

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0013.6
(937 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
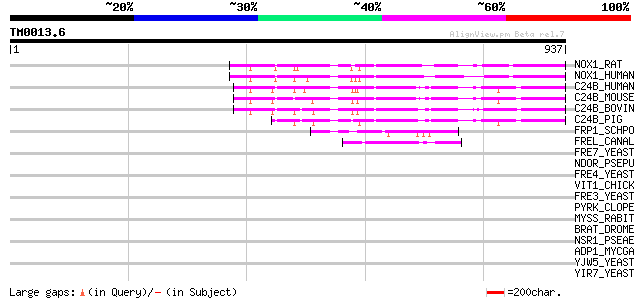
NOX1_RAT (Q9WV87) NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NADH/... 263 2e-69
NOX1_HUMAN (Q9Y5S8) NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NAD... 262 4e-69
C24B_HUMAN (P04839) Cytochrome B-245 heavy chain (P22 phagocyte ... 258 5e-68
C24B_MOUSE (Q61093) Cytochrome B-245 heavy chain (P22 phagocyte ... 254 1e-66
C24B_BOVIN (O46522) Cytochrome B-245 heavy chain (P22 phagocyte ... 252 4e-66
C24B_PIG (P52649) Cytochrome B-245 heavy chain (P22 phagocyte B-... 218 8e-56
FRP1_SCHPO (Q04800) Ferric reductase transmembrane component (EC... 62 9e-09
FREL_CANAL (P78588) Probable ferric reductase transmembrane comp... 50 2e-05
FRE7_YEAST (Q12333) Ferric reductase transmembrane component 7 (... 45 0.001
NDOR_PSEPU (Q52126) Naphthalene 1,2-dioxygenase system ferredoxi... 43 0.004
FRE4_YEAST (P53746) Ferric reductase transmembrane component 4 p... 43 0.004
VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin... 39 0.084
FRE3_YEAST (Q08905) Ferric reductase transmembrane component 3 p... 39 0.084
PYRK_CLOPE (Q8XL63) Dihydroorotate dehydrogenase electron transf... 38 0.11
MYSS_RABIT (P02562) Myosin heavy chain, skeletal muscle (Fragments) 37 0.25
BRAT_DROME (Q8MQJ9) Brain tumor protein 37 0.32
NSR1_PSEAE (Q9HYL3) Regulatory protein nosR 35 0.71
ADP1_MYCGA (Q49379) Adhesin P1 precursor (Cytadhesin P1) (Attach... 35 0.71
YJW5_YEAST (P40889) Hypothetical 197.6 kDa protein in FSP2 5'region 35 0.93
YIR7_YEAST (P40434) Hypothetical 197.5 kDa protein in SDL1 5'region 35 0.93
>NOX1_RAT (Q9WV87) NADPH oxidase homolog 1 (NOX-1) (NOH-1)
(NADH/NADPH mitogenic oxidase subunit P65-MOX)
(Mitogenic oxidase 1) (MOX1)
Length = 563
Score = 263 bits (671), Expect = 2e-69
Identities = 198/633 (31%), Positives = 275/633 (43%), Gaps = 142/633 (22%)
Query: 372 LEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSF----QVMGYCLPVAKGAAETLKLNMA 427
+ +W L WL LF + F Y + +++G L +A+ +A L N
Sbjct: 6 VNHWLSVLFLVSWLGLNIFLFVYVFLNYEKSDKYYYTREILGTALALARASALCLNFNSM 65
Query: 428 LILLPVCRNTLTWLRSTRA------RKIVPFDDNINFHKIIACAIVVGVVVHAGNHL--- 478
+IL+PVCRN L++LR T + RK P D N+ FHK++A I + +H HL
Sbjct: 66 VILIPVCRNLLSFLRGTCSFCNHTLRK--PLDHNLTFHKLVAYMICIFTAIHIIAHLFNF 123
Query: 479 --------ACDFPL------LINSSPEEFSLIASDFNNKKPTYKTLLTGVEGVTGISMVT 524
A D L L + E+ L N Y T + G+TG+
Sbjct: 124 ERYSRSQQAMDGSLASVLSSLFHPEKEDSWLNPIQSPNVTVMYAAF-TSIAGLTGVVATV 182
Query: 525 LMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHG--------- 575
+ + T A RRN F FWY+HHL + I L IHG
Sbjct: 183 ALVLMVTSAMEFIRRNY------------FELFWYTHHLFIIYIICLGIHGLGGIVRGQT 230
Query: 576 --------------SFLNLTHRW--YNKT------------TWMYISVPLLLYIAERTLR 607
SF H W Y ++ +W +I P+ YI ER LR
Sbjct: 231 EESMSESHPRNCSYSF----HEWDKYERSCRSPHFVGQPPESWKWILAPIAFYIFERILR 286
Query: 608 TSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSA 667
RS+ V I KV + P V L M K GF GQYIF+ CP IS EWHPF++TSA
Sbjct: 287 FYRSRQKVV-ITKVVMHPCKVLELQMRK-RGFTMGIGQYIFVNCPSISFLEWHPFTLTSA 344
Query: 668 PGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGA 727
P +E+ S+HIR GDWT+ L F + P PR+ VDGP+G
Sbjct: 345 PEEEFFSIHIRAAGDWTENLIRTFEQQHSP------------------MPRIEVDGPFGT 386
Query: 728 PAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTS 787
++D ++V +L+G GIG TPF S L+ + W F
Sbjct: 387 VSEDVFQYEVAVLVGAGIGVTPFASFLKSI------------------------WYKFQR 422
Query: 788 SNLTPGGNKRSQRTTNAYFYWVTREPGSFEWFKGVMDEV-AEMDHRGQIELHNYLTSVYE 846
++ +T YFYW+ RE G+F WF +++ + EMD G+ + NY +
Sbjct: 423 AH-------NKLKTQKIYFYWICRETGAFAWFNNLLNSLEQEMDELGKPDFLNYRLFL-- 473
Query: 847 EGDARSTLITMIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYC 906
+ I ALN + D+L+G + +T F RP W F +IA+ HP S VGVF C
Sbjct: 474 --TGWDSNIAGHAALNFDR-ATDVLTGLKQKTSFGRPMWDNEFSRIATAHPKSVVGVFLC 530
Query: 907 GMPVLAKELKKLSLELS--HKTTTRFDFHKEYF 937
G P LAK L+K S +F F+KE F
Sbjct: 531 GPPTLAKSLRKCCRRYSSLDPRKVQFYFNKETF 563
>NOX1_HUMAN (Q9Y5S8) NADPH oxidase homolog 1 (NOX-1) (NOH-1)
(NADH/NADPH mitogenic oxidase subunit P65-MOX)
(Mitogenic oxidase 1) (MOX1)
Length = 564
Score = 262 bits (669), Expect = 4e-69
Identities = 201/631 (31%), Positives = 285/631 (44%), Gaps = 137/631 (21%)
Query: 372 LEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSF----QVMGYCLPVAKGAAETLKLNMA 427
+ +W L +WL LF F +Y + +++G L A+ +A L N
Sbjct: 6 VNHWFSVLFLVVWLGLNVFLFVDAFLKYEKADKYYYTRKILGSTLACARASALCLNFNST 65
Query: 428 LILLPVCRNTLTWLRSTRA------RKIVPFDDNINFHKIIACAIVVGVVVHAGNHL--- 478
LILLPVCRN L++LR T + RK D N+ FHK++A I + +H HL
Sbjct: 66 LILLPVCRNLLSFLRGTCSFCSRTLRK--QLDHNLTFHKLVAYMICLHTAIHIIAHLFNF 123
Query: 479 --------ACDFPL--LINSSPEEFSLIASDFN-----NKKPTYKTLLTGVEGVTGISMV 523
A D L +++S + S N N Y T T + G+TG+ M
Sbjct: 124 DCYSRSRQATDGSLASILSSLSHDEKKGGSWLNPIQSRNTTVEYVTF-TSIAGLTGVIMT 182
Query: 524 TLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYIL-LFIHG------- 575
+ + T AT RR+ F FWY+HHL + YIL L IHG
Sbjct: 183 IALILMVTSATEFIRRSY------------FEVFWYTHHLF-IFYILGLGIHGIGGIVRG 229
Query: 576 ---SFLNLTH---------RWYNKTT--------------WMYISVPLLLYIAERTLRTS 609
+N +H W ++ + W +I P++LYI ER LR
Sbjct: 230 QTEESMNESHPRKCAESFEMWDDRDSHCRRPKFEGHPPESWKWILAPVILYICERILRFY 289
Query: 610 RSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPG 669
RS+ V I KV + P V L M+K GF + GQYIF+ CP IS EWHPF++TSAP
Sbjct: 290 RSQQKVV-ITKVVMHPSKVLELQMNK-RGFSMEVGQYIFVNCPSISLLEWHPFTLTSAPE 347
Query: 670 DEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPA 729
+++ S+HIR GDWT+ L F E P+ PR+ VDGP+G +
Sbjct: 348 EDFFSIHIRAAGDWTENLIRAF-EQQYSPI-----------------PRIEVDGPFGTAS 389
Query: 730 QDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSN 789
+D ++V +L+G GIG TPF SIL+ + + +D ++
Sbjct: 390 EDVFQYEVAVLVGAGIGVTPFASILKSIWYKFQCADHNL--------------------- 428
Query: 790 LTPGGNKRSQRTTNAYFYWVTREPGSFEWFKGVMDEV-AEMDHRGQIELHNYLTSVYEEG 848
+T YFYW+ RE G+F WF ++ + EM+ G++ NY +
Sbjct: 429 ----------KTKKIYFYWICRETGAFSWFNNLLTSLEQEMEELGKVGFLNYRLFL---- 474
Query: 849 DARSTLITMIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGM 908
+ I ALN K DI++G + +T F RP W F IA+ HP S VGVF CG
Sbjct: 475 TGWDSNIVGHAALNFDK-ATDIVTGLKQKTSFGRPMWDNEFSTIATSHPKSVVGVFLCGP 533
Query: 909 PVLAKELKKLSLELS--HKTTTRFDFHKEYF 937
LAK L+K S +F F+KE F
Sbjct: 534 RTLAKSLRKCCHRYSSLDPRKVQFYFNKENF 564
>C24B_HUMAN (P04839) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 569
Score = 258 bits (659), Expect = 5e-68
Identities = 190/625 (30%), Positives = 284/625 (45%), Gaps = 133/625 (21%)
Query: 379 WVLFLWLVTTASLFAWKFYQYRNRTSF----QVMGYCLPVAKGAAETLKLNMALILLPVC 434
+V+ +WL LF W + Y F +++G L +A+ A L N LILLPVC
Sbjct: 12 FVILVWLGLNVFLFVWYYRVYDIPPKFFYTRKLLGSALALARAPAACLNFNCMLILLPVC 71
Query: 435 RNTLTWLR------STRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLA----CDFPL 484
RN L++LR STR R+ D N+ FHK++A I + +H HL C
Sbjct: 72 RNLLSFLRGSSACCSTRVRR--QLDRNLTFHKMVAWMIALHSAIHTIAHLFNVEWCVNAR 129
Query: 485 LINSSPEEFSL--------------IASDFNNKKPTYKTLLTGVEGVTGISMVTLMAISF 530
+ NS P +L N + +T + G+TG+ + + +
Sbjct: 130 VNNSDPYSVALSELGDRQNESYLNFARKRIKNPEGGLYLAVTLLAGITGVVITLCLILII 189
Query: 531 TLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGS-------------F 577
T +T RR+ F FWY+HHL + +I L IHG+
Sbjct: 190 TSSTKTIRRSY------------FEVFWYTHHLFVIFFIGLAIHGAERIVRGQTAESLAV 237
Query: 578 LNLT------HRWYN-------------KTTWMYISVPLLLYIAERTLRTSRSKHYAVKI 618
N+T W TW +I P+ LY+ ER +R RS+ V I
Sbjct: 238 HNITVCEQKISEWGKIKECPIPQFAGNPPMTWKWIVGPMFLYLCERLVRFWRSQQKVV-I 296
Query: 619 QKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIR 678
KV P L M K GFK + GQYIF++CPK+S EWHPF++TSAP +++ S+HIR
Sbjct: 297 TKVVTHPFKTIELQMKK-KGFKMEVGQYIFVKCPKVSKLEWHPFTLTSAPEEDFFSIHIR 355
Query: 679 TVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVL 738
VGDWT+ LF C E D + P++ VDGP+G ++D +++V+
Sbjct: 356 IVGDWTEG---LFNA-------CGCDKQEF--QDAWKLPKIAVDGPFGTASEDVFSYEVV 403
Query: 739 LLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNKRS 798
+L+G GIG TPF SIL+ + W + + N +
Sbjct: 404 MLVGAGIGVTPFASILKSV------------------------WYKYCN-------NATN 432
Query: 799 QRTTNAYFYWVTREPGSFEWFKGVM----DEVAEMDHRGQIELHNYLTSVYEEGDARSTL 854
+ YFYW+ R+ +FEWF ++ ++ E ++ G + + YLT ++E A
Sbjct: 433 LKLKKIYFYWLCRDTHAFEWFADLLQLLESQMQERNNAGFLSYNIYLTG-WDESQANHFA 491
Query: 855 ITMIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKE 914
+ H D+++G + +T + RPNW F IAS+HP + +GVF CG LA+
Sbjct: 492 V-------HHDEEKDVITGLKQKTLYGRPNWDNEFKTIASQHPNTRIGVFLCGPEALAET 544
Query: 915 LKKLSLELSHK--TTTRFDFHKEYF 937
L K S+ S F F+KE F
Sbjct: 545 LSKQSISNSESGPRGVHFIFNKENF 569
>C24B_MOUSE (Q61093) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 570
Score = 254 bits (648), Expect = 1e-66
Identities = 188/626 (30%), Positives = 286/626 (45%), Gaps = 135/626 (21%)
Query: 379 WVLFLWLVTTASLFAWKFYQYRNRTSF----QVMGYCLPVAKGAAETLKLNMALILLPVC 434
+V+ +WL LF + Y + + +++G L +A+ A L N LILLPVC
Sbjct: 13 FVILVWLGLNVFLFINYYKVYDDGPKYNYTRKLLGSALALARAPAACLNFNCMLILLPVC 72
Query: 435 RNTLTWLR------STRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINS 488
RN L++LR STR R+ D N+ FHK++A I + +H HL + +N+
Sbjct: 73 RNLLSFLRGSSACCSTRIRR--QLDRNLTFHKMVAWMIALHTAIHTIAHLF-NVEWCVNA 129
Query: 489 S---PEEFSLIASDF-NNKKPTYKTL---------------LTGVEGVTGISMVTLMAIS 529
+ +S+ SD +N+ Y +T + G+TGI + + +
Sbjct: 130 RVGISDRYSIALSDIGDNENEEYLNFAREKIKNPEGGLYVAVTRLAGITGIVITLCLILI 189
Query: 530 FTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGS------------- 576
T +T RR+ F FWY+HHL + +I L IHG+
Sbjct: 190 ITSSTKTIRRSY------------FEVFWYTHHLFVIFFIGLAIHGAERIVRGQTAESLE 237
Query: 577 ------FLNLTHRWYN-------------KTTWMYISVPLLLYIAERTLRTSRSKHYAVK 617
+ W TW +I P+ LY+ ER +R RS+ V
Sbjct: 238 EHNLDICADKIEEWGKIKECPVPKFAGNPPMTWKWIVGPMFLYLCERLVRFWRSQQKVV- 296
Query: 618 IQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHI 677
I KV P L M K GFK + GQYIF++CPK+S EWHPF++TSAP +++ S+HI
Sbjct: 297 ITKVVTHPFKTIELQMKK-KGFKMEVGQYIFVKCPKVSKLEWHPFTLTSAPEEDFFSIHI 355
Query: 678 RTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDV 737
R VGDWT+ LF C E D + P++ VDGP+G ++D +++V
Sbjct: 356 RIVGDWTEG---LFNA-------CGCDKQEF--QDAWKLPKIAVDGPFGTASEDVFSYEV 403
Query: 738 LLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNKR 797
++L+G GIG TPF SIL+ + W + N
Sbjct: 404 VMLVGAGIGVTPFASILKSV------------------------WYKYCD-------NAT 432
Query: 798 SQRTTNAYFYWVTREPGSFEWFKGVMD----EVAEMDHRGQIELHNYLTSVYEEGDARST 853
S + YFYW+ R+ +FEWF ++ ++ E ++ + + YLT ++E A
Sbjct: 433 SLKLKKIYFYWLCRDTHAFEWFADLLQLLETQMQERNNANFLSYNIYLTG-WDESQANHF 491
Query: 854 LITMIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAK 913
+ H D+++G + +T + RPNW F IAS+HP +T+GVF CG LA+
Sbjct: 492 AV-------HHDEEKDVITGLKQKTLYGRPNWDNEFKTIASEHPNTTIGVFLCGPEALAE 544
Query: 914 ELKKLSLELSHK--TTTRFDFHKEYF 937
L K S+ S F F+KE F
Sbjct: 545 TLSKQSISNSESGPRGVHFIFNKENF 570
>C24B_BOVIN (O46522) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 570
Score = 252 bits (643), Expect = 4e-66
Identities = 188/625 (30%), Positives = 285/625 (45%), Gaps = 133/625 (21%)
Query: 379 WVLFLWLVTTASLFAWKFYQYRNRTSF----QVMGYCLPVAKGAAETLKLNMALILLPVC 434
+V+ +WL LF W + Y F +++G L +A+ A L N LILLPVC
Sbjct: 13 FVILVWLGMNVFLFVWYYRVYDIPDKFFYTRKLLGSALALARAPAACLNFNCMLILLPVC 72
Query: 435 RNTLTWLR------STRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLA----CDFPL 484
RN L++LR STR R+ D N+ FHK++A I + +H HL C
Sbjct: 73 RNLLSFLRGSSACCSTRIRR--QLDRNLTFHKMVAWMIALHTAIHTIAHLFNVEWCVNAR 130
Query: 485 LINSSPEEFSLIASDFNNK-KPTYKTLL---------------TGVEGVTGISMVTLMAI 528
+ NS P +S+ SD +K TY + T + G+TG+ + + +
Sbjct: 131 VNNSDP--YSIALSDIGDKPNETYLNFVRQRIKNPEGGLYVAVTRLAGITGVVITLCLIL 188
Query: 529 SFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGS------------ 576
T +T RR+ F FWY+HHL + +I L IHG+
Sbjct: 189 IITSSTKTIRRSY------------FEVFWYTHHLFVIFFIGLAIHGAQRIVRGQTAESL 236
Query: 577 --------FLNLTHRWYN-------------KTTWMYISVPLLLYIAERTLRTSRSKHYA 615
+ N++ +W TW +I P+ LY+ ER +R RS+
Sbjct: 237 LKHQPRNCYQNIS-QWGKIENCPIPEFSGNPPMTWKWIVGPMFLYLCERLVRFWRSQQKV 295
Query: 616 VKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSV 675
V I KV P L M K GFK + GQYIF++CP +S EWHPF++TSAP +++ S+
Sbjct: 296 V-ITKVVTHPFKTIELQMKK-KGFKMEVGQYIFVKCPVVSKLEWHPFTLTSAPEEDFFSI 353
Query: 676 HIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNF 735
HIR VGDWT+ L C E D + P++ VDGP+G ++D ++
Sbjct: 354 HIRIVGDWTEGLFKA----------CGCDKQEF--QDAWKLPKIAVDGPFGTASEDVFSY 401
Query: 736 DVLLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGN 795
+V++L+G GIG TPF SIL+ + W + N P
Sbjct: 402 EVVMLVGAGIGVTPFASILKSV------------------------WYKY--CNKAP--- 432
Query: 796 KRSQRTTNAYFYWVTREPGSFEWFKGVMDEV-AEMDHRGQIELHNYLTSVYEEGDARSTL 854
+ R YFYW+ R+ +FEWF ++ + +M + + +Y + +++++
Sbjct: 433 --NLRLKKIYFYWLCRDTHAFEWFADLLQLLETQMQEKNNTDFLSYNICLTGWDESQASH 490
Query: 855 ITMIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKE 914
M H D+++G + +T + RPNW F I S+HP + +GVF CG LA
Sbjct: 491 FAM-----HHDEEKDVITGLKQKTLYGRPNWDNEFKTIGSQHPNTRIGVFLCGPEALADT 545
Query: 915 LKK--LSLELSHKTTTRFDFHKEYF 937
L K +S S F F+KE F
Sbjct: 546 LNKQCISNSDSGPRGVHFIFNKENF 570
>C24B_PIG (P52649) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generatin
Length = 484
Score = 218 bits (554), Expect = 8e-56
Identities = 163/554 (29%), Positives = 248/554 (44%), Gaps = 129/554 (23%)
Query: 443 STRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLA----CDFPLLINSSPEEFSLIAS 498
STR R+ D N+ FHK++A I + +H HL C + NS P +S+ S
Sbjct: 1 STRVRR--QLDRNLTFHKMVAWMIALHATIHTIAHLFNVEWCVNARVNNSDP--YSIALS 56
Query: 499 DFNNK-KPTYKTLL---------------TGVEGVTGISMVTLMAISFTLATHHFRRNAV 542
D +K TY + T + G+TG+ + + + T +T RR+
Sbjct: 57 DIGDKPNETYLNFVRQRIKNPEGGLYVAVTRLAGITGVVITLCLILIITSSTKTIRRSY- 115
Query: 543 RLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKT-------------- 588
F FWY+HHL + +I L IHG+ + R K+
Sbjct: 116 -----------FEVFWYTHHLFVIFFIGLAIHGAE-RIVRRQTPKSLLVHDPKACAQNIS 163
Query: 589 -------------------TWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVF 629
TW +I P+ LY+ ER +R RS+ V I KV P
Sbjct: 164 QWGKIKDCPIPEFAGNPPMTWKWIVGPMFLYLCERLVRFWRSQQKVV-ITKVVTHPFKTI 222
Query: 630 SLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKL 689
L M K GF+ + GQYIF++ P +S EWHPF++TSAP +++ S+HIR VGDWT+ L
Sbjct: 223 ELQMKK-KGFRMEVGQYIFVKRPAVSKLEWHPFTLTSAPEEDFFSIHIRIVGDWTEGLFK 281
Query: 690 LFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATP 749
C E D + P++ VDGP+G ++D ++ V++L+G GIG TP
Sbjct: 282 A----------CGCDKQEF--QDAWKLPKIAVDGPFGTASEDVFSYQVVMLVGAGIGVTP 329
Query: 750 FISILRDLLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTNAYFYWV 809
F SIL+ + W + + N + R YFYW+
Sbjct: 330 FASILKSV------------------------WYKYCN-------NATNLRLKKIYFYWL 358
Query: 810 TREPGSFEWFKGVMD----EVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHAK 865
R+ +FEWF ++ ++ E ++ G + + YLT ++E A + H
Sbjct: 359 CRDTHAFEWFADLLQLLETQMQERNNAGFLSYNIYLTG-WDESQANHFAV-------HHD 410
Query: 866 HGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHK 925
D+++G + +T + RPNW F IAS+HP + +GVF CG LA+ L K + S
Sbjct: 411 EEKDVITGLKQKTLYGRPNWDNEFKTIASQHPTTRIGVFLCGPEALAETLNKQCISNSDS 470
Query: 926 TT--TRFDFHKEYF 937
+ F F+KE F
Sbjct: 471 SPRGVHFIFNKENF 484
>FRP1_SCHPO (Q04800) Ferric reductase transmembrane component (EC
1.16.1.7) (Ferric-chelate reductase)
Length = 564
Score = 61.6 bits (148), Expect = 9e-09
Identities = 59/280 (21%), Positives = 114/280 (40%), Gaps = 55/280 (19%)
Query: 508 KTLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVV 567
+ LLT + G ++ LM I + FRR + F+ HH+ +
Sbjct: 184 RALLTARVTIIGYVILGLMVIMIVSSLPFFRRRF------------YEWFFVLHHMCSIG 231
Query: 568 YIL-LFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPG 626
+++ +++H ++ +Y+ V + +Y+ +R R RS K V V
Sbjct: 232 FLITIWLH-----------HRRCVVYMKVCVAVYVFDRGCRMLRSFLNRSKFDVVLVEDD 280
Query: 627 NVFSLIMSKPN------GFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTV 680
++ M P G + +G ++++ P +S ++ HPF+I S P D+++ + +
Sbjct: 281 LIY---MKGPRPKKSFFGLPWGAGNHMYINIPSLSYWQIHPFTIASVPSDDFIELFVAVR 337
Query: 681 GDWTQEL------KLLFTEDDI----PPVNCNATFG------------ELVQMDQREHPR 718
+T+ L K L DI + N G +LV
Sbjct: 338 AGFTKRLAKKVSSKSLSDVSDINISDEKIEKNGDVGIEVMERHSLSQEDLVFESSAAKVS 397
Query: 719 LFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLL 758
+ +DGPYG + +K++ L L G+G + + I+ D +
Sbjct: 398 VLMDGPYGPVSNPYKDYSYLFLFAGGVGVSYILPIILDTI 437
>FREL_CANAL (P78588) Probable ferric reductase transmembrane
component (EC 1.16.1.7) (Ferric-chelate reductase)
Length = 669
Score = 50.4 bits (119), Expect = 2e-05
Identities = 42/201 (20%), Positives = 85/201 (41%), Gaps = 21/201 (10%)
Query: 563 LLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVS 622
L+ +V ++ F+ G + +L + Y W I+V + +R +R R + + VS
Sbjct: 343 LIHIVLVVFFVVGGYYHLESQGYGDFMWAAIAV----WAFDRVVRLGRIFFFGARKATVS 398
Query: 623 VLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPF-EWHPFSITSAPGDEYLSVHIRTVG 681
+ + + + KP +K +G + F+ K + F + HPF+ T+ ++ + ++ +
Sbjct: 399 IKGDDTLKIEVPKPKYWKSVAGGHAFIHFLKPTLFLQSHPFTFTTTESNDKIVLYAKIKN 458
Query: 682 DWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLI 741
T + + P AT R+ V+GPYG P+ +N ++ +
Sbjct: 459 GITSNIAKYLS----PLPGNTATI------------RVLVEGPYGEPSSAGRNCKNVVFV 502
Query: 742 GLGIGATPFISILRDLLNNTR 762
G G S DL ++
Sbjct: 503 AGGNGIPGIYSECVDLAKKSK 523
>FRE7_YEAST (Q12333) Ferric reductase transmembrane component 7 (EC
1.16.1.7) (Ferric-chelate reductase 7)
Length = 629
Score = 45.1 bits (105), Expect = 0.001
Identities = 59/240 (24%), Positives = 94/240 (38%), Gaps = 29/240 (12%)
Query: 547 PLNRLTGFNAFWYSHHLLGV-VYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERT 605
P+ R + F H +L V YI LF H +H + T ++ + +
Sbjct: 257 PIARRHFYEIFLQLHWILAVGFYISLFYHVYPELNSHMYLVATIVVWFAQLFYRLAVKGY 316
Query: 606 LRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSIT 665
LR RS A I VS++ LI+ K Y GQ+IF++ HPFSI
Sbjct: 317 LRPGRS-FMASTIANVSIVGEGCVELIV-KDVEMAYSPGQHIFVRTIDKGIISNHPFSIF 374
Query: 666 SAPGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPY 725
P +Y +K+L + F + + + ++ +DGPY
Sbjct: 375 --PSAKYPG-----------GIKML--------IRAQKGFSKRLYESNDDMKKILIDGPY 413
Query: 726 GAPAQDFKNFDVLLLIGLGIGAT---PFISILRDLLNNTRASDESMDQSNTETTRSDESW 782
G +D ++F + LI G G + PF+ +L+ T ++D R D SW
Sbjct: 414 GGIERDIRSFTNVYLICSGSGISTCLPFLQKYGPILHKTNLEVITLDW--VVRHREDISW 471
>NDOR_PSEPU (Q52126) Naphthalene 1,2-dioxygenase system
ferredoxin--NAD(+) reductase component (EC 1.18.1.3)
Length = 328
Score = 42.7 bits (99), Expect = 0.004
Identities = 36/128 (28%), Positives = 51/128 (39%), Gaps = 25/128 (19%)
Query: 633 MSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFT 692
+SKP F++ GQY LQ SP P+S+ P D+ + HIR V
Sbjct: 120 LSKP--FEFSPGQYATLQ---FSPEHARPYSMAGLPDDQEMEFHIRKVP----------- 163
Query: 693 EDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFIS 752
E V RE + + GP G K+ +L +G G G P +S
Sbjct: 164 ---------GGRVTEYVFEHVREGTSIKLSGPLGTAYLRQKHTGPMLCVGGGTGLAPVLS 214
Query: 753 ILRDLLNN 760
I+R L +
Sbjct: 215 IVRGALKS 222
>FRE4_YEAST (P53746) Ferric reductase transmembrane component 4
precursor (EC 1.16.1.7) (Ferric-chelate reductase 4)
Length = 719
Score = 42.7 bits (99), Expect = 0.004
Identities = 48/223 (21%), Positives = 99/223 (43%), Gaps = 24/223 (10%)
Query: 551 LTGFNAFWYSHHL-LGVVYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTS 609
+ F +Y + L +V+ LF++ + ++T+ ++ W+Y ++ + + +R +R +
Sbjct: 364 IAAFRRHYYETFMALHIVFAALFLYTCWEHVTN--FSGIEWIYAAIAI--WGVDRIVRIT 419
Query: 610 RSKHYAVKIQKVSVLPGNVFSLIMSKPNGF-KYKSGQYIFLQCPKISPF-EWHPFSITSA 667
R + ++ ++ + + KP F K K GQY+F+ + F + HPF++ +
Sbjct: 420 RIALLGFPKADLQLVGSDLVRVTVKKPKKFWKAKPGQYVFVSFLRPLCFWQSHPFTVMDS 479
Query: 668 PGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGA 727
++ V + +L F E + + RL ++GPYG+
Sbjct: 480 CVNDRELVIVLKAKKGVTKLVRNFVE----------------RKGGKASMRLAIEGPYGS 523
Query: 728 PAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESMDQ 770
+ + FD +LL+ G G IS +L T AS ++ Q
Sbjct: 524 KSTAHR-FDNVLLLAGGSGLPGPISHALELGKTTAASGKNFVQ 565
>VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP42]
Length = 1912
Score = 38.5 bits (88), Expect = 0.084
Identities = 35/120 (29%), Positives = 57/120 (47%), Gaps = 8/120 (6%)
Query: 78 ASSGAGTSSSSSHATGDEGTSA--SGAGAVARSLSKNLSITSRIRRKFPWLRSMSMRTSA 135
+SSG+ +SSSSS ++ + +S+ S + + + S S + S +S + K S S R+S+
Sbjct: 1151 SSSGSSSSSSSSSSSSSDSSSSSRSSSSSDSSSSSSSSSSSSSSKSKSSSRSSKSNRSSS 1210
Query: 136 SSTESVEDPVESARNAR---RMQAKLERTRSSAQRALKGLRFISKSGEAGEELWRKVEER 192
SS S N++ +K TR A++ K F S GE R V E+
Sbjct: 1211 SSNSKDSSSSSSKSNSKGSSSSSSKASGTRQKAKKQSKTTSFPHASAAEGE---RSVHEQ 1267
>FRE3_YEAST (Q08905) Ferric reductase transmembrane component 3
precursor (EC 1.16.1.7) (Ferric-chelate reductase 3)
Length = 711
Score = 38.5 bits (88), Expect = 0.084
Identities = 62/301 (20%), Positives = 127/301 (41%), Gaps = 63/301 (20%)
Query: 450 VPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIASDFN-NKKPTYK 508
V + I FHK + + + V+H + + +S+ D+ +K+ TY
Sbjct: 307 VKYTSFIMFHKWLGRMMFLDAVIHGAAYTS-------------YSVFYKDWAASKEETYW 353
Query: 509 TLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVY 568
GV + +V +M + F+LA + R + AF + H +LG
Sbjct: 354 QF-----GVAALCIVGVM-VFFSLA--------------MFRKFFYEAFLFLHIVLGA-- 391
Query: 569 ILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNV 628
LF + + ++ + W+Y ++ + + +R +R R ++ + ++ ++
Sbjct: 392 --LFFYTCWEHVVE--LSGIEWIYAAIAI--WTIDRLIRIVRVSYFGFPKASLQLVGDDI 445
Query: 629 FSLIMSKP-NGFKYKSGQYIFLQ-CPKISPFEWHPFSIT-SAPGDEYLSVHIRTVGDWTQ 685
+ + +P +K K GQY+F+ + ++ HPF++ S D L++ ++ T+
Sbjct: 446 IRVTVKRPVRLWKAKPGQYVFVSFLHHLYFWQSHPFTVLDSIIKDGELTIILKEKKGVTK 505
Query: 686 ELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLLIGLGI 745
+K V CN + RL ++GPYG+ + N+D +LL+ G
Sbjct: 506 LVKKY--------VCCNGGKASM---------RLAIEGPYGS-SSPVNNYDNVLLLTGGT 547
Query: 746 G 746
G
Sbjct: 548 G 548
>PYRK_CLOPE (Q8XL63) Dihydroorotate dehydrogenase electron transfer
subunit
Length = 246
Score = 38.1 bits (87), Expect = 0.11
Identities = 35/158 (22%), Positives = 68/158 (42%), Gaps = 30/158 (18%)
Query: 612 KHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDE 671
+++ K+++ L ++SL++ + K +GQ+ ++ P + F P S+ G++
Sbjct: 4 EYFKGKVKENIELVEGIYSLVVE--HEAKINAGQFYMIKTP--NTFLGRPISVCEVNGND 59
Query: 672 YLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQD 731
V+ TVG T E+K + + D+I + GP G
Sbjct: 60 VRFVYA-TVGAGTNEMKKMISGDEIEII-----------------------GPLGNGFDI 95
Query: 732 FKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESMD 769
K++ + L+ GIG P + + + L N + D MD
Sbjct: 96 NKDYGRVALVSGGIGTAPMLELAKSLRKNNK--DIKMD 131
>MYSS_RABIT (P02562) Myosin heavy chain, skeletal muscle (Fragments)
Length = 1084
Score = 37.0 bits (84), Expect = 0.25
Identities = 52/254 (20%), Positives = 115/254 (44%), Gaps = 28/254 (11%)
Query: 130 SMRTSASSTESVEDPVESARNARRMQAKLERTRSSAQRALKGL-RFISKSGEAGE---EL 185
++ T+ E +E+ +E+ R +R AK E+ RS R L+ + + ++G A E+
Sbjct: 256 ALMTNLQRIEELEEEIEAERASR---AKAEKQRSDLSRELEEISERLEEAGGATSAQIEM 312
Query: 186 WRKVEERFGSLAKDGLLAREDFGGCIGMDDSKEFAVGVFDALARRKGRKASKITKE--EL 243
+K E F + +D ++++ AL ++ +++ ++ L
Sbjct: 313 NKKREAEFEKMRRD-------------LEEATLQHEATAAALRKKHADSVAELGEQIDNL 359
Query: 244 HHFWQQISDQSFDARLQIFFDMADSNEDGRITREEVQELIMLSASANKLSKLKEQAEGYA 303
Q++ + + +++I D+A + E + +++ M ++LS++K + E +
Sbjct: 360 QRVKQKLEKEKSELKMEID-DLAGNMETVSKAKGNLEK--MCRTLEDQLSEVKTKEEEHQ 416
Query: 304 ALIMEELDPENLGYIELWQLEMLLLQKDRYMNYSRQLSTSSVSWSQNMTGLKHKNEIQRL 363
LI E + + E + L +KD ++ QLS +++Q + GLK + E +
Sbjct: 417 RLINELSAQKARLHTESGEFSRQLDEKDAMVS---QLSRGGQAFTQQIEGLKRQLEEETK 473
Query: 364 CRTLQCLALEYWRR 377
++ AL+ RR
Sbjct: 474 AKSALAHALQSSRR 487
>BRAT_DROME (Q8MQJ9) Brain tumor protein
Length = 1037
Score = 36.6 bits (83), Expect = 0.32
Identities = 34/137 (24%), Positives = 55/137 (39%), Gaps = 7/137 (5%)
Query: 4 SSFGRYSTGDIETDSPENLRTSSPPASEGHAPSHGYSSMLPVFLNDLRTNHHKELVEITL 63
+S Y +DSP L SSPPAS+ S Y+ V E+ I
Sbjct: 17 NSIESYEHAGYLSDSPLTLSGSSPPASDSAICSDEYTGGSSV-------KSRSEVTVING 69
Query: 64 ELEDDAVVLCNVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSRIRRKF 123
A V + + +SS +SSSSS ++ +S SG + S +S +
Sbjct: 70 HHPISASVSSSSSASSSSCSSSSSSSSSSSSSSSSTSGLSGCGSTSSSVISANNVASSNG 129
Query: 124 PWLRSMSMRTSASSTES 140
P + ++++S + S
Sbjct: 130 PGVIGSNLQSSNNGGNS 146
>NSR1_PSEAE (Q9HYL3) Regulatory protein nosR
Length = 715
Score = 35.4 bits (80), Expect = 0.71
Identities = 32/93 (34%), Positives = 44/93 (46%), Gaps = 20/93 (21%)
Query: 516 GVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRL-TGFNAF------WYSHHLLGVVY 568
GV ++V L+AI F + VR P + RL TG+ AF WYS L VV
Sbjct: 423 GVLCTALVLLLAILF------LQDRLVRRPRLMQRLRTGYLAFTLVYLGWYSLGQLSVVN 476
Query: 569 ILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYI 601
+L F+H F RW +++S PLL +
Sbjct: 477 VLTFVHALFEGF--RWE-----LFLSDPLLFIL 502
>ADP1_MYCGA (Q49379) Adhesin P1 precursor (Cytadhesin P1)
(Attachment protein)
Length = 1122
Score = 35.4 bits (80), Expect = 0.71
Identities = 26/89 (29%), Positives = 45/89 (50%), Gaps = 3/89 (3%)
Query: 76 APASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSRIRRKFPWLRSMSMRTSA 135
APA+ S++ A G G+SA+G GAVA + + S T+R+ + +M +A
Sbjct: 180 APAAGDTSAEGSATPAGGGSGSSAAGGGAVAPAAA---SSTARLVEEGNSAGMGTMTPTA 236
Query: 136 SSTESVEDPVESARNARRMQAKLERTRSS 164
S++E+V D + + L+ + SS
Sbjct: 237 STSETVIDYNSDQNKIPKPKTLLDSSESS 265
>YJW5_YEAST (P40889) Hypothetical 197.6 kDa protein in FSP2 5'region
Length = 1758
Score = 35.0 bits (79), Expect = 0.93
Identities = 22/78 (28%), Positives = 40/78 (51%), Gaps = 8/78 (10%)
Query: 74 NVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSRIRRKFPWLRSMSMRT 133
NV +++ +++SS++AT E T++S S + N S T+ S+++RT
Sbjct: 1202 NVRTSATTTESTNSSTNATTTESTNSSTNATTTESTNSNTSATTTA--------SINVRT 1253
Query: 134 SASSTESVEDPVESARNA 151
SA++TES + A
Sbjct: 1254 SATTTESTNSSTSATTTA 1271
Score = 32.7 bits (73), Expect = 4.6
Identities = 23/94 (24%), Positives = 47/94 (49%), Gaps = 4/94 (4%)
Query: 71 VLCNVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSRIRRKFPWLRSMS 130
V N +S T+++S++A+ + T+AS S + + ++ +R S++
Sbjct: 1143 VRTNATTNASTNATTNASTNASTNATTNASTNATTNSSTNATTTASTNVRTSATTTASIN 1202
Query: 131 MRTSASSTESVEDPVESARNARRMQAKLERTRSS 164
+RTSA++TES S+ NA ++ T ++
Sbjct: 1203 VRTSATTTES----TNSSTNATTTESTNSSTNAT 1232
>YIR7_YEAST (P40434) Hypothetical 197.5 kDa protein in SDL1 5'region
Length = 1758
Score = 35.0 bits (79), Expect = 0.93
Identities = 22/78 (28%), Positives = 40/78 (51%), Gaps = 8/78 (10%)
Query: 74 NVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSRIRRKFPWLRSMSMRT 133
NV +++ +++SS++AT E T++S S + N S T+ S+++RT
Sbjct: 1202 NVRTSATTTESTNSSTNATTTESTNSSTNATTTESTNSNTSATTTA--------SINVRT 1253
Query: 134 SASSTESVEDPVESARNA 151
SA++TES + A
Sbjct: 1254 SATTTESTNSSTSATTTA 1271
Score = 32.7 bits (73), Expect = 4.6
Identities = 23/94 (24%), Positives = 47/94 (49%), Gaps = 4/94 (4%)
Query: 71 VLCNVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSRIRRKFPWLRSMS 130
V N +S T+++S++A+ + T+AS S + + ++ +R S++
Sbjct: 1143 VRTNATTNASTNATTNASTNASTNATTNASTNATTNSSTNATTTASTNVRTSATTTASIN 1202
Query: 131 MRTSASSTESVEDPVESARNARRMQAKLERTRSS 164
+RTSA++TES S+ NA ++ T ++
Sbjct: 1203 VRTSATTTES----TNSSTNATTTESTNSSTNAT 1232
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 107,868,867
Number of Sequences: 164201
Number of extensions: 4494704
Number of successful extensions: 15388
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 15223
Number of HSP's gapped (non-prelim): 130
length of query: 937
length of database: 59,974,054
effective HSP length: 120
effective length of query: 817
effective length of database: 40,269,934
effective search space: 32900536078
effective search space used: 32900536078
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0013.6


