

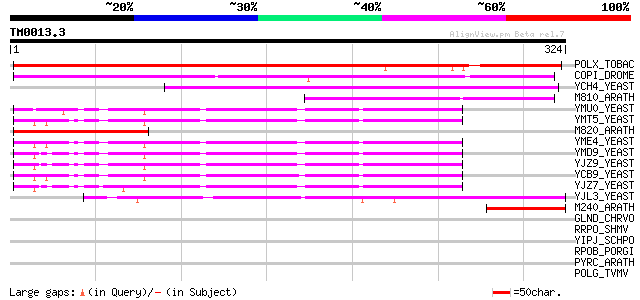
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0013.3
(324 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 231 1e-60
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 191 2e-48
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 125 2e-28
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 98 2e-20
YMU0_YEAST (Q04670) Transposon Ty1 protein B 82 2e-15
YMT5_YEAST (Q04214) Transposon Ty1 protein B 81 3e-15
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 81 3e-15
YME4_YEAST (Q04711) Transposon Ty1 protein B 79 2e-14
YMD9_YEAST (Q03434) Transposon Ty1 protein B 79 2e-14
YJZ9_YEAST (P47100) Transposon Ty1 protein B 79 2e-14
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 79 2e-14
YJZ7_YEAST (P47098) Transposon Ty1 protein B 77 8e-14
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 74 6e-13
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 45 3e-04
GLND_CHRVO (Q7NTY6) [Protein-PII] uridylyltransferase (EC 2.7.7.... 32 2.8
RRPO_SHMV (P89202) RNA-directed RNA polymerase (EC 2.7.7.48) (18... 31 4.8
YIPJ_SCHPO (Q9UT42) Hypothetical protein C343.19 in chromosome I 30 6.3
RPOB_PORGI (Q7MX27) DNA-directed RNA polymerase beta chain (EC 2... 30 6.3
PYRC_ARATH (O04904) Dihydroorotase, mitochondrial precursor (EC ... 30 6.3
POLG_TVMV (P09814) Genome polyprotein [Contains: P1 proteinase (... 30 6.3
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 231 bits (590), Expect = 1e-60
Identities = 135/337 (40%), Positives = 203/337 (60%), Gaps = 23/337 (6%)
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDYF 62
E+ +L+KN T+K+V+LP+G + + KWV+++K++ D L RYKARLV KG+ Q +G+D+
Sbjct: 833 EMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFD 892
Query: 63 DTFSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAGVPTM-KPNQ 121
+ FSPV K+T++R +L+LAA+ + + QLDV AFLHG L+E++YM P G K +
Sbjct: 893 EIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHM 952
Query: 122 VCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHSLFVK-FQDTLFAGLLVYVDDV 180
VCKL KSLYGLKQA R+WY K S +++ Y + SD ++ K F + F LL+YVDD+
Sbjct: 953 VCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDM 1012
Query: 181 ILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVA--HSSLGISLCQRKYCLDLLTE 238
++ G +K L +F +KDL + LG+++ +S + L Q KY +L
Sbjct: 1013 LIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLER 1072
Query: 239 TGTLGSKPVSTPLDPSCKL--------VVNQGE----PYSDATAYRRLVGRLLY-LTTTR 285
+KPVSTPL KL V +G PYS A VG L+Y + TR
Sbjct: 1073 FNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSA------VGSLMYAMVCTR 1126
Query: 286 PDISFATQQLSQFMSNPMTDHYKAALRVLRFLKTSPG 322
PDI+ A +S+F+ NP +H++A +LR+L+ + G
Sbjct: 1127 PDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTTG 1163
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 191 bits (486), Expect = 2e-48
Identities = 111/320 (34%), Positives = 179/320 (55%), Gaps = 7/320 (2%)
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDYF 62
E+ A + N TW I P+ + ++WV+ +K N G RYKARLVA+G+ Q +DY
Sbjct: 913 ELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYE 972
Query: 63 DTFSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAGVPTMKPNQV 122
+TF+PVA++++ R +L+L N +HQ+DV AFL+GTL E++YM +P G+ N V
Sbjct: 973 ETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGISCNSDN-V 1031
Query: 123 CKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHSLFVKFQDTLFAGL--LVYVDDV 180
CKL K++YGLKQA+R W+E L+ + ++ D +++ + + + L+YVDDV
Sbjct: 1032 CKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDV 1091
Query: 181 ILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISLCQRKYCLDLLTETG 240
++ + K L F + DL +K+F+G+ + I L Q Y +L++
Sbjct: 1092 VIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFN 1151
Query: 241 TLGSKPVSTPLDPSCKL-VVNQGEPYSDATAYRRLVGRLLY-LTTTRPDISFATQQLSQF 298
VSTPL ++N E + T R L+G L+Y + TRPD++ A LS++
Sbjct: 1152 MENCNAVSTPLPSKINYELLNSDEDCN--TPCRSLIGCLMYIMLCTRPDLTTAVNILSRY 1209
Query: 299 MSNPMTDHYKAALRVLRFLK 318
S ++ ++ RVLR+LK
Sbjct: 1210 SSKNNSELWQNLKRVLRYLK 1229
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 125 bits (313), Expect = 2e-28
Identities = 80/233 (34%), Positives = 123/233 (52%), Gaps = 3/233 (1%)
Query: 91 LDVDNAFLHGTLDEDVYMNIPAG-VPTMKPNQVCKLQKSLYGLKQASRKWYEKLSSHLRN 149
+DVD AFL+ T+DE +Y+ P G V P+ V +L +YGLKQA W E +++ L+
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 150 LGYSQATSDHSLFVKFQDTLFAGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVIL 209
+G+ + +H L+ + + VYVDD+++ + VK L + +KDL +
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 210 KYFLGLEVAHSSLG-ISLCQRKYCLDLLTETGTLGSKPVSTPLDPSCKLVVNQGEPYSDA 268
FLGL + S+ G I+L + Y +E+ K TPL S L D
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 269 TAYRRLVGRLLYLTTT-RPDISFATQQLSQFMSNPMTDHYKAALRVLRFLKTS 320
T Y+ +VG+LL+ T RPDIS+ LS+F+ P H ++A RVLR+L T+
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTT 233
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 98.2 bits (243), Expect = 2e-20
Identities = 56/146 (38%), Positives = 81/146 (55%), Gaps = 1/146 (0%)
Query: 173 LLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISLCQRKYC 232
LL+YVDD++L G+ ++ L + F +KDL + YFLG+++ G+ L Q KY
Sbjct: 3 LLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYA 62
Query: 233 LDLLTETGTLGSKPVSTPLDPSCKLVVNQGEPYSDATAYRRLVGRLLYLTTTRPDISFAT 292
+L G L KP+STPL V+ + Y D + +R +VG L YLT TRPDIS+A
Sbjct: 63 EQILNNAGMLDCKPMSTPLPLKLNSSVSTAK-YPDPSDFRSIVGALQYLTLTRPDISYAV 121
Query: 293 QQLSQFMSNPMTDHYKAALRVLRFLK 318
+ Q M P + RVLR++K
Sbjct: 122 NIVCQRMHEPTLADFDLLKRVLRYVK 147
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 82.0 bits (201), Expect = 2e-15
Identities = 80/275 (29%), Positives = 132/275 (47%), Gaps = 31/275 (11%)
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWV----YRIKRNADGTLARYKARLVAKGYNQIEG 58
E+ L K +TW D K I K V + R DGT +KAR VA+G
Sbjct: 830 EVNQLLKMKTWD-TDRYYDRKEIDPKRVINSMFIFNRKRDGT---HKARFVARG-----D 880
Query: 59 LDYFDTFSPVAKLTTVRMV-----LALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAG 113
+ + DT+ P + TV L+LA N+++ QLD+ +A+L+ + E++Y+ P
Sbjct: 881 IQHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP-- 938
Query: 114 VPTMKPN-QVCKLQKSLYGLKQASRKWYEKLSSHL-RNLGYSQATSDHSLFVKFQDTLFA 171
P + N ++ +L+KSLYGLKQ+ WYE + S+L + G + +F Q T
Sbjct: 939 -PHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT--- 994
Query: 172 GLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISL-CQRK 230
+ ++VDD+ILF L + + +L + K +I E+ + LG+ + QR
Sbjct: 995 -ICLFVDDMILFSKDLNANKKIITTLKKQYDTK--IINLGESDNEIQYDILGLEIKYQRG 1051
Query: 231 YCLDLLTETGTLGSKP-VSTPLDPSCKLVVNQGEP 264
+ L E P ++ PL+P + + G+P
Sbjct: 1052 KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQP 1086
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 81.3 bits (199), Expect = 3e-15
Identities = 78/276 (28%), Positives = 135/276 (48%), Gaps = 33/276 (11%)
Query: 3 EIAALEKNETW---KIVDLPQ--GVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIE 57
E+ L K +TW K D + + I + +++ KR DGT +KAR VA+G
Sbjct: 830 EVNQLLKMKTWDTDKYYDRKEIDPKRVINSMFIFNRKR--DGT---HKARFVARG----- 879
Query: 58 GLDYFDTFSPVAKLTTVRMV-----LALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPA 112
+ + DT+ + TV L+LA N+++ QLD+ +A+L+ + E++Y+ P
Sbjct: 880 DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP- 938
Query: 113 GVPTMKPN-QVCKLQKSLYGLKQASRKWYEKLSSHL-RNLGYSQATSDHSLFVKFQDTLF 170
P + N ++ +L+KSLYGLKQ+ WYE + S+L + G + +F Q T
Sbjct: 939 --PHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFENSQVT-- 994
Query: 171 AGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISL-CQR 229
+ ++VDD++LF L + + D L + K +I E+ + LG+ + QR
Sbjct: 995 --ICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTK--IINLGESDEEIQYDILGLEIKYQR 1050
Query: 230 KYCLDLLTETGTLGSKP-VSTPLDPSCKLVVNQGEP 264
+ L E P ++ PL+P + + G+P
Sbjct: 1051 GKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQP 1086
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 81.3 bits (199), Expect = 3e-15
Identities = 37/79 (46%), Positives = 56/79 (70%)
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDYF 62
E+ AL +N+TW +V P +G KWV++ K ++DGTL R KARLVAKG++Q EG+ +
Sbjct: 47 ELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFV 106
Query: 63 DTFSPVAKLTTVRMVLALA 81
+T+SPV + T+R +L +A
Sbjct: 107 ETYSPVVRTATIRTILNVA 125
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 79.0 bits (193), Expect = 2e-14
Identities = 78/276 (28%), Positives = 134/276 (48%), Gaps = 33/276 (11%)
Query: 3 EIAALEKNETW---KIVDLPQ--GVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIE 57
E+ L K TW K D + + I + +++ KR DGT +KAR VA+G
Sbjct: 830 EVNQLLKMNTWDTDKYYDRKEIDPKRVINSMFIFNRKR--DGT---HKARFVARG----- 879
Query: 58 GLDYFDTFSPVAKLTTVRMV-----LALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPA 112
+ + DT+ + TV L+LA N+++ QLD+ +A+L+ + E++Y+ P
Sbjct: 880 DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP- 938
Query: 113 GVPTMKPN-QVCKLQKSLYGLKQASRKWYEKLSSHL-RNLGYSQATSDHSLFVKFQDTLF 170
P + N ++ +L+KSLYGLKQ+ WYE + S+L + G + +F Q T
Sbjct: 939 --PHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT-- 994
Query: 171 AGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISL-CQR 229
+ ++VDD+ILF L + + +L + K +I E+ + LG+ + QR
Sbjct: 995 --ICLFVDDMILFSKDLNANKKIITTLKKQYDTK--IINLGESDNEIQYDILGLEIKYQR 1050
Query: 230 KYCLDLLTETGTLGSKP-VSTPLDPSCKLVVNQGEP 264
+ L E P ++ PL+P + + G+P
Sbjct: 1051 GKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQP 1086
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 78.6 bits (192), Expect = 2e-14
Identities = 80/279 (28%), Positives = 137/279 (48%), Gaps = 39/279 (13%)
Query: 3 EIAALEKNETW--------KIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYN 54
E+ L K +TW K +D P+ V I + +++ KR DGT +KAR VA+G
Sbjct: 830 EVNQLLKMKTWDTDEYYDRKEID-PKRV--INSMFIFNKKR--DGT---HKARFVARG-- 879
Query: 55 QIEGLDYFDTFSPVAKLTTVRMV-----LALAAAQNWHLHQLDVDNAFLHGTLDEDVYMN 109
+ + DT+ + TV L+LA N+++ QLD+ +A+L+ + E++Y+
Sbjct: 880 ---DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 936
Query: 110 IPAGVPTMKPN-QVCKLQKSLYGLKQASRKWYEKLSSHL-RNLGYSQATSDHSLFVKFQD 167
P P + N ++ +L+KSLYGLKQ+ WYE + S+L + G + +F Q
Sbjct: 937 PP---PHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQV 993
Query: 168 TLFAGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISL- 226
T + ++VDD+ILF L + + +L + K +I E+ + LG+ +
Sbjct: 994 T----ICLFVDDMILFSKDLNANKKIITTLKKQYDTK--IINLGESDNEIQYDILGLEIK 1047
Query: 227 CQRKYCLDLLTETGTLGSKP-VSTPLDPSCKLVVNQGEP 264
QR + L E P ++ PL+P + + G+P
Sbjct: 1048 YQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQP 1086
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 78.6 bits (192), Expect = 2e-14
Identities = 79/279 (28%), Positives = 137/279 (48%), Gaps = 39/279 (13%)
Query: 3 EIAALEKNETW--------KIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYN 54
E+ L K +TW K +D P+ V I + +++ KR DGT +KAR VA+G
Sbjct: 1257 EVNQLLKMKTWDTDEYYDRKEID-PKRV--INSMFIFNKKR--DGT---HKARFVARG-- 1306
Query: 55 QIEGLDYFDTFSPVAKLTTVRMV-----LALAAAQNWHLHQLDVDNAFLHGTLDEDVYMN 109
+ + DT+ + TV L+LA N+++ QLD+ +A+L+ + E++Y+
Sbjct: 1307 ---DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 1363
Query: 110 IPAGVPTMKPN-QVCKLQKSLYGLKQASRKWYEKLSSHL-RNLGYSQATSDHSLFVKFQD 167
P P + N ++ +L+KSLYGLKQ+ WYE + S+L + G + +F Q
Sbjct: 1364 PP---PHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVFKNSQV 1420
Query: 168 TLFAGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISL- 226
T + ++VDD++LF L + + + L + K +I E+ + LG+ +
Sbjct: 1421 T----ICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTK--IINLGESDEEIQYDILGLEIK 1474
Query: 227 CQRKYCLDLLTETGTLGSKP-VSTPLDPSCKLVVNQGEP 264
QR + L E P ++ PL+P + + G+P
Sbjct: 1475 YQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQP 1513
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 78.6 bits (192), Expect = 2e-14
Identities = 79/275 (28%), Positives = 131/275 (46%), Gaps = 31/275 (11%)
Query: 3 EIAALEKNETW---KIVDLPQ--GVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIE 57
EI+ L K TW K D K I + +++ KR DGT +KAR VA+G
Sbjct: 1272 EISQLLKMNTWDTNKYYDRNDIDPKKVINSMFIFNKKR--DGT---HKARFVARG----- 1321
Query: 58 GLDYFDTFSPVAKLTTVRMV-----LALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPA 112
+ + DT+ + TV L++A ++++ QLD+ +A+L+ + E++Y+ P
Sbjct: 1322 DIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEELYIRPP- 1380
Query: 113 GVPTMKPN-QVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHSLFVKFQDTLFA 171
P + N ++ +L+KSLYGLKQ+ WYE + S+L N Q S K
Sbjct: 1381 --PHLGLNDKLLRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSCVFKNSQVT-- 1436
Query: 172 GLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISL-CQRK 230
+ ++VDD+ILF L + + +L + K +I E+ + LG+ + QR
Sbjct: 1437 -ICLFVDDMILFSKDLNANKKIITTLKKQYDTK--IINLGESDNEIQYDILGLEIKYQRS 1493
Query: 231 YCLDLLTETGTLGSKP-VSTPLDPSCKLVVNQGEP 264
+ L E P ++ PL+P K + G+P
Sbjct: 1494 KYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQP 1528
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 76.6 bits (187), Expect = 8e-14
Identities = 80/276 (28%), Positives = 136/276 (48%), Gaps = 33/276 (11%)
Query: 3 EIAALEKNETW--------KIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYN 54
E+ L K +TW K +D P+ V I + +++ KR DGT +KAR VA+G
Sbjct: 1257 EVNQLLKMKTWDTDEYYDRKEID-PKRV--INSMFIFNKKR--DGT---HKARFVARG-- 1306
Query: 55 QIEGLDYFDTF--SPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPA 112
I+ D +DT S + L+LA N+++ QLD+ +A+L+ + E++Y+ P
Sbjct: 1307 DIQHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP- 1365
Query: 113 GVPTMKPN-QVCKLQKSLYGLKQASRKWYEKLSSHL-RNLGYSQATSDHSLFVKFQDTLF 170
P + N ++ +L+KS YGLKQ+ WYE + S+L + G + +F Q T
Sbjct: 1366 --PHLGMNDKLIRLKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT-- 1421
Query: 171 AGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISL-CQR 229
+ ++VDD+ILF L + + +L + K +I E+ + LG+ + QR
Sbjct: 1422 --ICLFVDDMILFSKDLNANKKIITTLKKQYDTK--IINLGESDNEIQYDILGLEIKYQR 1477
Query: 230 KYCLDLLTETGTLGSKP-VSTPLDPSCKLVVNQGEP 264
+ L E P ++ PL+P + + G+P
Sbjct: 1478 GKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQP 1513
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 73.6 bits (179), Expect = 6e-13
Identities = 71/303 (23%), Positives = 139/303 (45%), Gaps = 34/303 (11%)
Query: 44 YKARLVAKGYNQIEGLDYFDTFSPVAKLTT----VRMVLALAAAQNWHLHQLDVDNAFLH 99
YKAR+V +G Q DT+S + + +++ L +A +N + LD+++AFL+
Sbjct: 1337 YKARIVCRGDTQSP-----DTYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLY 1391
Query: 100 GTLDEDVYMNIPAGVPTMKPNQVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDH 159
L+E++Y+ P V KL K+LYGLKQ+ ++W + L +L +G +
Sbjct: 1392 AKLEEEIYIPHPHD-----RRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTP 1446
Query: 160 SLFVKFQDTLFAGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIK------DLVILKYFL 213
L+ L + VYVDD ++ + + L + F +K D V+ L
Sbjct: 1447 GLYQTEDKNLM--IAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDIL 1504
Query: 214 GLEVAHSS-LG---------ISLCQRKYCLDLLTETGTLGSKPVSTPLDPSCKLVVNQGE 263
G+++ ++ LG I+ +KY +L + + +DP ++ E
Sbjct: 1505 GMDLVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEE 1564
Query: 264 PYSDAT-AYRRLVGRLLYLT-TTRPDISFATQQLSQFMSNPMTDHYKAALRVLRFLKTSP 321
+ ++L+G L Y+ R DI FA +++++ ++ P + +++++L
Sbjct: 1565 EFRQGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYK 1624
Query: 322 GLG 324
+G
Sbjct: 1625 DIG 1627
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 44.7 bits (104), Expect = 3e-04
Identities = 21/46 (45%), Positives = 30/46 (64%)
Query: 279 LYLTTTRPDISFATQQLSQFMSNPMTDHYKAALRVLRFLKTSPGLG 324
+YLT TRPD++FA +LSQF S T +A +VL ++K + G G
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQG 46
>GLND_CHRVO (Q7NTY6) [Protein-PII] uridylyltransferase (EC 2.7.7.59)
(PII uridylyl-transferase) (Uridylyl removing enzyme)
(UTase)
Length = 856
Score = 31.6 bits (70), Expect = 2.8
Identities = 15/51 (29%), Positives = 27/51 (52%)
Query: 203 IKDLVILKYFLGLEVAHSSLGISLCQRKYCLDLLTETGTLGSKPVSTPLDP 253
+ + L + +GLE+ HS + C R+ D+ ET L ++ V+ P +P
Sbjct: 97 VSHFIGLMWDIGLEIGHSVRTLDECLREAAGDITIETNLLENRLVAGPAEP 147
>RRPO_SHMV (P89202) RNA-directed RNA polymerase (EC 2.7.7.48) (186 kDa
protein) [Contains: Methyltransferase/RNA helicase
(MT/HEL) (128 kDa protein)]
Length = 1629
Score = 30.8 bits (68), Expect = 4.8
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Query: 258 VVNQGEPYSDATAYRRLVGRLLYLTTTRPDISFATQQLSQFMSNPMTDHYKAALRVLRFL 317
+V+ G P+ A R R +Y T PD+ T Q +Q +SN + D Y A L
Sbjct: 1076 IVSAGSPHV-TVALSRHTNRFVYYTVV-PDVVMTTVQKTQCVSNFLLDMYAVAYTQKXQL 1133
Query: 318 KTSP 321
+ SP
Sbjct: 1134 QISP 1137
>YIPJ_SCHPO (Q9UT42) Hypothetical protein C343.19 in chromosome I
Length = 624
Score = 30.4 bits (67), Expect = 6.3
Identities = 20/74 (27%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query: 93 VDNAFLHGTLDEDVYMNIPAG---VP-TMKPNQVCKLQKSLYGLKQASRKWYEKLSSHLR 148
+DN+ D + + P G +P ++ + + L+ K SR+W+EKLS LR
Sbjct: 410 IDNSLAFPYKHPDSWRSFPYGWLSLPRSLFTQPFTEFTRQLFLHKLTSREWWEKLSDDLR 469
Query: 149 NLGYSQATSDHSLF 162
N+ D +F
Sbjct: 470 NVFNQDLDFDEKMF 483
>RPOB_PORGI (Q7MX27) DNA-directed RNA polymerase beta chain (EC
2.7.7.6) (RNAP beta subunit) (Transcriptase beta chain)
(RNA polymerase beta subunit)
Length = 1269
Score = 30.4 bits (67), Expect = 6.3
Identities = 18/60 (30%), Positives = 27/60 (45%)
Query: 78 LALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAGVPTMKPNQVCKLQKSLYGLKQASR 137
L A A N+ H +++ LDE + +PAG+ M + K +K G K A R
Sbjct: 993 LIKAVAVNYLKHSKEIEAELRRRKLDETIGDELPAGIVQMAKVYIAKKRKIQVGDKMAGR 1052
>PYRC_ARATH (O04904) Dihydroorotase, mitochondrial precursor (EC
3.5.2.3) (DHOase)
Length = 377
Score = 30.4 bits (67), Expect = 6.3
Identities = 20/60 (33%), Positives = 25/60 (41%), Gaps = 5/60 (8%)
Query: 62 FDTFSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAGV--PTMKP 119
FD S K VRM L + +WHLH D D LH + N + P +KP
Sbjct: 19 FDRSSEKVKTRAVRMELTITQPDDWHLHLRDGD--LLHAVVPHSA-SNFKRAIVMPNLKP 75
>POLG_TVMV (P09814) Genome polyprotein [Contains: P1 proteinase
(N-terminal protein); Helper component proteinase (EC
3.4.22.45) (HC-pro); Protein P3; 6 kDa protein 1 (6K1);
Cytoplasmic inclusion protein (CI); 6 kDa protein 2
(6K2); Viral genome-linked pr
Length = 3023
Score = 30.4 bits (67), Expect = 6.3
Identities = 16/75 (21%), Positives = 38/75 (50%), Gaps = 5/75 (6%)
Query: 101 TLDEDVYMNIPAGVPTMKPNQVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHS 160
T ++D++ +I + + ++C+++ +K AS EK+++ + L D +
Sbjct: 27 TAEKDIFSSIKERLERKRHGKICRMKNGSIYIKAASSTKVEKINAAAKKL-----ADDKA 81
Query: 161 LFVKFQDTLFAGLLV 175
F+K Q T+ ++V
Sbjct: 82 AFLKAQPTIVDKIIV 96
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,396,631
Number of Sequences: 164201
Number of extensions: 1517464
Number of successful extensions: 3310
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 3271
Number of HSP's gapped (non-prelim): 21
length of query: 324
length of database: 59,974,054
effective HSP length: 110
effective length of query: 214
effective length of database: 41,911,944
effective search space: 8969156016
effective search space used: 8969156016
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0013.3


