAGPAPPAAKLRPPP 130
Score = 32.3 bits (72), Expect = 6.0
Identities = 27/100 (27%), Positives = 35/100 (35%), Gaps = 3/100 (3%)
Query: 829 QPPPSPAAAARARHQAPPPP--PASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPP 886
Q P AA + Q PPPP A+ A A + Q AA + Q P
Sbjct: 664 QLTPDSAAFIQQSQQQPPPPTSQATTALTAVVLSSSVQRTAGKTAATVTSALQPPVLSLT 723
Query: 887 SPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
P + +G+P QP KP P T+ P
Sbjct: 724 QPTQVGVGK-QGQPTPLVIQQPPKPGALIRPPQVTLTQTP 762
>XP2_XENLA (P17437) Skin secretory protein xP2 precursor (APEG
protein)
Length = 439
Score = 65.1 bits (157), Expect = 8e-10
Identities = 47/128 (36%), Positives = 61/128 (46%), Gaps = 8/128 (6%)
Query: 812 PTPSQPAAAAIATNQAPQPPPSPA---AAARARHQAPPPPPAS--PAAAARARHQAPQPP 866
P P++ A A A + P P+PA A A A AP P PA A A A +AP P
Sbjct: 83 PAPAEGGAPAPAPAEGGAPAPAPAEGGAPAPAEGGAPAPAPAEGEAPAPAPAEGEAPAPA 142
Query: 867 PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSK---PAAQPT 923
PA A A A +AP P P A A A EGE A + ++ + PA P++ PA P
Sbjct: 143 PAEGEAPAPAEGEAPAPAPAEVEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPA 202
Query: 924 KKPATKPS 931
+ A P+
Sbjct: 203 EGEAPAPA 210
Score = 64.7 bits (156), Expect = 1e-09
Identities = 47/130 (36%), Positives = 60/130 (46%), Gaps = 10/130 (7%)
Query: 812 PTPSQPAAAAIATNQAPQPPPSPAAA---ARARHQAPPPPPASPAAAARARHQAPQPPPA 868
P P++ A A A AP P P+ A A A +AP P PA A A A +AP P PA
Sbjct: 103 PAPAEGGAPAPAEGGAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAEGEAPAPAPA 162
Query: 869 S--PAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPS-----KPAAQ 921
A A A +AP P P A A A EGE A + ++ + PA P+ PA
Sbjct: 163 EVEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAPAEGEAPAPA 222
Query: 922 PTKKPATKPS 931
P + A P+
Sbjct: 223 PAEGEAPAPA 232
Score = 60.1 bits (144), Expect = 3e-08
Identities = 50/135 (37%), Positives = 64/135 (47%), Gaps = 14/135 (10%)
Query: 811 QPTPSQPAAAAIATNQAPQPPPSPAAA-ARARHQAPPPPPASPAAAARARHQAPQPPPAS 869
+P P++ A A A AP P P+ A A A AP P PA A A A AP P PA
Sbjct: 28 EPAPAEGVAPAPAEGGAPAPAPAEGEAPAPAEGGAPAPAPAEGAEPAPADGGAPAPAPAE 87
Query: 870 PAAAARARHQ--APQPPP-----PSPA---AAARARHEGEPAARSASQPKKPAVQPSK-- 917
A A A + AP P P P+PA A A A EGE A + ++ + PA P++
Sbjct: 88 GGAPAPAPAEGGAPAPAPAEGGAPAPAEGGAPAPAPAEGEAPAPAPAEGEAPAPAPAEGE 147
Query: 918 -PAAQPTKKPATKPS 931
PA + PA P+
Sbjct: 148 APAPAEGEAPAPAPA 162
Score = 60.1 bits (144), Expect = 3e-08
Identities = 52/147 (35%), Positives = 70/147 (47%), Gaps = 18/147 (12%)
Query: 802 KCPLPPAKKQPTPS----QPAAAAIATNQAPQPPP----SPAAAARARHQAPPPPPA--- 850
+ P P + P P+ + A A A +AP P P +PA A A +AP P PA
Sbjct: 147 EAPAPAEGEAPAPAPAEVEAPAPAPAEGEAPAPAPAEGEAPAPAP-AEGEAPAPAPAEGE 205
Query: 851 SPA-AAARARHQAPQPPPA--SPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQ 907
+PA A A A +AP P PA A A A +AP P P A A A EGE A + ++
Sbjct: 206 APAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAE 265
Query: 908 PKKPAVQPSK---PAAQPTKKPATKPS 931
+ PA P++ PA P + A P+
Sbjct: 266 GEAPAPAPAEGEAPAPAPAEGEAPAPA 292
Score = 59.7 bits (143), Expect = 4e-08
Identities = 46/136 (33%), Positives = 57/136 (41%), Gaps = 8/136 (5%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAA---ARARHQAPPPPPASPAAAARARH 860
P P + P P++ A A A + +P P+ A A A AP P PA A A A
Sbjct: 47 PAPAEGEAPAPAEGGAPAPAPAEGAEPAPADGGAPAPAPAEGGAPAPAPAEGGAPAPAPA 106
Query: 861 Q--APQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGE---PAARSASQPKKPAVQP 915
+ AP P A A A +AP P P A A A EGE PA A P V+
Sbjct: 107 EGGAPAPAEGGAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAEGEAPAPAPAEVEA 166
Query: 916 SKPAAQPTKKPATKPS 931
PA + PA P+
Sbjct: 167 PAPAPAEGEAPAPAPA 182
Score = 59.7 bits (143), Expect = 4e-08
Identities = 52/153 (33%), Positives = 67/153 (42%), Gaps = 14/153 (9%)
Query: 793 CHQKGHNQRKCPLPPAKKQPTPSQ--PAAAAIATNQAPQP----PPSPAAA-----ARAR 841
C+ +G + P P P P++ A A A +AP P P+PA A A A
Sbjct: 18 CYIQGQDAGGEPAPAEGVAPAPAEGGAPAPAPAEGEAPAPAEGGAPAPAPAEGAEPAPAD 77
Query: 842 HQAPPPPPASPAAAARARHQ--APQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGE 899
AP P PA A A A + AP P PA A A A AP P P A A A EGE
Sbjct: 78 GGAPAPAPAEGGAPAPAPAEGGAPAPAPAEGGAPAPAEGGAPAPAPAEGEAPAPAPAEGE 137
Query: 900 PAARSASQPKKPA-VQPSKPAAQPTKKPATKPS 931
A + ++ + PA + PA P + A P+
Sbjct: 138 APAPAPAEGEAPAPAEGEAPAPAPAEVEAPAPA 170
Score = 59.3 bits (142), Expect = 5e-08
Identities = 49/141 (34%), Positives = 62/141 (43%), Gaps = 12/141 (8%)
Query: 796 KGHNQRKCPLPPAKKQPTPS----QPAAAAIATNQAPQPPPSPA---AAARARHQAPPPP 848
+G P P + P P+ + A A A +AP P P+ A A A +AP P
Sbjct: 203 EGEAPAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPA 262
Query: 849 PA--SPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSAS 906
PA A A A +AP P PA A A A +AP P P A A A EG A S +
Sbjct: 263 PAEGEAPAPAPAEGEAPAPAPAEGEAPAPAEGEAPAPAPAEGEAPAPAPAEG--GAPSPA 320
Query: 907 QPKKPAVQPSKPAAQPTKKPA 927
+ PA P++ A P PA
Sbjct: 321 EGGAPAAAPAEGGA-PAPAPA 340
Score = 58.2 bits (139), Expect = 1e-07
Identities = 51/160 (31%), Positives = 68/160 (41%), Gaps = 32/160 (20%)
Query: 804 PLPPAKKQPTPS----QPAAAAIATNQAPQPPPSPA-----AAARARHQAPPPPPA---- 850
P P + P P+ + A A A +AP P P+PA A A A +AP P PA
Sbjct: 179 PAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEA 238
Query: 851 ---SPA-----AAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAA 902
+PA A A A +AP P PA A A A + P P A A EGE A
Sbjct: 239 PAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAEGEAPA 298
Query: 903 RSASQPKKPAVQPSK-----------PAAQPTKKPATKPS 931
+ ++ + PA P++ PAA P + A P+
Sbjct: 299 PAPAEGEAPAPAPAEGGAPSPAEGGAPAAAPAEGGAPAPA 338
Score = 56.6 bits (135), Expect = 3e-07
Identities = 54/174 (31%), Positives = 75/174 (43%), Gaps = 18/174 (10%)
Query: 770 PDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLP-PAKKQ---PTPSQPAAAAIATN 825
P E P + G + G + + P P PA+ + P P++ A A A
Sbjct: 95 PAEGGAPAPAPAEGGAPAPAEGGAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAEG 154
Query: 826 QAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPAS--PAAAARARHQAPQP 883
+AP P P+ A AP P A A A +AP P PA A A A +AP
Sbjct: 155 EAPAPAPAEVEAP-----APAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAP-A 208
Query: 884 PPPSPA---AAARARHEGEPAARSASQPKKPAVQPSK---PAAQPTKKPATKPS 931
P P+PA A A A EGE A + ++ + PA P++ PA P + A P+
Sbjct: 209 PAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPA 262
Score = 51.6 bits (122), Expect = 1e-05
Identities = 43/123 (34%), Positives = 52/123 (41%), Gaps = 10/123 (8%)
Query: 804 PLPPAKKQPTPS----QPAAAAIATNQAPQPPP----SPAAAARARHQAPPPPPASPAAA 855
P P + P P+ + A A A +AP P P +PA A A +AP P PA A
Sbjct: 231 PAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAP-AEGEAPAPAPAEGEAP 289
Query: 856 ARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHE-GEPAARSASQPKKPAVQ 914
A A +AP P PA A A A + P P A A A E G PA A P +
Sbjct: 290 APAEGEAPAPAPAEGEAPAPAPAEGGAPSPAEGGAPAAAPAEGGAPAPAPAPVEVGPKTE 349
Query: 915 PSK 917
K
Sbjct: 350 DCK 352
Score = 40.8 bits (94), Expect = 0.017
Identities = 37/112 (33%), Positives = 47/112 (41%), Gaps = 14/112 (12%)
Query: 804 PLPPAKKQPTPS----QPAAAAIATNQAPQPPP----SPAAAARARHQAPPPPPASPAAA 855
P P + P P+ + A A A +AP P P +PA A +AP P PA A
Sbjct: 251 PAPAEGEAPAPAPAEGEAPAPAPAEGEAPAPAPAEGEAPAP---AEGEAPAPAPAEGEAP 307
Query: 856 ARARHQ--APQPPPASPAAAARARHQAPQP-PPPSPAAAARARHEGEPAARS 904
A A + AP P AAA A AP P P P +G+P R+
Sbjct: 308 APAPAEGGAPSPAEGGAPAAAPAEGGAPAPAPAPVEVGPKTEDCKGDPFKRT 359
>FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity
protein)
Length = 1468
Score = 64.7 bits (156), Expect = 1e-09
Identities = 52/194 (26%), Positives = 74/194 (37%), Gaps = 21/194 (10%)
Query: 733 ISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREP--DEPDKKNPTKLKRGGSSYTC 790
+ P + E+K + SQ I+P ++ L + P D D P + + G SS +
Sbjct: 814 LKPCDAESKATRSSQ--IVPKKLTIS------LTQLSPSKDSKDIHAPFQTREGTSSSS- 864
Query: 791 GRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPA 850
Q+ + P PP P P P PP P + PPPPP
Sbjct: 865 ----QQKISPPAPPTPPPLPPPLIPPPPPLPPGLGPLPPAPPIPPVCPVSPPPPPPPPPP 920
Query: 851 SPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEP------AARS 904
+P + P PPP P A P PPPP P P ++ S
Sbjct: 921 TPVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPPPPGLSFGLSSSS 980
Query: 905 ASQPKKPAVQPSKP 918
+ P+KPA++PS P
Sbjct: 981 SQYPRKPAIEPSCP 994
Score = 47.4 bits (111), Expect = 2e-04
Identities = 32/113 (28%), Positives = 40/113 (35%), Gaps = 5/113 (4%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP PTP P+ P PPP P A PPPPP P AP
Sbjct: 912 PPPPPPPPPTPVPPSDGP--PPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGL---AP 966
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPS 916
PPP + + Q P+ P P+ + + SQ P + S
Sbjct: 967 PPPPGLSFGLSSSSSQYPRKPAIEPSCPMKPLYWTRIQINDKSQDAAPTLWDS 1019
Score = 38.5 bits (88), Expect = 0.084
Identities = 37/154 (24%), Positives = 49/154 (31%), Gaps = 43/154 (27%)
Query: 807 PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPP 866
P + + S + I+ P PPP P PPPPP P AP P
Sbjct: 853 PFQTREGTSSSSQQKISPPAPPTPPPLPPPLI------PPPPPLPPGLGPLP--PAPPIP 904
Query: 867 PASPAAAARARHQAPQPPPPSPAAAARA-------------------RHEGEPAARSASQ 907
P P + P PPPP P + + G P
Sbjct: 905 PVCPVSPP------PPPPPPPPTPVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPP 958
Query: 908 PKKPAVQPSKP----------AAQPTKKPATKPS 931
P P + P P ++Q +KPA +PS
Sbjct: 959 PPPPGLAPPPPPGLSFGLSSSSSQYPRKPAIEPS 992
>FM14_MOUSE (Q05859) Formin 1 isoform IV (Limb deformity protein)
Length = 1206
Score = 64.7 bits (156), Expect = 1e-09
Identities = 52/194 (26%), Positives = 74/194 (37%), Gaps = 21/194 (10%)
Query: 733 ISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREP--DEPDKKNPTKLKRGGSSYTC 790
+ P + E+K + SQ I+P ++ L + P D D P + + G SS +
Sbjct: 588 LKPCDAESKATRSSQ--IVPKKLTIS------LTQLSPSKDSKDIHAPFQTREGTSSSS- 638
Query: 791 GRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPA 850
Q+ + P PP P P P PP P + PPPPP
Sbjct: 639 ----QQKISPPAPPTPPPLPPPLIPPPPPLPPGLGPLPPAPPIPPVCPVSPPPPPPPPPP 694
Query: 851 SPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEP------AARS 904
+P + P PPP P A P PPPP P P ++ S
Sbjct: 695 TPVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPPPPGLSFGLSSSS 754
Query: 905 ASQPKKPAVQPSKP 918
+ P+KPA++PS P
Sbjct: 755 SQYPRKPAIEPSCP 768
Score = 47.4 bits (111), Expect = 2e-04
Identities = 32/113 (28%), Positives = 40/113 (35%), Gaps = 5/113 (4%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP PTP P+ P PPP P A PPPPP P AP
Sbjct: 686 PPPPPPPPPTPVPPSDGP--PPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGL---AP 740
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPS 916
PPP + + Q P+ P P+ + + SQ P + S
Sbjct: 741 PPPPGLSFGLSSSSSQYPRKPAIEPSCPMKPLYWTRIQINDKSQDAAPTLWDS 793
Score = 38.5 bits (88), Expect = 0.084
Identities = 37/154 (24%), Positives = 49/154 (31%), Gaps = 43/154 (27%)
Query: 807 PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPP 866
P + + S + I+ P PPP P PPPPP P AP P
Sbjct: 627 PFQTREGTSSSSQQKISPPAPPTPPPLPPPLI------PPPPPLPPGLGPLP--PAPPIP 678
Query: 867 PASPAAAARARHQAPQPPPPSPAAAARA-------------------RHEGEPAARSASQ 907
P P + P PPPP P + + G P
Sbjct: 679 PVCPVSPP------PPPPPPPPTPVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPP 732
Query: 908 PKKPAVQPSKP----------AAQPTKKPATKPS 931
P P + P P ++Q +KPA +PS
Sbjct: 733 PPPPGLAPPPPPGLSFGLSSSSSQYPRKPAIEPS 766
>MYPH_CHICK (Q05623) Myosin-binding protein H (MyBP-H) (H-protein)
(86 kDa protein)
Length = 537
Score = 64.3 bits (155), Expect = 1e-09
Identities = 45/134 (33%), Positives = 56/134 (41%), Gaps = 8/134 (5%)
Query: 795 QKGHNQRKCPLPPAKK--QPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASP 852
+K +K P P +KK +P P + A AP P A + H PP +P
Sbjct: 11 KKAPAAKKAPAPASKKAPEPAPKEKPAPTPKEGHAPTPKEEHAPPPKEEHAPPPKEEHAP 70
Query: 853 AAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPA 912
A AA AP+ PP + AA A AP P AA A EG P A A P P
Sbjct: 71 APAAET-PPAPEHPPDAEQPAAPAAEHAPTP----THEAAPAHEEGPPPAAPAEAP-APE 124
Query: 913 VQPSKPAAQPTKKP 926
+P KP +P P
Sbjct: 125 PEPEKPKEEPPSVP 138
Score = 48.9 bits (115), Expect = 6e-05
Identities = 38/122 (31%), Positives = 46/122 (37%), Gaps = 11/122 (9%)
Query: 817 PAAA--AIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAA 874
PAAA A A +AP P A + + P P A + H PPP A
Sbjct: 7 PAAAKKAPAAKKAPAPASKKAPEPAPKEKPAPTPKEGHAPTPKEEH---APPPKEEHAPP 63
Query: 875 RARHQAPQP---PPPSPAAAARARHEGEPAARSASQP---KKPAVQPSKPAAQPTKKPAT 928
AP P PP+P A PAA A P PA + P A P + PA
Sbjct: 64 PKEEHAPAPAAETPPAPEHPPDAEQPAAPAAEHAPTPTHEAAPAHEEGPPPAAPAEAPAP 123
Query: 929 KP 930
+P
Sbjct: 124 EP 125
Score = 48.9 bits (115), Expect = 6e-05
Identities = 33/96 (34%), Positives = 41/96 (42%), Gaps = 3/96 (3%)
Query: 833 SPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAA 892
+PAAA +A P PAS A A + P P P A AP PP A
Sbjct: 6 APAAAKKAPAAKKAPAPASKKAPEPAPKEKPAPTPKEGHAPTPKEEHAP-PPKEEHAPPP 64
Query: 893 RARHEGEPAARSASQPKKP--AVQPSKPAAQPTKKP 926
+ H PAA + P+ P A QP+ PAA+ P
Sbjct: 65 KEEHAPAPAAETPPAPEHPPDAEQPAAPAAEHAPTP 100
Score = 46.6 bits (109), Expect = 3e-04
Identities = 42/150 (28%), Positives = 57/150 (38%), Gaps = 23/150 (15%)
Query: 752 PPEIKRGPGR---PKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPA 808
P K+ P P ++ P+ K+ P + G + T H + P P
Sbjct: 7 PAAAKKAPAAKKAPAPASKKAPEPAPKEKPAPTPKEGHAPTPKEEHAPPPKEEHAPPPKE 66
Query: 809 KKQPTPSQPAAAAIATNQAPQPPPS---PAAAARARHQAPPPPPASPAAAARARHQAPQP 865
+ P P A T AP+ PP PAA A A H P A+PA P
Sbjct: 67 EHAPAP------AAETPPAPEHPPDAEQPAAPA-AEHAPTPTHEAAPA-------HEEGP 112
Query: 866 PPASPAAAARARHQAPQP---PPPSPAAAA 892
PPA+PA A + +P PP P + A
Sbjct: 113 PPAAPAEAPAPEPEPEKPKEEPPSVPLSLA 142
Score = 37.4 bits (85), Expect = 0.19
Identities = 26/82 (31%), Positives = 34/82 (40%), Gaps = 9/82 (10%)
Query: 851 SPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPS-PAAAARARHEGEPAARSASQPK 909
+PAAA +A PA+ A A A +AP+P P PA + H P A PK
Sbjct: 6 APAAAKKA--------PAAKKAPAPASKKAPEPAPKEKPAPTPKEGHAPTPKEEHAPPPK 57
Query: 910 KPAVQPSKPAAQPTKKPATKPS 931
+ P K P T P+
Sbjct: 58 EEHAPPPKEEHAPAPAAETPPA 79
>ALGP_PSEAE (P15276) Transcriptional regulatory protein algP
(Alginate regulatory protein algR3)
Length = 352
Score = 64.3 bits (155), Expect = 1e-09
Identities = 48/126 (38%), Positives = 64/126 (50%), Gaps = 10/126 (7%)
Query: 806 PPAKKQPTPS-QPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
P AK P+ +PAA A A +P P A A+ P PA+ AAA+ +
Sbjct: 158 PAAKPAAKPAAKPAAKPAAKTAAAKPAAKPTAKPAAK---PAAKPAAKTAAAKPAAKPAA 214
Query: 865 PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTK 924
P A PAA A+ A +P PAA A+ +PAA++A+ KPA +KPAA+P
Sbjct: 215 KPVAKPAAKPAAKTAAAKPAA-KPAAKPVAKPTAKPAAKTAA--AKPA---AKPAAKPAA 268
Query: 925 KPATKP 930
KPA KP
Sbjct: 269 KPAAKP 274
Score = 63.5 bits (153), Expect = 2e-09
Identities = 55/146 (37%), Positives = 69/146 (46%), Gaps = 22/146 (15%)
Query: 806 PPAKKQPTPS-QPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
P AK P+ +PAA A A +P PAA A+ A P A AAA A A +
Sbjct: 183 PAAKPTAKPAAKPAAKPAAKTAAAKPAAKPAAKPVAKPAAKPA--AKTAAAKPAAKPAAK 240
Query: 865 P---PPASPAAAARARHQAPQP---PPPSPAA-----AARARHEGEPAARSASQP-KKPA 912
P P A PAA A A +P P PAA +A A+ +PAA+ A++P KPA
Sbjct: 241 PVAKPTAKPAAKTAAAKPAAKPAAKPAAKPAAKPVAKSAAAKPAAKPAAKPAAKPAAKPA 300
Query: 913 VQP-------SKPAAQPTKKPATKPS 931
+P +KPA P KPA PS
Sbjct: 301 AKPVAAKPAATKPATAPAAKPAATPS 326
Score = 62.4 bits (150), Expect = 5e-09
Identities = 47/125 (37%), Positives = 62/125 (49%), Gaps = 7/125 (5%)
Query: 808 AKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPP- 866
++K ++PAA A A PAA A+ A P A PAA A A +P
Sbjct: 132 SRKAKPATKPAAKAAAKPAVKTVAAKPAAKPAAKPAAKPA--AKPAAKTAAAKPAAKPTA 189
Query: 867 -PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQP-SKPAAQPTK 924
PA+ AA A A P PAA A+ +PAA++A+ KPA +P +KP A+PT
Sbjct: 190 KPAAKPAAKPAAKTAAAKPAAKPAAKPVAKPAAKPAAKTAA--AKPAAKPAAKPVAKPTA 247
Query: 925 KPATK 929
KPA K
Sbjct: 248 KPAAK 252
Score = 61.2 bits (147), Expect = 1e-08
Identities = 55/148 (37%), Positives = 70/148 (47%), Gaps = 29/148 (19%)
Query: 806 PPAKKQPTPS-QPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
P AK P+ +PAA A A +P PAA A+ P A AAA A A +
Sbjct: 162 PAAKPAAKPAAKPAAKTAAAKPAAKPTAKPAAKPAAK------PAAKTAAAKPAAKPAAK 215
Query: 865 P--PPAS-PAAAARARHQAPQP-------PPPSPAA-AARARHEGEPAARSASQP----- 908
P PA+ PAA A A +P P PAA A A+ +PAA+ A++P
Sbjct: 216 PVAKPAAKPAAKTAAAKPAAKPAAKPVAKPTAKPAAKTAAAKPAAKPAAKPAAKPAAKPV 275
Query: 909 -----KKPAVQP-SKPAAQPTKKPATKP 930
KPA +P +KPAA+P KPA KP
Sbjct: 276 AKSAAAKPAAKPAAKPAAKPAAKPAAKP 303
Score = 57.4 bits (137), Expect = 2e-07
Identities = 46/134 (34%), Positives = 59/134 (43%), Gaps = 14/134 (10%)
Query: 806 PPAKKQPTP-SQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
P AK P ++PAA A A +P PAA A+ P PA+ AAA+ +
Sbjct: 208 PAAKPAAKPVAKPAAKPAAKTAAAKPAAKPAAKPVAK---PTAKPAAKTAAAKPAAKPAA 264
Query: 865 PPPASPAAAARARHQAPQP---PPPSPAAAARARHEGEPAARSASQPK-------KPAVQ 914
P A PAA A+ A +P P PAA A+ +P A + K KPA
Sbjct: 265 KPAAKPAAKPVAKSAAAKPAAKPAAKPAAKPAAKPAAKPVAAKPAATKPATAPAAKPAAT 324
Query: 915 PSKPAAQPTKKPAT 928
PS PAA + AT
Sbjct: 325 PSAPAAASSAASAT 338
Score = 56.2 bits (134), Expect = 4e-07
Identities = 38/98 (38%), Positives = 52/98 (52%), Gaps = 4/98 (4%)
Query: 837 AARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARH 896
A +R P PA+ AAA A P A PAA A+ A +P + AA A+
Sbjct: 129 ALESRKAKPATKPAAKAAAKPAVKTVAAKPAAKPAAKPAAK-PAAKPAAKTAAAKPAAKP 187
Query: 897 EGEPAARSASQP--KKPAVQP-SKPAAQPTKKPATKPS 931
+PAA+ A++P K A +P +KPAA+P KPA KP+
Sbjct: 188 TAKPAAKPAAKPAAKTAAAKPAAKPAAKPVAKPAAKPA 225
Score = 53.1 bits (126), Expect = 3e-06
Identities = 39/118 (33%), Positives = 52/118 (44%), Gaps = 12/118 (10%)
Query: 806 PPAKKQPTP-SQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
P AK P ++P A A A +P PAA A+ P P + +AAA+ +
Sbjct: 233 PAAKPAAKPVAKPTAKPAAKTAAAKPAAKPAAKPAAK---PAAKPVAKSAAAKPAAKPAA 289
Query: 865 PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
P A PAA A+ A +P PA A PAA+ A+ P PA S +A P
Sbjct: 290 KPAAKPAAKPAAKPVAAKPAATKPATA--------PAAKPAATPSAPAAASSAASATP 339
Score = 50.8 bits (120), Expect = 2e-05
Identities = 41/122 (33%), Positives = 58/122 (46%), Gaps = 16/122 (13%)
Query: 818 AAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARAR 877
AA ++ +P PAA A A+ PA AA+ + P A PAA A+
Sbjct: 125 AAGKALESRKAKPATKPAAKAAAK-------PAVKTVAAKPAAKPAAKPAAKPAAKPAAK 177
Query: 878 HQAPQPPPPSPAAAARARHEGEPAARSASQPK--KPAVQP-SKPAAQP-----TKKPATK 929
A +P P A A+ +PAA++A+ KPA +P +KPAA+P KPA K
Sbjct: 178 TAAAKPAA-KPTAKPAAKPAAKPAAKTAAAKPAAKPAAKPVAKPAAKPAAKTAAAKPAAK 236
Query: 930 PS 931
P+
Sbjct: 237 PA 238
Score = 42.7 bits (99), Expect = 0.004
Identities = 31/88 (35%), Positives = 40/88 (45%), Gaps = 3/88 (3%)
Query: 806 PPAKKQPTPS-QPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
P AK P+ +PAA +A + A +P PAA A+ A P A P AA A +
Sbjct: 258 PAAKPAAKPAAKPAAKPVAKSAAAKPAAKPAAKPAAKPAAKPA--AKPVAAKPAATKPAT 315
Query: 865 PPPASPAAAARARHQAPQPPPPSPAAAA 892
P A PAA A A +PAA +
Sbjct: 316 APAAKPAATPSAPAAASSAASATPAAGS 343
>SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor
(SSG 185)
Length = 485
Score = 63.9 bits (154), Expect = 2e-09
Identities = 43/128 (33%), Positives = 53/128 (40%), Gaps = 24/128 (18%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P+ PA +P P+ A+++ P PPPSP R +PPPP SP P
Sbjct: 219 PIGPAPNN-SPLPPSPQPTASSRPPSPPPSP------RPPSPPPPSPSPPPPPPPPPPPP 271
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPT 923
PPP SP P PPPP P P S S P+KP PS P P
Sbjct: 272 PPPPPSPP---------PPPPPPPPPPP-------PPPPPSPSPPRKPP-SPSPPVPPPP 314
Query: 924 KKPATKPS 931
P+ P+
Sbjct: 315 SPPSVLPA 322
Score = 50.1 bits (118), Expect = 3e-05
Identities = 29/96 (30%), Positives = 38/96 (39%), Gaps = 9/96 (9%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
PLPP+ + S+P + + PPPSP+ PPPPP P+ P
Sbjct: 228 PLPPSPQPTASSRPPSPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPP 287
Query: 864 QPPPASPAAAARARHQA---------PQPPPPSPAA 890
PPP P + + R P PP PAA
Sbjct: 288 PPPPPPPPSPSPPRKPPSPSPPVPPPPSPPSVLPAA 323
>IF2_CORGL (Q8NP40) Translation initiation factor IF-2
Length = 1004
Score = 63.9 bits (154), Expect = 2e-09
Identities = 52/152 (34%), Positives = 69/152 (45%), Gaps = 27/152 (17%)
Query: 797 GHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAA 856
G ++ PA +P +PAA+A AP P +PAA P PA+P+AA+
Sbjct: 55 GSDKSDTAAKPAAAKPAAPKPAASAAPKPGAPAKPAAPAAK---------PAPAAPSAAS 105
Query: 857 RARHQAPQPP--PASPAAAAR----ARHQAPQPP-------PPSPAAAARARHEGE---P 900
A+ A P A PAAAA+ A+ AP P P PAAAA+ G
Sbjct: 106 AAKPGAAPKPGVQAKPAAAAKPGAPAKPAAPAAPSAAKSGSAPKPAAAAKPAFSGPTPGD 165
Query: 901 AARSASQPKKPAVQPSKPAA--QPTKKPATKP 930
AA+ A KP + +P +P KPA KP
Sbjct: 166 AAKKAEPAAKPGAEAPRPGGMPRPMGKPAPKP 197
Score = 59.3 bits (142), Expect = 5e-08
Identities = 55/203 (27%), Positives = 90/203 (44%), Gaps = 28/203 (13%)
Query: 732 MISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCG 791
+++ + + ++ K + I PP +KR ++ K+ T K +
Sbjct: 21 LLATLKDKGEFVKTASSTIEPPVVKR-------MQEHYGSSGSDKSDTAAKPAAAKPAAP 73
Query: 792 RCHQKGHNQRKCPLPPAKK--QPTPSQPAAAAIAT-NQAPQP--PPSPAAAARARHQAPP 846
+ + P PA +P P+ P+AA+ A AP+P PAAAA+ A P
Sbjct: 74 KPAASAAPKPGAPAKPAAPAAKPAPAAPSAASAAKPGAAPKPGVQAKPAAAAKPGAPAKP 133
Query: 847 PPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSAS 906
PA+P+AA AP+ PAAAA+ P P+P AA+ + EPAA+ +
Sbjct: 134 AAPAAPSAAKSG--SAPK-----PAAAAK-----PAFSGPTPGDAAK---KAEPAAKPGA 178
Query: 907 QPKKPAVQPSKPAAQPTKKPATK 929
+ +P P +P +P KP +
Sbjct: 179 EAPRPGGMP-RPMGKPAPKPGAR 200
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 63.2 bits (152), Expect = 3e-09
Identities = 42/129 (32%), Positives = 52/129 (39%), Gaps = 25/129 (19%)
Query: 803 CPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQA 862
CP PP K QP P AP P P P+ + + + P PPPA P + + +
Sbjct: 88 CPSPPPKPQPKPPP----------APSPSPCPSPPPKPQPK-PVPPPACPPTPPKPQPK- 135
Query: 863 PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
P PPP A A P P PP P A +P P P KPA P
Sbjct: 136 PAPPPEPKPAPPPAPKPVPCPSPPKPPA-------------PTPKPVPPHGPPPKPAPAP 182
Query: 923 TKKPATKPS 931
T P+ KP+
Sbjct: 183 TPAPSPKPA 191
Score = 57.0 bits (136), Expect = 2e-07
Identities = 37/109 (33%), Positives = 44/109 (39%), Gaps = 19/109 (17%)
Query: 803 CPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQA 862
CP PP K QP P P A T PQP P+P + APPP P + + A
Sbjct: 108 CPSPPPKPQPKPVPPPACP-PTPPKPQPKPAPPPEPKP---APPPAPKPVPCPSPPKPPA 163
Query: 863 PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP 911
P P P P H P P P+P A P+ + A P KP
Sbjct: 164 PTPKPVPP-------HGPPPKPAPAPTPA--------PSPKPAPSPPKP 197
Score = 47.4 bits (111), Expect = 2e-04
Identities = 38/131 (29%), Positives = 46/131 (35%), Gaps = 27/131 (20%)
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQP 865
P QP P P P P P P A + PP SP + + Q P+P
Sbjct: 49 PRPYPQPWPMNPPT--------PDPSPKPVAPPGPSSKPVAPPGPSPCPSPPPKPQ-PKP 99
Query: 866 PPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPS-----KPAA 920
PPA + + PQP P P A P KP +P+ KPA
Sbjct: 100 PPAPSPSPCPSPPPKPQPKPVPPPACPPT-------------PPKPQPKPAPPPEPKPAP 146
Query: 921 QPTKKPATKPS 931
P KP PS
Sbjct: 147 PPAPKPVPCPS 157
Score = 33.5 bits (75), Expect = 2.7
Identities = 22/66 (33%), Positives = 24/66 (36%), Gaps = 13/66 (19%)
Query: 801 RKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARH 860
+ P P K P PS P AP P P P PPP PA A +
Sbjct: 143 KPAPPPAPKPVPCPSPPKP------PAPTPKPVPP-------HGPPPKPAPAPTPAPSPK 189
Query: 861 QAPQPP 866
AP PP
Sbjct: 190 PAPSPP 195
Score = 32.0 bits (71), Expect = 7.8
Identities = 23/85 (27%), Positives = 27/85 (31%), Gaps = 10/85 (11%)
Query: 856 ARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSA---------- 905
A+ H+ P P P + P P PSP A +P A
Sbjct: 35 AQVMHRRLWPWPLWPRPYPQPWPMNPPTPDPSPKPVAPPGPSSKPVAPPGPSPCPSPPPK 94
Query: 906 SQPKKPAVQPSKPAAQPTKKPATKP 930
QPK P P P KP KP
Sbjct: 95 PQPKPPPAPSPSPCPSPPPKPQPKP 119
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 62.0 bits (149), Expect = 7e-09
Identities = 39/127 (30%), Positives = 48/127 (37%), Gaps = 17/127 (13%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP K QP P AP P P P + + + PP P SP Q P
Sbjct: 6 PKPPPKPQPKPPP----------APTPSPCPPQPPKPQPKPPPAPTPSPCPP-----QPP 50
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPT 923
+P P P A + P P PP P + + P+ P KP KP+ P
Sbjct: 51 KPQPKPPPAPGPSPKPGPSPSPPKPPPSPAPKPVPPPSPSPKPSPPKPPAPSPKPS--PP 108
Query: 924 KKPATKP 930
K PA P
Sbjct: 109 KPPAPSP 115
Score = 59.3 bits (142), Expect = 5e-08
Identities = 34/115 (29%), Positives = 42/115 (35%), Gaps = 10/115 (8%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP P+P P P P PSP + Q PPP P+ P
Sbjct: 14 PKPPPAPTPSPCPPQPPKPQPKPPPAPTPSPCPPQPPKPQPKPPPAPGPSPKPGPSPSPP 73
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKP 918
+PPP+ +P+P PP P A + S PK PA P KP
Sbjct: 74 KPPPSPAPKPVPPPSPSPKPSPPKPPA----------PSPKPSPPKPPAPSPPKP 118
Score = 48.9 bits (115), Expect = 6e-05
Identities = 31/97 (31%), Positives = 40/97 (40%), Gaps = 17/97 (17%)
Query: 803 CPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQ-----APPPPPASPAA--- 854
CP P K QP P + Q P+P P P A + +PP PP SPA
Sbjct: 25 CPPQPPKPQPKPPPAPTPSPCPPQPPKPQPKPPPAPGPSPKPGPSPSPPKPPPSPAPKPV 84
Query: 855 ---AARARHQAPQPPPASPAAAARARHQAPQPPPPSP 888
+ + P+PP SP + P+PP PSP
Sbjct: 85 PPPSPSPKPSPPKPPAPSP------KPSPPKPPAPSP 115
Score = 35.4 bits (80), Expect = 0.71
Identities = 26/89 (29%), Positives = 33/89 (36%), Gaps = 20/89 (22%)
Query: 845 PPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARS 904
PP P P + P+PPPA + + PQP PP PA
Sbjct: 1 PPKPQPKPPPKPQ-----PKPPPAPTPSPCPPQPPKPQPKPP-------------PAPTP 42
Query: 905 ASQPKKPAVQPSK--PAAQPTKKPATKPS 931
+ P +P K PA P+ KP PS
Sbjct: 43 SPCPPQPPKPQPKPPPAPGPSPKPGPSPS 71
>PRB2_HUMAN (P02812) Basic salivary proline-rich protein 2 (Salivary
proline-rich protein) (Con1 glycoprotein) [Contains:
Basic peptide P-F] (Fragment)
Length = 382
Score = 61.6 bits (148), Expect = 9e-09
Identities = 47/181 (25%), Positives = 64/181 (34%), Gaps = 10/181 (5%)
Query: 752 PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQ 811
PP+ P P + + P N ++ R G Q G+ + P PP K Q
Sbjct: 171 PPQGGNKPQGPPPPGKPQGPPPQGDNKSQSARSPPGKPQGPPPQGGNQPQGPPPPPGKPQ 230
Query: 812 PTPSQPAAAAIATNQAPQPP-----PSPAAAARARH-QAPPPPPASPAAAARARHQAPQP 865
P Q + Q P PP P P +++R ++PP P P + Q P P
Sbjct: 231 GPPPQGGNKS----QGPPPPGKPQGPPPQGGSKSRSSRSPPGKPQGPPPQGGNQPQGPPP 286
Query: 866 PPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKK 925
PP P PQ PPP G + + S P KP P + P
Sbjct: 287 PPGKPQGPPPQGGNKPQGPPPPGKPQGPPPQGGSKSRSARSPPGKPQGPPQQEGNNPQGP 346
Query: 926 P 926
P
Sbjct: 347 P 347
Score = 60.1 bits (144), Expect = 3e-08
Identities = 51/193 (26%), Positives = 75/193 (38%), Gaps = 23/193 (11%)
Query: 735 PINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCH 794
P G+ + P D P + PG+P+ + ++P P K G
Sbjct: 59 PPPGKPQGPPPQGDKSRSP--RSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPP------- 109
Query: 795 QKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAA 854
+G N+ + P PP K Q P Q + ++ P P P + Q PPPPP P
Sbjct: 110 PQGGNKPQGPPPPGKPQGPPPQGDNKSRSSRSPPGKPQGPPPQGGNQPQGPPPPPGKPQG 169
Query: 855 AARARHQAPQ--PPPASPAAAARARHQAPQPPPPSPAAAARA---RHEGEPAARSASQPK 909
PQ PPP P Q P P + + +AR+ + +G P + +QP+
Sbjct: 170 PPPQGGNKPQGPPPPGKP--------QGPPPQGDNKSQSARSPPGKPQG-PPPQGGNQPQ 220
Query: 910 KPAVQPSKPAAQP 922
P P KP P
Sbjct: 221 GPPPPPGKPQGPP 233
Score = 54.3 bits (129), Expect = 1e-06
Identities = 54/206 (26%), Positives = 73/206 (35%), Gaps = 38/206 (18%)
Query: 735 PINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTC---- 790
P G+NK SQ PP +GP + + P P K +GG+
Sbjct: 191 PPQGDNK----SQSARSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPPPQGGNKSQGPPPP 246
Query: 791 ----GRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPP 846
G Q G R PP K Q P Q NQ PPP P + Q PP
Sbjct: 247 GKPQGPPPQGGSKSRSSRSPPGKPQGPPPQGG------NQPQGPPPPPG-----KPQGPP 295
Query: 847 PP-------------PASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAAR 893
P P P ++ ++ + PP P + PQ PPP PA
Sbjct: 296 PQGGNKPQGPPPPGKPQGPPPQGGSKSRSARSPPGKPQGPPQQEGNNPQGPPP-PAGGNP 354
Query: 894 ARHEGEPAARSASQPKKP-AVQPSKP 918
+ + PA + P+ P +PS+P
Sbjct: 355 QQPQAPPAGQPQGPPRPPQGGRPSRP 380
Score = 53.1 bits (126), Expect = 3e-06
Identities = 39/144 (27%), Positives = 50/144 (34%), Gaps = 9/144 (6%)
Query: 795 QKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPP--PASP 852
Q G+ + P PP K Q P Q P P P + Q PPPP P P
Sbjct: 8 QGGNKPQGPPSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPPPQGGNKPQGPPPPGKPQGP 67
Query: 853 AAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPA 912
+ ++P+ PP P PQ PPP P +G + P KP
Sbjct: 68 PPQGD-KSRSPRSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPPPQGGNKPQGPPPPGKPQ 126
Query: 913 VQP------SKPAAQPTKKPATKP 930
P S+ + P KP P
Sbjct: 127 GPPPQGDNKSRSSRSPPGKPQGPP 150
Score = 52.8 bits (125), Expect = 4e-06
Identities = 48/199 (24%), Positives = 65/199 (32%), Gaps = 31/199 (15%)
Query: 752 PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQ 811
PP+ P P + + P N ++ R G Q G+ + P PP K Q
Sbjct: 109 PPQGGNKPQGPPPPGKPQGPPPQGDNKSRSSRSPPGKPQGPPPQGGNQPQGPPPPPGKPQ 168
Query: 812 PTPSQPAAAAIATNQAPQPPPSP-------------AAAARARHQAPPPPPASPAAAARA 858
P Q Q P PP P A + + Q PPP +
Sbjct: 169 GPPPQGGNKP----QGPPPPGKPQGPPPQGDNKSQSARSPPGKPQGPPPQGGN------- 217
Query: 859 RHQAPQPPPASPAA---AARARHQAPQPP----PPSPAAAARARHEGEPAARSASQPKKP 911
+ Q P PPP P + Q P PP P P +++R P + P +
Sbjct: 218 QPQGPPPPPGKPQGPPPQGGNKSQGPPPPGKPQGPPPQGGSKSRSSRSPPGKPQGPPPQG 277
Query: 912 AVQPSKPAAQPTKKPATKP 930
QP P P K P
Sbjct: 278 GNQPQGPPPPPGKPQGPPP 296
Score = 48.5 bits (114), Expect = 8e-05
Identities = 36/128 (28%), Positives = 46/128 (35%), Gaps = 23/128 (17%)
Query: 817 PAAAAIATNQAPQPPPSPAAAARA-------RHQAPPPPPASPAAAARARHQAPQ--PPP 867
P A PQ PPSP + + Q PPPPP P PQ PPP
Sbjct: 2 PQGAPPQGGNKPQGPPSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPPPQGGNKPQGPPPP 61
Query: 868 ASPAAAARA--RHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTK- 924
P + ++P+ PP P P + +QP+ P P KP P +
Sbjct: 62 GKPQGPPPQGDKSRSPRSPPGKPQG---------PPPQGGNQPQGPPPPPGKPQGPPPQG 112
Query: 925 --KPATKP 930
KP P
Sbjct: 113 GNKPQGPP 120
>PRP1_HUMAN (P04280) Basic salivary proline-rich protein 1 precursor
(Salivary proline-rich protein) [Contains: Basic peptide
IB-6; Peptide P-H]
Length = 392
Score = 61.2 bits (147), Expect = 1e-08
Identities = 55/207 (26%), Positives = 78/207 (37%), Gaps = 33/207 (15%)
Query: 735 PINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCH 794
P G+ + P D P + PG+P+ + ++P P K G
Sbjct: 194 PPPGKPQGPPPQGDKSQSP--RSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPP------- 244
Query: 795 QKGHNQRKCPLPPAKKQPTPSQ----------------PAAAAIATNQAPQPPPS----P 834
Q+G N+ + P PP K Q P Q P Q P PPP P
Sbjct: 245 QQGGNRPQGPPPPGKPQGPPPQGDKSRSPQSPPGKPQGPPPQGGNQPQGPPPPPGKPQGP 304
Query: 835 AAAARARHQAPPPP--PASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAA 892
+ Q PPPP P P A ++ Q+ + PP P + PQ PPP PA
Sbjct: 305 PPQGGNKPQGPPPPGKPQGPPAQGGSKSQSARAPPGKPQGPPQQEGNNPQGPPP-PAGGN 363
Query: 893 RARHEGEPAARSASQPKKP-AVQPSKP 918
+ + PA + P+ P +PS+P
Sbjct: 364 PQQPQAPPAGQPQGPPRPPQGGRPSRP 390
Score = 57.8 bits (138), Expect = 1e-07
Identities = 46/173 (26%), Positives = 58/173 (32%), Gaps = 16/173 (9%)
Query: 759 PGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPA 818
PG+P+ P + DK + G G Q G+ + P PP K Q P Q
Sbjct: 196 PGKPQG----PPPQGDKSQSPRSPPGKPQ---GPPPQGGNQPQGPPPPPGKPQGPPQQGG 248
Query: 819 AAAIATNQAPQPP-----PSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAA 873
Q P PP P P Q+PP P P + Q P PPP P
Sbjct: 249 NRP----QGPPPPGKPQGPPPQGDKSRSPQSPPGKPQGPPPQGGNQPQGPPPPPGKPQGP 304
Query: 874 ARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
PQ PPP G + + + P KP P + P P
Sbjct: 305 PPQGGNKPQGPPPPGKPQGPPAQGGSKSQSARAPPGKPQGPPQQEGNNPQGPP 357
Score = 53.9 bits (128), Expect = 2e-06
Identities = 54/218 (24%), Positives = 74/218 (33%), Gaps = 34/218 (15%)
Query: 735 PINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCH 794
P G+ + P D P + PG+P+ + ++P P K G
Sbjct: 72 PPPGKPQGPPPQGDKSRSP--RSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPP------- 122
Query: 795 QKGHNQRKCPLPPAKKQPTPSQ----------------PAAAAIATNQAPQPPPS----P 834
+G N+ + P PP K Q P Q P Q P PPP P
Sbjct: 123 PQGGNKPQGPPPPGKPQGPPPQGDKSQSPRSPPGKPQGPPPQGGNQPQGPPPPPGKPQGP 182
Query: 835 AAAARARHQAPPPP--PASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAA 892
+ Q PPPP P P + Q+P+ PP P PQ PPP P
Sbjct: 183 PPQGGNKPQGPPPPGKPQGPPPQGD-KSQSPRSPPGKPQGPPPQGGNQPQGPPPPPGKPQ 241
Query: 893 RARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
+G + P KP P P ++ P + P
Sbjct: 242 GPPQQGGNRPQGPPPPGKPQGPP--PQGDKSRSPQSPP 277
Score = 51.2 bits (121), Expect = 1e-05
Identities = 52/206 (25%), Positives = 74/206 (35%), Gaps = 32/206 (15%)
Query: 735 PINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCH 794
P G+ + P D P + PG+P+ + ++P P K G
Sbjct: 133 PPPGKPQGPPPQGDKSQSP--RSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPP------- 183
Query: 795 QKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAA 854
+G N+ + P PP K Q P Q + + P P P + Q PPPPP P
Sbjct: 184 PQGGNKPQGPPPPGKPQGPPPQGDKSQ-SPRSPPGKPQGPPPQGGNQPQGPPPPPGKPQG 242
Query: 855 AARARHQAPQ--PPPASPAAAARA--RHQAPQPP---PPSPAAAARARHEGEPAARSASQ 907
+ PQ PPP P + ++PQ P P P + +Q
Sbjct: 243 PPQQGGNRPQGPPPPGKPQGPPPQGDKSRSPQSPPGKPQG------------PPPQGGNQ 290
Query: 908 PKKPAVQPSKPAAQPTK---KPATKP 930
P+ P P KP P + KP P
Sbjct: 291 PQGPPPPPGKPQGPPPQGGNKPQGPP 316
Score = 47.0 bits (110), Expect = 2e-04
Identities = 38/141 (26%), Positives = 49/141 (33%), Gaps = 21/141 (14%)
Query: 791 GRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPA 850
G Q G+ + P PP K Q P Q PQ PP P + Q PPP
Sbjct: 38 GPSPQGGNKPQGPPPPPGKPQGPPPQGG-------NKPQGPPPPG-----KPQGPPPQGD 85
Query: 851 SPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKK 910
+ ++P+ PP P PQ PPP P +G + P K
Sbjct: 86 --------KSRSPRSPPGKPQGPPPQGGNQPQGPPPPPGKPQGPPPQGGNKPQGPPPPGK 137
Query: 911 P-AVQPSKPAAQPTKKPATKP 930
P P +Q + P KP
Sbjct: 138 PQGPPPQGDKSQSPRSPPGKP 158
Score = 45.4 bits (106), Expect = 7e-04
Identities = 30/107 (28%), Positives = 40/107 (37%), Gaps = 16/107 (14%)
Query: 831 PPSPAAAARARHQAPPPPPASPAAAARARHQAPQ--PPPASPAAAARA--RHQAPQPPPP 886
P P+ + Q PPPPP P PQ PPP P + ++P+ PP
Sbjct: 36 PQGPSPQGGNKPQGPPPPPGKPQGPPPQGGNKPQGPPPPGKPQGPPPQGDKSRSPRSPPG 95
Query: 887 SPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTK---KPATKP 930
P P + +QP+ P P KP P + KP P
Sbjct: 96 KPQG---------PPPQGGNQPQGPPPPPGKPQGPPPQGGNKPQGPP 133
>EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precursor
(PELP)
Length = 426
Score = 61.2 bits (147), Expect = 1e-08
Identities = 38/130 (29%), Positives = 52/130 (39%), Gaps = 10/130 (7%)
Query: 802 KCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQ 861
K P PP P+P +P+ + Q PQPPP A+ PPPPP + + A+
Sbjct: 147 KPPPPP----PSPCKPSPPDQSAKQPPQPPP-----AKQPSPPPPPPPVKAPSPSPAKQP 197
Query: 862 APQPPPA-SPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAA 920
P PPP +P+ + + QPPPP A + P A P+ P
Sbjct: 198 PPPPPPVKAPSPSPATQPPTKQPPPPPRAKKSPLLPPPPPVAYPPVMTPSPSPAAEPPII 257
Query: 921 QPTKKPATKP 930
P P P
Sbjct: 258 APFPSPPANP 267
Score = 60.8 bits (146), Expect = 2e-08
Identities = 43/129 (33%), Positives = 49/129 (37%), Gaps = 11/129 (8%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP + P QP A Q PPP P A + A PPP P A + A
Sbjct: 158 PSPPDQSAKQPPQPPPA----KQPSPPPPPPPVKAPSPSPAKQPPPPPPPVKAPSPSPAT 213
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSA------SQPKKPAVQPSK 917
QPP P RA+ PPPP A PAA S P P + P +
Sbjct: 214 QPPTKQPPPPPRAKKSPLLPPPPPVAYPPVMTPSPSPAAEPPIIAPFPSPPANPPLIPRR 273
Query: 918 PAAQPTKKP 926
P A P KP
Sbjct: 274 P-APPVVKP 281
Score = 60.8 bits (146), Expect = 2e-08
Identities = 42/127 (33%), Positives = 51/127 (40%), Gaps = 14/127 (11%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P P K P P P + A QPP P A+ +PPPPP P A + A
Sbjct: 142 PSPLVKPPPPPPSPCKPSPPDQSAKQPPQPPP----AKQPSPPPPP--PPVKAPSPSPAK 195
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPT 923
QPPP P A + A QPP P RA+ +S P P V P P+
Sbjct: 196 QPPPPPPPVKAPSPSPATQPPTKQPPPPPRAK-------KSPLLPPPPPV-AYPPVMTPS 247
Query: 924 KKPATKP 930
PA +P
Sbjct: 248 PSPAAEP 254
Score = 56.6 bits (135), Expect = 3e-07
Identities = 48/181 (26%), Positives = 69/181 (37%), Gaps = 34/181 (18%)
Query: 752 PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQ 811
PP + P P L + P P P+ + ++ P PP KQ
Sbjct: 132 PPVNQPKPSSPSPLVKPPPPPPSPCKPSP---------------PDQSAKQPPQPPPAKQ 176
Query: 812 PTPSQPAAAAIATNQAP--QPPP--------SPAAAARARHQAPPPPPASPAAAARARHQ 861
P+P P A + +P QPPP SP+ A + + PPPPP A ++
Sbjct: 177 PSPPPPPPPVKAPSPSPAKQPPPPPPPVKAPSPSPATQPPTKQPPPPP----RAKKSPLL 232
Query: 862 APQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQ 921
P PP A P + A +PP +P + A P +P P V+P P +
Sbjct: 233 PPPPPVAYPPVMTPSPSPAAEPPIIAPFPSPPANPPLIP-----RRPAPPVVKPLPPLGK 287
Query: 922 P 922
P
Sbjct: 288 P 288
Score = 55.1 bits (131), Expect = 9e-07
Identities = 37/123 (30%), Positives = 47/123 (38%), Gaps = 36/123 (29%)
Query: 807 PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPP 866
P QP PS P+ +PPP PPP P P+ ++ Q PQPP
Sbjct: 132 PPVNQPKPSSPSPLV-------KPPP------------PPPSPCKPSPPDQSAKQPPQPP 172
Query: 867 PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
PA + +P PPPP A P+ A QP P P+ P +P
Sbjct: 173 PA--------KQPSPPPPPPPVKA---------PSPSPAKQPPPPPPPVKAPSPSPATQP 215
Query: 927 ATK 929
TK
Sbjct: 216 PTK 218
Score = 51.6 bits (122), Expect = 1e-05
Identities = 39/155 (25%), Positives = 54/155 (34%), Gaps = 29/155 (18%)
Query: 805 LPPAKKQPTPSQPAAAAIATNQAPQPPPS-------------PAAAARARHQA------- 844
LPP P+P P+ + + +P PP + P ++ A
Sbjct: 61 LPPPSPLPSPPPPSPSPPPPSPSPPPPSTIPLIPPFTGGFLPPLPGSKLPDFAGLLPLIP 120
Query: 845 -----PP----PPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARAR 895
PP PP P ++ + P PPP SP + A QPP P PA
Sbjct: 121 NLPDVPPIGGGPPVNQPKPSSPSPLVKPPPPPPSPCKPSPPDQSAKQPPQPPPAKQPSPP 180
Query: 896 HEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
P + P K P P P+ PAT+P
Sbjct: 181 PPPPPVKAPSPSPAKQPPPPPPPVKAPSPSPATQP 215
Score = 43.1 bits (100), Expect = 0.003
Identities = 47/174 (27%), Positives = 60/174 (34%), Gaps = 60/174 (34%)
Query: 804 PLP---PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARH 860
PLP P + P P P+ T P PPSP + +PPPP SP +
Sbjct: 35 PLPFDWPPAEIPLPDIPSPFDGPTFVLP--PPSPLPSPPPPSPSPPPPSPSPPPPSTIPL 92
Query: 861 QAP----------------------------------------QPPPASPAAAARARHQA 880
P QP P+SP+ +
Sbjct: 93 IPPFTGGFLPPLPGSKLPDFAGLLPLIPNLPDVPPIGGGPPVNQPKPSSPSPLVK----- 147
Query: 881 PQPPPPSPAAAARARHEGEPAARSASQPKK--PAVQPS-KPAAQPTKKPATKPS 931
P PPPPSP + P +SA QP + PA QPS P P K P+ P+
Sbjct: 148 PPPPPPSPC-------KPSPPDQSAKQPPQPPPAKQPSPPPPPPPVKAPSPSPA 194
>IF2_STRCO (Q8CJQ8) Translation initiation factor IF-2
Length = 1033
Score = 60.8 bits (146), Expect = 2e-08
Identities = 54/142 (38%), Positives = 64/142 (45%), Gaps = 23/142 (16%)
Query: 795 QKGHNQRKC--PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASP 852
Q G N R P P K P PS P+ A +QA +PAA RA A P P A+P
Sbjct: 52 QGGGNGRSAGRPAAPKKAAPRPSAPSPAQAGPSQA-----APAAGDRA---AAPRPSAAP 103
Query: 853 AAAARARHQAPQPPPASPAAAARARHQAPQPPP-PSPA---AAARARHEGEPAARSASQP 908
A A + PA+P+A A A Q P+P P P PA A A PAA +A P
Sbjct: 104 KAPAAQQ-------PAAPSAPAPAPSQGPRPTPGPKPAPRPAPAAPEFTAPPAAPAA--P 154
Query: 909 KKPAVQPSKPAAQPTKKPATKP 930
PA PS P + A KP
Sbjct: 155 STPAPAPSGPKPGGARPGAPKP 176
Score = 58.2 bits (139), Expect = 1e-07
Identities = 43/137 (31%), Positives = 57/137 (41%), Gaps = 8/137 (5%)
Query: 784 GGSSYTCGRCHQKGHNQRKCPLP--PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARAR 841
GG+ + GR ++ P P P+ Q PSQ A AA AP+P +P A A +
Sbjct: 54 GGNGRSAGR---PAAPKKAAPRPSAPSPAQAGPSQAAPAAGDRAAAPRPSAAPKAPAAQQ 110
Query: 842 HQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPA 901
AP P +P+ R P P PA A A AP P +P+ A A +P
Sbjct: 111 PAAPSAPAPAPSQGPRP---TPGPKPAPRPAPAAPEFTAPPAAPAAPSTPAPAPSGPKPG 167
Query: 902 ARSASQPKKPAVQPSKP 918
PK +PS P
Sbjct: 168 GARPGAPKPGGARPSGP 184
Score = 49.7 bits (117), Expect = 4e-05
Identities = 47/175 (26%), Positives = 67/175 (37%), Gaps = 24/175 (13%)
Query: 757 RGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAK-KQPTPS 815
R GRP ++ P P +P + ++ G ++ P P A K P
Sbjct: 58 RSAGRPAAPKKAAP-RPSAPSPAQAGPSQAAPAAG-------DRAAAPRPSAAPKAPAAQ 109
Query: 816 QPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAAR 875
QPAA + P P+P+ R P P PA A A AP PA+P+ A
Sbjct: 110 QPAAPSA-------PAPAPSQGPR---PTPGPKPAPRPAPAAPEFTAPPAAPAAPSTPAP 159
Query: 876 ARHQAPQPPPPSPAA----AARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
A P+P P A AR G+ + Q + +P PA +P +P
Sbjct: 160 A-PSGPKPGGARPGAPKPGGARPSGPGQDRGQQGGQGRPGGQRPGAPAQRPGGRP 213
Score = 43.9 bits (102), Expect = 0.002
Identities = 37/99 (37%), Positives = 46/99 (46%), Gaps = 24/99 (24%)
Query: 834 PAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAAR 893
PAA +A AP P SPA A P A+PAA RA P P+AA +
Sbjct: 63 PAAPKKA---APRPSAPSPAQAG--------PSQAAPAAGDRA-------AAPRPSAAPK 104
Query: 894 ARHEGEPAARSASQPKKPAVQPSK-PAAQPTKKPATKPS 931
A PAA+ + P PA PS+ P P KPA +P+
Sbjct: 105 A-----PAAQQPAAPSAPAPAPSQGPRPTPGPKPAPRPA 138
Score = 37.7 bits (86), Expect = 0.14
Identities = 34/119 (28%), Positives = 41/119 (33%), Gaps = 21/119 (17%)
Query: 829 QPPPSPAAAARARHQAPPP---PPASPAAAARARHQAPQP--------PPASPAAAARA- 876
Q P +PA R P P P S A AR AP+P P +P R
Sbjct: 200 QRPGAPAQRPGGRPGGPRPGNNPFTSGGNAGMARPSAPRPQGGPRPGGPGGAPGGGPRPQ 259
Query: 877 ---------RHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
R QAP PSP + R + G P + P +PAA P P
Sbjct: 260 GPGGQGGGPRPQAPGGNRPSPGSMPRPQGGGAGPRPGGGPRPNPGMMPQRPAAGPRPGP 318
>IF2_STRAW (Q82K53) Translation initiation factor IF-2
Length = 1046
Score = 60.8 bits (146), Expect = 2e-08
Identities = 57/202 (28%), Positives = 75/202 (36%), Gaps = 51/202 (25%)
Query: 751 LPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKK 810
L +++G G R+ P +P +P + R + P PPA K
Sbjct: 46 LTDALQQGNGGKAAPRKAAPAKPGAPSPAQAARPAA-----------------PRPPAPK 88
Query: 811 QPTPSQPAAAA--IATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPA 868
+PAAA A AP P P P A P P PA+PA AA AP PA
Sbjct: 89 PAAAERPAAAERPAAAPAAPGPRPGPKPA-------PRPAPAAPAPAA-PEFTAPPSAPA 140
Query: 869 SPAAAARARHQAPQPPPPSPAAAARA----------------------RHEGEPAARSAS 906
+PAAAA +P P P A A R + + AAR
Sbjct: 141 APAAAASGPRPGARPGAPKPGGARPATPGQGQGGQVRGERTDRGDRGDRGDRQGAARPGG 200
Query: 907 QPKKPAVQPSKPAAQPTKKPAT 928
Q +P +P+ P +P P T
Sbjct: 201 QAPRPGARPAGP--RPGNNPFT 220
Score = 50.4 bits (119), Expect = 2e-05
Identities = 32/82 (39%), Positives = 43/82 (52%), Gaps = 18/82 (21%)
Query: 850 ASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPK 909
A+P AA A+ P +P+ A AR AP+PP P PAAA R PAA +
Sbjct: 58 AAPRKAAPAK-------PGAPSPAQAARPAAPRPPAPKPAAAER------PAA-----AE 99
Query: 910 KPAVQPSKPAAQPTKKPATKPS 931
+PA P+ P +P KPA +P+
Sbjct: 100 RPAAAPAAPGPRPGPKPAPRPA 121
Score = 47.8 bits (112), Expect = 1e-04
Identities = 41/111 (36%), Positives = 48/111 (42%), Gaps = 26/111 (23%)
Query: 827 APQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAAR--ARHQAPQP- 883
AP P +P+ A AR AP PP PAAA R PAAA R A AP P
Sbjct: 64 APAKPGAPSPAQAARPAAPRPPAPKPAAAER------------PAAAERPAAAPAAPGPR 111
Query: 884 ----PPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
P P PA AA A PAA + P P+ A+ P +P +P
Sbjct: 112 PGPKPAPRPAPAAPA-----PAAPEFTAPPSAPAAPAAAASGP--RPGARP 155
Score = 33.5 bits (75), Expect = 2.7
Identities = 38/158 (24%), Positives = 48/158 (30%), Gaps = 15/158 (9%)
Query: 756 KRGPGRPKKLRRREPDEPDKKNP--TKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPT 813
++G RP R P P GGS+ G + P P P
Sbjct: 192 RQGAARPGGQAPRPGARPAGPRPGNNPFTSGGST---GMARPQAPRPGGAPRPGGAGAPG 248
Query: 814 PSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAA 873
+P A ++AP+P P A P P P A P+P P A
Sbjct: 249 GPRPQGAG-GQDRAPRPQGGPGGA--------PRPQGGPGGARPTPGGMPRPQAPRPGGA 299
Query: 874 ARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP 911
P P PAA R G P R +P
Sbjct: 300 PGGNRPNPGMMPQRPAAGPRP-GGGGPGGRGPGGGGRP 336
>SEPA_EMENI (P78621) Cytokinesis protein sepA (FH1/2 protein) (Forced
expression inhibition of growth A)
Length = 1790
Score = 59.7 bits (143), Expect = 4e-08
Identities = 38/134 (28%), Positives = 45/134 (33%), Gaps = 22/134 (16%)
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQP 865
PP P P+ P + A P PPP P A A PPPPP P P P
Sbjct: 1016 PPPPPPPPPAHPGLSGAAPPPPPPPPPPPPGAGAAPPPPPPPPPPPPGGLGGPPPPPPPP 1075
Query: 866 PPAS-----PAAAARARHQAPQPPPPSPAAAA-----------------RARHEGEPAAR 903
PP P P PPPP P A RA + A
Sbjct: 1076 PPGGFGGPPPPPPPPGGFGGPPPPPPPPPGGAFGVPPPPPPPGTVIGGWRANYLASQGAP 1135
Query: 904 SASQPKKPAVQPSK 917
S + P +++P K
Sbjct: 1136 SHAIPVMSSIRPKK 1149
Score = 56.2 bits (134), Expect = 4e-07
Identities = 33/104 (31%), Positives = 33/104 (31%)
Query: 823 ATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQ 882
AT P PPP P A APPPPP P A P PPP P P
Sbjct: 1012 ATAAPPPPPPPPPAHPGLSGAAPPPPPPPPPPPPGAGAAPPPPPPPPPPPPGGLGGPPPP 1071
Query: 883 PPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
PPPP P P P P P P P
Sbjct: 1072 PPPPPPGGFGGPPPPPPPPGGFGGPPPPPPPPPGGAFGVPPPPP 1115
Score = 52.4 bits (124), Expect = 6e-06
Identities = 30/80 (37%), Positives = 34/80 (42%), Gaps = 2/80 (2%)
Query: 811 QPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPA--AAARARHQAPQPPPA 868
+PT S+ A AT Q A +A PPPPP PA + A P PPP
Sbjct: 983 KPTESEQPAEGAATKGDEQGVDDTVAVDKATAAPPPPPPPPPAHPGLSGAAPPPPPPPPP 1042
Query: 869 SPAAAARARHQAPQPPPPSP 888
P A A P PPPP P
Sbjct: 1043 PPPGAGAAPPPPPPPPPPPP 1062
Score = 38.5 bits (88), Expect = 0.084
Identities = 26/82 (31%), Positives = 28/82 (33%), Gaps = 5/82 (6%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP P P P PPP P PPPPP P A P
Sbjct: 1059 PPPPGGLGGPPPPPPPPPPGGFGGPPPPPPPPGGFGG---PPPPPPPPPGGAFGV--PPP 1113
Query: 864 QPPPASPAAAARARHQAPQPPP 885
PPP + RA + A Q P
Sbjct: 1114 PPPPGTVIGGWRANYLASQGAP 1135
Score = 33.9 bits (76), Expect = 2.1
Identities = 25/91 (27%), Positives = 30/91 (32%), Gaps = 8/91 (8%)
Query: 840 ARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGE 899
A+ + P PA A + + A +A P PPPP PA H G
Sbjct: 978 AKDEGKPTESEQPAEGAATK--GDEQGVDDTVAVDKATAAPPPPPPPPPA------HPGL 1029
Query: 900 PAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
A P P P AA P P P
Sbjct: 1030 SGAAPPPPPPPPPPPPGAGAAPPPPPPPPPP 1060
>PMP3_MOUSE (P05143) Proline-rich protein MP-3 (Fragment)
Length = 296
Score = 59.3 bits (142), Expect = 5e-08
Identities = 58/207 (28%), Positives = 74/207 (35%), Gaps = 31/207 (14%)
Query: 735 PINGENKWPKM-SQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRC 793
P+NG + P PP+ PG P+ + P P P + G
Sbjct: 23 PVNGSQQGPPPPGGPQPRPPQGPPPPGGPQPRPPQGPPPPGGPQPRPPQ--------GPP 74
Query: 794 HQKGHNQR--KCPLPPAKKQPTPSQ-PAAAAIATNQAPQPPPSPAAAARARHQAPPPP-- 848
G R + P PP QP P Q P + PQ PP P + Q PPPP
Sbjct: 75 PPGGPQPRPPQGPPPPGGPQPRPPQGPPPPGGPQQRPPQGPPPPGGPQQRPPQGPPPPGG 134
Query: 849 -----PASPAAAARARHQAPQ--PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPA 901
P P A + + PQ PPPA P H P PP P + R+ P
Sbjct: 135 PQPRPPQGPPPPAGPQPRPPQGPPPPAGP-------HLRPTQGPP-PTGGPQQRYPQSPP 186
Query: 902 ARSASQPKKP--AVQPSKPAAQPTKKP 926
QP+ P P P +PT+ P
Sbjct: 187 PPGGPQPRPPQGPPPPGGPHPRPTQGP 213
Score = 53.9 bits (128), Expect = 2e-06
Identities = 42/131 (32%), Positives = 49/131 (37%), Gaps = 8/131 (6%)
Query: 804 PLPPAKKQPTPSQ-PAAAAIATNQAPQPPPSPAAAARARHQAPPPP--PASPAAAARARH 860
P PP QP P Q P + PQ PP P Q PPPP P
Sbjct: 31 PPPPGGPQPRPPQGPPPPGGPQPRPPQGPPPPGGPQPRPPQGPPPPGGPQPRPPQGPPPP 90
Query: 861 QAPQP-PPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA 919
PQP PP P + + PQ PPP P + +G P QP+ P P P
Sbjct: 91 GGPQPRPPQGPPPPGGPQQRPPQGPPP-PGGPQQRPPQGPPPP-GGPQPRPPQGPP--PP 146
Query: 920 AQPTKKPATKP 930
A P +P P
Sbjct: 147 AGPQPRPPQGP 157
Score = 53.5 bits (127), Expect = 3e-06
Identities = 37/128 (28%), Positives = 45/128 (34%), Gaps = 7/128 (5%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PPA QP P Q + P P P + R+ PPPP P R P
Sbjct: 143 PPPPAGPQPRPPQGPPPPAGPHLRPTQGPPPTGGPQQRYPQSPPPPGGP--QPRPPQGPP 200
Query: 864 QPPPASPAAAARARHQAPQPPP---PSPAAAARARHEGEPAARSASQPKKP--AVQPSKP 918
P P PQP P P P + R P QP+ P P+ P
Sbjct: 201 PPGGPHPRPTQGPPPTGPQPRPTQGPPPTGGPQQRPPQGPPPPGGPQPRPPQGPPPPTGP 260
Query: 919 AAQPTKKP 926
+PT+ P
Sbjct: 261 QPRPTQGP 268
Score = 49.3 bits (116), Expect = 5e-05
Identities = 36/136 (26%), Positives = 49/136 (35%), Gaps = 18/136 (13%)
Query: 800 QRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPP-------PASP 852
Q + +P + P+ SQP + Q P PP P Q PPPP P P
Sbjct: 3 QNQIQIPNQRPPPSGSQPRPPVNGSQQGPPPPGGPQPRPP---QGPPPPGGPQPRPPQGP 59
Query: 853 AAAARARHQAPQ--PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKK 910
+ + PQ PPP P + + PQ PPP R P +P +
Sbjct: 60 PPPGGPQPRPPQGPPPPGGP------QPRPPQGPPPPGGPQPRPPQGPPPPGGPQQRPPQ 113
Query: 911 PAVQPSKPAAQPTKKP 926
P P +P + P
Sbjct: 114 GPPPPGGPQQRPPQGP 129
Score = 43.9 bits (102), Expect = 0.002
Identities = 44/174 (25%), Positives = 51/174 (29%), Gaps = 22/174 (12%)
Query: 753 PEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQR--KCPLPPAKK 810
P +GP P + R P P L+ T G G QR + P PP
Sbjct: 137 PRPPQGPPPPAGPQPRPPQGPPPPAGPHLRP-----TQGPPPTGGPQQRYPQSPPPPGGP 191
Query: 811 QPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPP--PPASPAAAARARHQAPQPPPA 868
QP P PQ PP P Q PPP P P Q PP
Sbjct: 192 QPRP-------------PQGPPPPGGPHPRPTQGPPPTGPQPRPTQGPPPTGGPQQRPPQ 238
Query: 869 SPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
P + + PQ PPP R P P P +P
Sbjct: 239 GPPPPGGPQPRPPQGPPPPTGPQPRPTQGPHPTGGPQQTPPLAGNPQGPPPGRP 292
>CAPU_DROME (Q24120) Cappuccino protein
Length = 1059
Score = 59.3 bits (142), Expect = 5e-08
Identities = 37/115 (32%), Positives = 45/115 (38%), Gaps = 15/115 (13%)
Query: 812 PTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPA 871
P P P A P PPP P A + APPPPP P + AP PPP +P
Sbjct: 487 PPPPPPPLHAFVAPPPPPPPPPPPPPPLANYGAPPPPPPPPPGSG----SAPPPPPPAPI 542
Query: 872 AAARARHQAPQPPPPSPAAAARARHEGEPAARSA--------SQPKKPAVQPSKP 918
P PPPP A+ ++ P A + +K AV P KP
Sbjct: 543 EGGGG---IPPPPPPMSASPSKTTISPAPLPDPAEGNWFHRTNTMRKSAVNPPKP 594
Score = 55.5 bits (132), Expect = 7e-07
Identities = 32/98 (32%), Positives = 39/98 (39%), Gaps = 7/98 (7%)
Query: 827 APQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPP 886
AP PPP P PPPPP P A + AP PPP P + A PPPP
Sbjct: 484 APPPPPPPPPLHAFVAPPPPPPPPPPPPPPLANYGAPPPPPPPPPGSGSA------PPPP 537
Query: 887 SPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTK 924
PA P ++ P K + P+ P P +
Sbjct: 538 PPAPIEGGGGIPPPPPPMSASPSKTTISPA-PLPDPAE 574
Score = 49.7 bits (117), Expect = 4e-05
Identities = 43/146 (29%), Positives = 52/146 (35%), Gaps = 25/146 (17%)
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP P P + AP PPP PA PPPPP S + + AP
Sbjct: 510 PPPPLANYGAPPPPPPPPPGSGSAPPPPP-PAPIEGGGGIPPPPPPMSASPSKTTISPAP 568
Query: 864 QPPPA----------------SPAAAAR----ARHQAPQPPPPSPAAAARARHEGEPAAR 903
P PA +P R R PP P P + A + E +
Sbjct: 569 LPDPAEGNWFHRTNTMRKSAVNPPKPMRPLYWTRIVTSAPPAPRPPSVANSTDSTENSGS 628
Query: 904 SASQPKKPAVQPSKPAAQPTKKPATK 929
S +P PA + A PT PATK
Sbjct: 629 SPDEP--PAANGAD--APPTAPPATK 650
Score = 48.5 bits (114), Expect = 8e-05
Identities = 30/88 (34%), Positives = 33/88 (37%), Gaps = 6/88 (6%)
Query: 844 APPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAAR 903
APPPPP P A P PPP P A + AP PPPP P + A PA
Sbjct: 484 APPPPPPPPPLHAFVAPPPPPPPPPPPPPPL-ANYGAPPPPPPPPPGSGSAPPPPPPAPI 542
Query: 904 SAS-----QPKKPAVQPSKPAAQPTKKP 926
P + PSK P P
Sbjct: 543 EGGGGIPPPPPPMSASPSKTTISPAPLP 570
Score = 33.9 bits (76), Expect = 2.1
Identities = 22/66 (33%), Positives = 24/66 (36%), Gaps = 9/66 (13%)
Query: 862 APQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQ 921
AP PPP P A P PPPP P P A + P P P +A
Sbjct: 484 APPPPPPPPPLHAFVA-PPPPPPPPPPPP--------PPLANYGAPPPPPPPPPGSGSAP 534
Query: 922 PTKKPA 927
P PA
Sbjct: 535 PPPPPA 540
>WAS1_HUMAN (Q92558) Wiskott-Aldrich syndrome protein family member
1 (WASP-family protein member 1) (Verprolin homology
domain-containing protein 1)
Length = 559
Score = 58.5 bits (140), Expect = 8e-08
Identities = 50/171 (29%), Positives = 63/171 (36%), Gaps = 44/171 (25%)
Query: 804 PLPPAKKQPTPSQPAAAAI-----ATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARA 858
P PP P PS + +++ +T P PPP P A + A PPPPA A
Sbjct: 322 PTPPPPPPPLPSALSTSSLRASMTSTPPPPVPPPPPPPATALQAPAVPPPPAPLQIAPGV 381
Query: 859 RHQAPQP--PP----------ASPAAAARARHQAPQ------PPPPSP------------ 888
H AP P PP A+P H PQ PPPP P
Sbjct: 382 LHPAPPPIAPPLVQPSPPVARAAPVCETVPVHPLPQGEVQGLPPPPPPPPLPPPGIRPSS 441
Query: 889 ----AAAARARHEGEPAARSASQPKKPAVQPSK-----PAAQPTKKPATKP 930
A A P +A P P + PS PA++P + P+T P
Sbjct: 442 PVTVTALAHPPSGLHPTPSTAPGPHVPLMPPSPPSQVIPASEPKRHPSTLP 492
Score = 42.4 bits (98), Expect = 0.006
Identities = 29/96 (30%), Positives = 36/96 (37%), Gaps = 8/96 (8%)
Query: 842 HQAPPPPPASPAAAAR-------ARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARA 894
H+ PPPPP A A+ + + P SPA +P PPPP P + A
Sbjct: 276 HEPPPPPPMHGAGDAKPIPTCISSATGLIENRPQSPATGRTPVFVSPTPPPPPPPLPS-A 334
Query: 895 RHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
A S P P P P A + PA P
Sbjct: 335 LSTSSLRASMTSTPPPPVPPPPPPPATALQAPAVPP 370
Score = 39.7 bits (91), Expect = 0.038
Identities = 35/130 (26%), Positives = 43/130 (32%), Gaps = 13/130 (10%)
Query: 804 PLPPAKKQPTPSQPAAAAI--ATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQ 861
P PP ++P I AT P SPA +P PPP P
Sbjct: 279 PPPPPMHGAGDAKPIPTCISSATGLIENRPQSPATGRTPVFVSPTPPPPPPPL------- 331
Query: 862 APQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQ 921
P A ++ RA + PPP P A PA P + A PA
Sbjct: 332 ----PSALSTSSLRASMTSTPPPPVPPPPPPPATALQAPAVPPPPAPLQIAPGVLHPAPP 387
Query: 922 PTKKPATKPS 931
P P +PS
Sbjct: 388 PIAPPLVQPS 397
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 120,690,211
Number of Sequences: 164201
Number of extensions: 5884663
Number of successful extensions: 71442
Number of sequences better than 10.0: 1924
Number of HSP's better than 10.0 without gapping: 622
Number of HSP's successfully gapped in prelim test: 1400
Number of HSP's that attempted gapping in prelim test: 32201
Number of HSP's gapped (non-prelim): 13846
length of query: 931
length of database: 59,974,054
effective HSP length: 120
effective length of query: 811
effective length of database: 40,269,934
effective search space: 32658916474
effective search space used: 32658916474
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0016.18


