

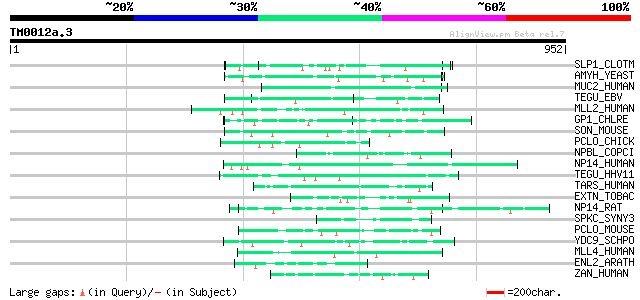
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012a.3
(952 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer... 67 2e-10
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 62 7e-09
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 57 3e-07
TEGU_EBV (P03186) Large tegument protein 56 5e-07
MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia p... 55 7e-07
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 54 2e-06
SON_MOUSE (Q9QX47) SON protein 54 3e-06
PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment) 53 3e-06
NPBL_COPCI (Q00333) Rad9 protein (SCC2 homolog) 52 8e-06
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 52 1e-05
TEGU_HHV11 (P10220) Large tegument protein (Virion protein UL36) 50 2e-05
TARS_HUMAN (Q7Z7G0) Target of Nesh-SH3 precursor (Tarsh) (Nesh b... 50 4e-05
EXTN_TOBAC (P13983) Extensin precursor (Cell wall hydroxyproline... 49 6e-05
NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 k... 49 8e-05
SPKC_SYNY3 (P74745) Serine/threonine-protein kinase C (EC 2.7.1.37) 48 1e-04
PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix prot... 48 1e-04
YDC9_SCHPO (Q10172) Hypothetical protein C25G10.09c in chromosome I 47 2e-04
MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia p... 47 2e-04
ENL2_ARATH (Q9T076) Early nodulin-like protein 2 precursor (Phyt... 47 2e-04
ZAN_HUMAN (Q9Y493) Zonadhesin precursor 47 3e-04
>SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer
layer protein B) (S-layer protein 1)
Length = 1664
Score = 67.4 bits (163), Expect = 2e-10
Identities = 94/420 (22%), Positives = 144/420 (33%), Gaps = 50/420 (11%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEE---SDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKK 425
P P P + + D S+E SD P + P D E P + ++
Sbjct: 1039 PTPSDEPTPSETPEEPIPTDTPSDEPTPSDEP-TPSDEPTPSDEPTPSDEPTPSETPEEP 1097
Query: 426 VRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSS 485
+ P+ E EP + + P+ T ++ + P +
Sbjct: 1098 IPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPI------PTDTPSDEPT 1151
Query: 486 NPLTPIPESQPA---EQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIP 542
P P +P E T S +P + +P PT+ P ++P+ SDEPT P
Sbjct: 1152 PSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTP-----SDEPTPSDEPT-----P 1201
Query: 543 YDPL--SSEPIIHEQPEPNQT--EP--------QPRTSDHSAPRASARPAARTTDTDSST 590
D S EP ++P P++T EP +P SD P P+ T +D T
Sbjct: 1202 SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT 1261
Query: 591 -AFTPVS-FPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERL 648
+ TP P + P+ + E ++ + +E PS P P
Sbjct: 1262 PSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE---PTPSDEPTPSDEPTPS 1318
Query: 649 VDPDEPILAN-PIQEADPLVQQA---HPVPDQQ-----EPIQPDPEPEQSVSNQSSVRSP 699
P+EPI + P E P + P P + EP P EP S S
Sbjct: 1319 ETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT-PSDEPTPSDEPTPSETPE 1377
Query: 700 HPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTSLSE 759
P T+ ++ P + G G P SP P + P + PT + E
Sbjct: 1378 EPTPTTTPTPTPSTTPTSGSGGSGGSGGGGGGGGGTVPTSPTPTPTSKPTSTPAPTEIEE 1437
Score = 62.0 bits (149), Expect = 7e-09
Identities = 87/398 (21%), Positives = 138/398 (33%), Gaps = 35/398 (8%)
Query: 371 PEFPSPPKRKAQRKMVLDESSEE-----SDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKK 425
PE P P + DE + SD P + P D E P + ++
Sbjct: 996 PEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEP-TPSDEPTPSDEPTPSDEPTPSETPEEP 1054
Query: 426 VRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSS 485
+ P+ E EP + + P+ T ++ + P +
Sbjct: 1055 IPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPI------PTDTPSDEPT 1108
Query: 486 NPLTPIPESQPA---EQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIP 542
P P +P E T S +P + +P PT+ P ++P+ SDEPT P
Sbjct: 1109 PSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTP-----SDEPTPSDEPT-----P 1158
Query: 543 YDPL--SSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPIN 600
D S EP ++P P++T +P +D P P+ T +D T P P
Sbjct: 1159 SDEPTPSDEPTPSDEPTPSETPEEPIPTD--TPSDEPTPSDEPTPSDEPT---PSDEPTP 1213
Query: 601 VVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGP--RPERLVDPDEPILAN 658
+ +PS E E + E P+P P P P P+EPI +
Sbjct: 1214 SDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTD 1273
Query: 659 PIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAP 718
+ + P + +P P E + S++ + ET + + T P+
Sbjct: 1274 TPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDE 1333
Query: 719 MINIGSPQGASEAHSSNHPA-SPAPNLSIIPYTHLQPT 755
P + E S+ P S P S P +PT
Sbjct: 1334 PTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT 1371
Score = 59.7 bits (143), Expect = 4e-08
Identities = 75/333 (22%), Positives = 119/333 (35%), Gaps = 49/333 (14%)
Query: 428 IVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNP 487
+V++P ++ A++ EP T S + T D+ D S S P
Sbjct: 758 VVIQPAPIKAASD-----EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETP 812
Query: 488 LTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLT----IPY 543
PIP P+++ T +P+P++ P + ++P+ SDEPT T IP
Sbjct: 813 EEPIPTDTPSDEPTPSD--------EPTPSDEPTPS---DEPTPSDEPTPSETPEEPIPT 861
Query: 544 DPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVD 603
D S EP ++P P+ +P SD P S P T + TP P +
Sbjct: 862 DTPSDEPTPSDEPTPSD---EPTPSDEPTP--SDEPTPSETPEEPIPTDTPSDEPTPSDE 916
Query: 604 SSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEA 663
+PS+ PS P P P+EPI + +
Sbjct: 917 PTPSDEPT-----------------------PSDEPTPSDEPTPSETPEEPIPTDTPSDE 953
Query: 664 DPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIG 723
+ P + +P P E + S++ + ET + + T P+
Sbjct: 954 PTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSD 1013
Query: 724 SPQGASEAHSSNHPA-SPAPNLSIIPYTHLQPT 755
P + E S+ P S P S P +PT
Sbjct: 1014 EPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT 1046
Score = 58.5 bits (140), Expect = 8e-08
Identities = 87/403 (21%), Positives = 129/403 (31%), Gaps = 46/403 (11%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRI 428
P P P + + D S+E +P D+ D P + + I
Sbjct: 886 PTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPI 945
Query: 429 VVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPL 488
EP P T S + T D+ D S S P
Sbjct: 946 PTDTPSDEPT--------PSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPE 997
Query: 489 TPIPESQPAEQTT-----------------SPPHSPRSSFFQPSPTEAPLWNLLQNQPSR 531
PIP P+++ T +P P S +P+P++ P + +P
Sbjct: 998 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSD-EPTPSDEPTPSETPEEPIP 1056
Query: 532 SDEPTSLLTIPYDPL-------SSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTT 584
+D P+ T +P S EP ++P P++T +P +D P P+ T
Sbjct: 1057 TDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTD--TPSDEPTPSDEPT 1114
Query: 585 DTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGP- 643
+D TP P + +PS E E + E P+P P P
Sbjct: 1115 PSDEP---TPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPS 1171
Query: 644 -RPERLVDPDEPILAN-PIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHP 701
P P+EPI + P E P P P + +P P + + P P
Sbjct: 1172 DEPTPSETPEEPIPTDTPSDEPTP---SDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIP 1228
Query: 702 LVETSDPHLGTSEPNAPMINIGS--PQGASEAHSSNHPASPAP 742
SD + EP S P + E S P P P
Sbjct: 1229 TDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIP 1271
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 62.0 bits (149), Expect = 7e-09
Identities = 88/401 (21%), Positives = 139/401 (33%), Gaps = 46/401 (11%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPL------VKKTKRKPDDGDDDDSEDGP---P 419
P P PS ++ V ++E S P+ ++ P +S P P
Sbjct: 457 PVPT-PSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTP 515
Query: 420 QKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSK-PALTSDDDLNLFDALPIS 478
+ PT P++ + P VT S+ SS P T + P++
Sbjct: 516 SSSTTESSSAPAPT---PSSSTTESSSAP--VTSSTTESSSAPVPTPSSSTTESSSTPVT 570
Query: 479 ALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSL 538
+ SS+ P P S E +++P +P SS + S AP PS S +S
Sbjct: 571 SSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPAPT-------PSSSTTESSS 623
Query: 539 LTIPYDPLSSEPIIHEQPEPNQTE----PQPRTSDHSAPRASAR-PAARTTDTDSSTAFT 593
+ S P + TE P P S + +SA P ++ T+SS+A
Sbjct: 624 APVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSA-- 681
Query: 594 PVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRR---------YPGPR 644
PV+ S+P +S + V S E P+P P P
Sbjct: 682 PVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPS 741
Query: 645 PERLVDPDEPILANPIQEADPLVQQAHPVPDQ---QEPIQPDPEPEQSVSNQSSVRSPHP 701
P+ ++ + + V P P + P P P S + SS P P
Sbjct: 742 SSTTESSSAPVTSSTTESSSAPV----PTPSSSTTESSSAPVPTPSSSTTESSSAPVPTP 797
Query: 702 LVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAP 742
T++ + + NI S +S SS+ +S P
Sbjct: 798 SSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVP 838
Score = 60.5 bits (145), Expect = 2e-08
Identities = 87/401 (21%), Positives = 142/401 (34%), Gaps = 48/401 (11%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRK---PDDGDDDDSEDGPPQKKKKK 425
P P PS ++ V ++E S P+ T P +S P +
Sbjct: 394 PVPT-PSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTE 452
Query: 426 VRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSK-PALTSDDDLNLFDALPISALLQHS 484
PT P++ + P VT S+ SS P T + P+++ S
Sbjct: 453 SSSAPVPT---PSSSTTESSSAP--VTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTES 507
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S+ P P S E +++P +P S S TE+ + + S P +
Sbjct: 508 SSAPVPTPSSSTTESSSAPAPTPSS-----STTESSSAPVTSSTTESSSAPVPTPSSSTT 562
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASA---RPAARTTDTDSSTAFTPVSFPINV 601
SS P+ E + + P P S + +SA P++ TT++ S+ A TP S
Sbjct: 563 ESSSTPVTSSTTE-SSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPAPTP-SSSTTE 620
Query: 602 VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRR---------YPGPRPERLVDPD 652
S+P +S + V S E P+P P P
Sbjct: 621 SSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSS 680
Query: 653 EPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSD----- 707
P+ ++ + + V + + P P P S + SS P P T++
Sbjct: 681 APVTSSTTESSSAPVTSS----TTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAP 736
Query: 708 ---PHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLS 745
P T+E ++ +P +S SS+ P P P+ S
Sbjct: 737 VPTPSSSTTESSS------APVTSSTTESSSAPV-PTPSSS 770
Score = 56.2 bits (134), Expect = 4e-07
Identities = 83/385 (21%), Positives = 134/385 (34%), Gaps = 39/385 (10%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRI 428
PAP PS ++ V ++E S P+ + +S P +
Sbjct: 526 PAPT-PSSSTTESSSAPVTSSTTESSSAPVPTPSS------STTESSSTPVTSSTTESSS 578
Query: 429 VVKPTRVEPAAEVVRRTEPPARVTRSSA--HSSKPALTSDDDLNLFDALPISALLQHSSN 486
PT P++ + P SS SS PA T + P+++ SS+
Sbjct: 579 APVPT---PSSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSS 635
Query: 487 PLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPL 546
P P S E +++P +P SS + S P S ++ ++ +T
Sbjct: 636 APVPTPSSSTTESSSAPVPTPSSSTTESSSAPVP-----TPSSSTTESSSAPVTSSTTES 690
Query: 547 SSEPIIHEQPEPNQTEPQPRTSDHSAPRASA---RPAARTTDTDSSTAFTPVSFPINVVD 603
SS P+ E + + P P S + +SA P++ TT++ S+ TP S
Sbjct: 691 SSAPVTSSTTE-SSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTP-SSSTTESS 748
Query: 604 SSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEA 663
S+P +S + V S E S P P P+ P +
Sbjct: 749 SAPVTSSTTESSSAPVPTPSSSTTES------SSAPVPTPSSSTTESSSAPV---PTPSS 799
Query: 664 DPLVQQAHPVPDQQEPIQ-----PDPEPEQSVSNQSSVRSPHP---LVETSDPHLGTSEP 715
PVP P P S + SSV P P E+S + +S
Sbjct: 800 STTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESSSAPVSSSTT 859
Query: 716 NAPMINIGSPQGASEAHSSNHPASP 740
+ + + +P +S SS + P
Sbjct: 860 ESSVAPVPTPSSSSNITSSAPSSIP 884
Score = 43.1 bits (100), Expect = 0.003
Identities = 73/356 (20%), Positives = 120/356 (33%), Gaps = 31/356 (8%)
Query: 402 TKRKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKP 461
TK KP D + P KKK T T P + ++ SS P
Sbjct: 281 TKEKPTPPHHDTT----PCTKKK--------TTTSKTCTKKTTTPVPTPSSSTTESSSAP 328
Query: 462 ALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHS----PRSSFFQPSPT 517
T + P+++ SS+ P P S E +++P S S+ S T
Sbjct: 329 VPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTT 388
Query: 518 EAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSD-HSAPRAS 576
E+ + S ++ ++ +T SS P+ E + T++ SAP S
Sbjct: 389 ESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTS 448
Query: 577 ARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPS 636
+ + + + ++ T S V S+ ++S + E SA T S
Sbjct: 449 STTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESS 508
Query: 637 PRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQ--QEPIQPDPEPEQSVSNQS 694
P P P P + + PV + P P P S + S
Sbjct: 509 SAPVPTPSSSTTESSSAPA---PTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESS 565
Query: 695 SVRSPHPLVETSD-----PHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLS 745
S E+S P T+E ++ + P +S S+ +P P+ S
Sbjct: 566 STPVTSSTTESSSAPVPTPSSSTTESSSAPV----PTPSSSTTESSSAPAPTPSSS 617
Score = 38.1 bits (87), Expect = 0.11
Identities = 61/296 (20%), Positives = 105/296 (34%), Gaps = 14/296 (4%)
Query: 452 TRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSF 511
T+SS +S + +S + ++ ++ SS + ES + TT+P +S
Sbjct: 208 TKSSTTTSSTSESSTTTSSTSESSTTTSSTSESSTTTSSTSESSTSSSTTAPATPTTTSC 267
Query: 512 FQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHS 571
+ PT + + +P+ T+ T +S+ + P T T S
Sbjct: 268 TKEKPTPPTTTSCTKEKPTPPHHDTTPCT-KKKTTTSKTCTKKTTTPVPTPSSSTTESSS 326
Query: 572 APRASARPAARTTDTDSS--TAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEE 629
AP + P++ TT++ S+ T+ T S V S S S E SA
Sbjct: 327 APVPT--PSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVT 384
Query: 630 YYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQS 689
T S P P P+ ++ + + V + E +
Sbjct: 385 SSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSA 444
Query: 690 VSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLS 745
S+ S V T P T+E ++ +P +S SS+ P P P+ S
Sbjct: 445 PVTSSTTESSSAPVPT--PSSSTTESSS------APVTSSTTESSSAPV-PTPSSS 491
Score = 37.7 bits (86), Expect = 0.15
Identities = 61/311 (19%), Positives = 108/311 (34%), Gaps = 19/311 (6%)
Query: 445 TEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPP 504
T + T S++ SS ++ + + S+ ++ P TP S E+ T PP
Sbjct: 217 TSESSTTTSSTSESSTTTSSTSESSTTTSSTSESSTSSSTTAPATPTTTSCTKEKPT-PP 275
Query: 505 HSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQ 564
+ + +P+P + + + S T T P P S P T
Sbjct: 276 TTTSCTKEKPTPPHHDTTPCTKKKTTTSKTCTKKTTTPV-PTPSSSTTESSSAPVPTPSS 334
Query: 565 PRTSDHSAPRASAR--------PAARTTDTDSSTAFTPVSFPINVVDSSP--SNNSESIR 614
T SAP S+ P ++ T+SS+A PV+ S+P S+ +ES
Sbjct: 335 STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSA--PVTSSTTESSSAPVTSSTTESSS 392
Query: 615 KFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVP 674
+ + +T + P + + + E+ +
Sbjct: 393 APVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTE 452
Query: 675 DQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSS 734
P+ P P S + SS E+S + T ++ + +P +S SS
Sbjct: 453 SSSAPV---PTPSSSTTESSSAPVTSSTTESSSAPVPTPS-SSTTESSSAPVTSSTTESS 508
Query: 735 NHPASPAPNLS 745
+ P P P+ S
Sbjct: 509 SAPV-PTPSSS 518
Score = 32.0 bits (71), Expect = 8.0
Identities = 50/259 (19%), Positives = 81/259 (30%), Gaps = 21/259 (8%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRK----------PDDGDDDDSEDGP 418
P P PS ++ V ++E S P+ + P + S
Sbjct: 736 PVPT-PSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPV 794
Query: 419 PQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSS-KPALTSDDDLNLFDALPI 477
P V P ++ + + P + SS SS P T + P+
Sbjct: 795 PTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESSSAPV 854
Query: 478 SALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTS 537
S+ SS P P S +++P P SS + T + PS S P S
Sbjct: 855 SSSTTESSVAPVPTPSSSSNITSSAPSSIPFSSTTESFSTGTTV------TPSSSKYPGS 908
Query: 538 LLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSS---TAFTP 594
+ I+ + + T P T + A TT S T P
Sbjct: 909 QTETSVSSTTETTIVPTKTTTSVTTPSTTTITTTVCSTGTNSAGETTSGCSPKTVTTTVP 968
Query: 595 VSFPINVVDSSPSNNSESI 613
+ +V SS + + ++
Sbjct: 969 TTTTTSVTTSSTTTITTTV 987
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 56.6 bits (135), Expect = 3e-07
Identities = 65/328 (19%), Positives = 108/328 (32%), Gaps = 19/328 (5%)
Query: 432 PTRVEPAAEVVRRTEPPARVTRS---SAHSSKPALTSDDDLNLFDALPISALLQHSSNPL 488
P P+ + T P T S S ++ P T+ + P + ++
Sbjct: 1433 PPTTTPSPPITTTTTPLPTTTPSPPISTTTTPPPTTTPSPPTTTPSPPTTTPSPPTTTTT 1492
Query: 489 TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSS 548
TP P + P+ T+P P S+ P T P + P+ T P P +S
Sbjct: 1493 TPPPTTTPSPPMTTPITPPASTTTLPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTS 1552
Query: 549 EPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSN 608
+ P T P P + + P + P+ TT T S T + P S P+
Sbjct: 1553 TTTL-----PPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPTT 1607
Query: 609 NSESIRKFMEVRKEKVSALEEYYLTCP-SPRRYPGPRPERLVDPDEPILANP---IQEAD 664
+ + + + T P P P P P +P +
Sbjct: 1608 TTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSP 1667
Query: 665 PLVQQAHPVPDQ--QEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINI 722
P+ P P PI P P + ++ +P P S P T+ P++
Sbjct: 1668 PITTTTTPPPTTTPSSPITTTPSPPTT-----TMTTPSPTTTPSSPITTTTTPSSTTTPS 1722
Query: 723 GSPQGASEAHSSNHPASPAPNLSIIPYT 750
P + + P+ P ++ +P T
Sbjct: 1723 PPPTTMTTPSPTTTPSPPTTTMTTLPPT 1750
Score = 47.4 bits (111), Expect = 2e-04
Identities = 69/314 (21%), Positives = 97/314 (29%), Gaps = 27/314 (8%)
Query: 432 PTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDL-NLFDALPISALLQH--SSNPL 488
P P+ T PP T S P T+ L + PIS ++ P
Sbjct: 1417 PPTTTPSPPTTTTTTPPPTTT-----PSPPITTTTTPLPTTTPSPPISTTTTPPPTTTPS 1471
Query: 489 TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSS 548
P P T SPP + ++ P PT P + P S T+P S
Sbjct: 1472 PPTTTPSPPTTTPSPPTTTTTT---PPPTTTP----SPPMTTPITPPASTTTLPPTTTPS 1524
Query: 549 EPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSN 608
P P T P P T+ P S TT T S T + P S P+
Sbjct: 1525 PPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTT-TPSPPPTTTTTPPPTTTPSPPTT 1583
Query: 609 NSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQ 668
+ S ++ T PSP P P P PI
Sbjct: 1584 TTPS--------PPTITTTTPPPTTTPSPPTTTTTTPPPTTTPSPP-TTTPITPPTSTTT 1634
Query: 669 QAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGA 728
P P + + + +P P + T+ T+ P++P+ SP
Sbjct: 1635 LPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTT 1694
Query: 729 SEAHSS--NHPASP 740
+ S P+SP
Sbjct: 1695 TMTTPSPTTTPSSP 1708
Score = 41.2 bits (95), Expect = 0.013
Identities = 41/187 (21%), Positives = 66/187 (34%), Gaps = 19/187 (10%)
Query: 432 PTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPI 491
PT P+ + T PP T S ++ + PI+ ++ P T
Sbjct: 1581 PTTTTPSPPTITTTTPPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTT 1640
Query: 492 PESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPI 551
P S P TT+PP P+ T +P + P + T P P+++ P
Sbjct: 1641 P-SPPPTTTTTPP---------PTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTP- 1689
Query: 552 IHEQPEPNQTEPQPRTSDHS------APRASARPAARTTDTDSSTAFTPVSFPINVVDSS 605
P T P P T+ S P ++ P+ T + + T S P + +
Sbjct: 1690 --SPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMTTL 1747
Query: 606 PSNNSES 612
P + S
Sbjct: 1748 PPTTTSS 1754
Score = 41.2 bits (95), Expect = 0.013
Identities = 55/275 (20%), Positives = 89/275 (32%), Gaps = 34/275 (12%)
Query: 489 TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSS 548
T P + P TT+ +P + Q +PT P+ P+ + PT T P+++
Sbjct: 4078 TQTPTTTPITTTTTVTPTPTPTGTQ-TPTTTPITTTTTVTPTPT--PTGTQTPTTTPITT 4134
Query: 549 EPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDS------STAFTPVSFPINVV 602
+ P P T+ T + + P T T + +T TP P
Sbjct: 4135 TTTVTPTPTPTGTQTPTTTPITTTTTVTPTPTPTGTQTPTTTPITTTTTVTPTPTPTGTQ 4194
Query: 603 DSSPSNNSESIRKFMEVRKEKVSALEEYYLTCP-SPRRYPGPRPERLVDPDEPILANPIQ 661
P++ S T P + P PE ++P+
Sbjct: 4195 TGPPTHTS----------------------TAPIAELTTSNPPPESSTPQTSRSTSSPLT 4232
Query: 662 EADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMIN 721
E+ L+ P + P + ++ SP P TS P T +
Sbjct: 4233 ESTTLLSTLPPAIEMTSTAPPSTPTAPTTTSGGHTLSPPPSTTTSPPGTPTRGTTTGSSS 4292
Query: 722 IGSPQGASEAHSSNHPASPAP--NLSIIPYTHLQP 754
+P +S +P P SII T L+P
Sbjct: 4293 APTPSTVQTTTTSAWTPTPTPLSTPSIIRTTGLRP 4327
Score = 40.4 bits (93), Expect = 0.023
Identities = 55/276 (19%), Positives = 83/276 (29%), Gaps = 37/276 (13%)
Query: 486 NPLTPIPESQPAEQTTSPPHS----PRSSFFQPSPTEAP---LWNLLQNQPSRSDEPTSL 538
+P T P P TT PP + P ++ P PT P + P+ + P
Sbjct: 1400 SPPTTTPSPPPTTTTTLPPTTTPSPPTTTTTTPPPTTTPSPPITTTTTPLPTTTPSPPIS 1459
Query: 539 LTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFP 598
T P ++ P P T P P T+ + P + P+ T + A T + P
Sbjct: 1460 TTTTPPPTTTPSPPTTTPSPPTTTPSPPTTTTTTPPPTTTPSPPMTTPITPPAST-TTLP 1518
Query: 599 INVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILAN 658
S P+ + T P P P P + P
Sbjct: 1519 PTTTPSPPTTTT----------------------TTPPPTTTPSPPTTTPITPPTSTTTL 1556
Query: 659 P---IQEADPLVQQAHP---VPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGT 712
P P P P P P P + + +P P T+ T
Sbjct: 1557 PPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSP-PTITTTTPPPTTTPSPPTTTTTTPPPT 1615
Query: 713 SEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIP 748
+ P+ P +P ++ SP P + P
Sbjct: 1616 TTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTP 1651
Score = 37.4 bits (85), Expect = 0.19
Identities = 47/222 (21%), Positives = 65/222 (29%), Gaps = 33/222 (14%)
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVR 427
PP PSPP S + P+ T S
Sbjct: 1596 PPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTT 1655
Query: 428 IVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNP 487
PT P+ + T PP T SS ++ P+ P + + S
Sbjct: 1656 TPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPS-------------PPTTTMTTPSPT 1702
Query: 488 LTPIPESQPAEQTTSP-----PHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIP 542
TP S P TT+P P P ++ PSPT P PT+ +T
Sbjct: 1703 TTP---SSPITTTTTPSSTTTPSPPPTTMTTPSPTTTP------------SPPTTTMTTL 1747
Query: 543 YDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTT 584
+S P+ P+ T P + P P T
Sbjct: 1748 PPTTTSSPLTTTPLPPSITPPTFSPFSTTTPTTPCVPLCNWT 1789
Score = 32.0 bits (71), Expect = 8.0
Identities = 37/125 (29%), Positives = 50/125 (39%), Gaps = 13/125 (10%)
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPS-PTEAPLWN---LLQNQPSR----SDEPT 536
+ P T + AE TTS P P SS Q S T +PL LL P S P
Sbjct: 4195 TGPPTHTSTAPIAELTTSNP-PPESSTPQTSRSTSSPLTESTTLLSTLPPAIEMTSTAPP 4253
Query: 537 SLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPR-ASARPAARTTDTDSSTAFTPV 595
S T P + P P+ T P T +S+ P T T +++A+TP
Sbjct: 4254 STPTAPTTTSGGHTL---SPPPSTTTSPPGTPTRGTTTGSSSAPTPSTVQTTTTSAWTPT 4310
Query: 596 SFPIN 600
P++
Sbjct: 4311 PTPLS 4315
>TEGU_EBV (P03186) Large tegument protein
Length = 3149
Score = 55.8 bits (133), Expect = 5e-07
Identities = 73/328 (22%), Positives = 106/328 (32%), Gaps = 38/328 (11%)
Query: 416 DGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDAL 475
+ PP P PA+ P + S+A +S +S LF +
Sbjct: 329 NSPPSSPASAAPASAAPASAAPASAAPASAAPASAAPASAAPASAAPASSPP---LF--I 383
Query: 476 PISALLQHSSNPLTPIPESQPAEQTTSPPHSPR--SSFFQPSPTEAPLWNLLQNQPSRSD 533
PI L P P P + P + + P +P+ + SP + P S +D
Sbjct: 384 PIPGL---GHTPGVPAPSTPPRASSGAAPQTPKRKKGLGKDSPHKKPTSGRRLPLSSTTD 440
Query: 534 EPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFT 593
L + P P P P P P S ++P P + + + +
Sbjct: 441 TEDDQLPRTHVPPHRPPSAARLPPP--VIPIPHQSPPASPTPHPAPVSTIAPSVTPSPRL 498
Query: 594 PVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDE 653
P+ PI + ++PSN + LT PSP P
Sbjct: 499 PLQIPIPLPQAAPSN-------------------PKIPLTTPSPSPTAAAAPTTTTLSPP 539
Query: 654 PILANPIQEA----DPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPH 709
P P Q A PL+ Q P P P P Q S+ R+P PL P
Sbjct: 540 PTQQQPPQSAAPAPSPLLPQQQPTPSAAPA--PSPLLPQQQPPPSAARAPSPLPPQQQP- 596
Query: 710 LGTSEPNAPMINIGSPQGASEAHSSNHP 737
L ++ P P P + NHP
Sbjct: 597 LPSATPAPPPAQQLPPSATTLEPEKNHP 624
Score = 47.8 bits (112), Expect = 1e-04
Identities = 58/231 (25%), Positives = 84/231 (36%), Gaps = 13/231 (5%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKT--KRKPDDGDDDDSEDGPPQKKKKKV 426
PAP P A + + D P K T +R P D +D P+
Sbjct: 395 PAPSTPPRASSGAAPQTPKRKKGLGKDSPHKKPTSGRRLPLSSTTDTEDDQLPRTHVPPH 454
Query: 427 RIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSK--PALTSDDDLNLFDALPI-SALLQH 483
R R+ P + PPA T A S P++T L L +P+ A +
Sbjct: 455 R-PPSAARLPPPVIPIPHQSPPASPTPHPAPVSTIAPSVTPSPRLPLQIPIPLPQAAPSN 513
Query: 484 SSNPLT-----PIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSL 538
PLT P + P T SPP + + +P +PL Q PS + P+ L
Sbjct: 514 PKIPLTTPSPSPTAAAAPTTTTLSPPPTQQQPPQSAAPAPSPLLPQQQPTPSAAPAPSPL 573
Query: 539 LTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSS 589
L P S+ P P Q +P P + P P+A T + + +
Sbjct: 574 LPQQQPPPSAARA--PSPLPPQQQPLPSATPAPPPAQQLPPSATTLEPEKN 622
Score = 36.2 bits (82), Expect = 0.43
Identities = 60/291 (20%), Positives = 101/291 (34%), Gaps = 23/291 (7%)
Query: 432 PTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSD-DDLNLFDALPISALLQHSSN---P 487
P PA+ P + S+A +S P L L +P + +S+ P
Sbjct: 350 PASAAPASAAPASAAPASAAPASAAPASSPPLFIPIPGLGHTPGVPAPSTPPRASSGAAP 409
Query: 488 LTP-----IPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQP-SRSDEPTSLLTI 541
TP + + P ++ TS P SS + P ++ ++P S + P ++ I
Sbjct: 410 QTPKRKKGLGKDSPHKKPTSGRRLPLSSTTDTEDDQLPRTHVPPHRPPSAARLPPPVIPI 469
Query: 542 PYDPLSSEPIIHEQP----EPNQTEPQPRTS-DHSAPRASARPA-ARTTDTDSSTAFTPV 595
P+ + P H P P+ T P PR P A P+ + T S + T
Sbjct: 470 PHQSPPASPTPHPAPVSTIAPSVT-PSPRLPLQIPIPLPQAAPSNPKIPLTTPSPSPTAA 528
Query: 596 SFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSP-----RRYPGPRPERLVD 650
+ P S P + + + + P+P ++ P P R
Sbjct: 529 AAPTTTTLSPPPTQQQPPQSAAPAPSPLLPQQQPTPSAAPAPSPLLPQQQPPPSAARAPS 588
Query: 651 PDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHP 701
P P P+ A P A +P ++P+ + + SP P
Sbjct: 589 PLPP-QQQPLPSATPAPPPAQQLPPSATTLEPEKNHPPAADRAGTEISPSP 638
>MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia
protein 2 (ALL1-related protein)
Length = 5262
Score = 55.5 bits (132), Expect = 7e-07
Identities = 114/486 (23%), Positives = 170/486 (34%), Gaps = 83/486 (17%)
Query: 313 PAHEDTSSEIRGRRMPLNDYPLWTQADNPEAIRCYVEDLRAQGFEID------LDDFVSR 366
P E + + + P + Q P ++C + L G +++ L++ +
Sbjct: 381 PTDEPDALYVACQGQPKGGHVTSMQPKEPGPLQCEAKPLGKAGVQLEPQLEAPLNEEMPL 440
Query: 367 LPPAPEFP-SPPKRK--------AQRKMVLDESSEESDVP----LVKKTKRKPDDGDDDD 413
LPP E P SPP + A R E S +P L + + P ++
Sbjct: 441 LPPPEESPLSPPPEESPTSPPPEASRLSPPPEELPASPLPEALHLSRPLEESPLSPPPEE 500
Query: 414 SEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFD 473
S PP + P P + PA T S +
Sbjct: 501 SPLSPPPESSP-----FSPLEESPLSPPEESPPSPALETPLSPPP--------------E 541
Query: 474 ALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSD 533
A P+S + S PL+P PE P TSPP P +S P P E+P+ + P
Sbjct: 542 ASPLSPPFEES--PLSPPPEELP----TSPP--PEASRLSPPPEESPMSPPPEESPMSPP 593
Query: 534 EPTSLLTIPYD--PLSSEPI---IHEQPEPNQTEPQPRTSDHSAP-------------RA 575
S L P++ PLS P + PE ++ P P S S P R
Sbjct: 594 PEASRLFPPFEESPLSPPPEESPLSPPPEASRLSPPPEDSPMSPPPEESPMSPPPEVSRL 653
Query: 576 SARPAAR--TTDTDSSTAFTPVSFPINVVD---SSPSNNSESIRKFMEVRKEKVSALEEY 630
S P + + S P P+ ++ + ++ + + +S E
Sbjct: 654 SPLPVVSRLSPPPEESPLSPPALSPLGELEYPFGAKGDSDPESPLAAPILETPISPPPEA 713
Query: 631 YLTCPSP--RRYPGPRPERLVDPDEPILANPI-QEADPLVQQAHPVPDQQEPIQPDPEPE 687
T P P P P V P PIL P+ + PL+Q + VP P Q P P
Sbjct: 714 NCTDPEPVPPMILPPSPGSPVGPASPILMEPLPPQCSPLLQHS-LVPQNSPPSQCSP-PA 771
Query: 688 QSVSNQSSVRSPHPLVETSDP----HLGT---SEPNAPMI--NIGSPQGASEAHSSNHPA 738
+S S + +V SD + T SEP P + + SP + S
Sbjct: 772 LPLSVPSPLSPIGKVVGVSDEAELHEMETEKVSEPECPALEPSATSPLPSPMGDLSCPAP 831
Query: 739 SPAPNL 744
SPAP L
Sbjct: 832 SPAPAL 837
Score = 36.6 bits (83), Expect = 0.33
Identities = 62/306 (20%), Positives = 121/306 (39%), Gaps = 48/306 (15%)
Query: 641 PGPRPERLVDPDEPILANPIQEADPLVQQA--HPVPDQQEPIQPDPEPEQSVSNQS-SVR 697
P RP D EP LA+ + E P V++ HP P Q P EP + ++ ++
Sbjct: 2821 PEERPPPAADASEPRLASVLPEVKPKVEEGGRHPSPCQFTIATPKVEPAPAANSLGLGLK 2880
Query: 698 SPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTSL 757
++ + D +GT P + G A ++ A+ P ++ + L
Sbjct: 2881 PGQSMMGSRDTRMGT----GPFSSSG-----HTAEKASFGATGGPPAHLLTPSPLSGPGG 2931
Query: 758 SECINIFYHEASLRLRNVHGQTDLSENAENVADEWNNLSTWLVA-QVPIMMQLLHAEGSQ 816
S + F E+ G L DE + + + LVA ++P++++ L
Sbjct: 2932 SSLLEKFELES--------GALTLPGGPAASGDELDKMESSLVASELPLLIEDL------ 2977
Query: 817 SIEAAKQRFARRVALHELEQRNKLLEAIEEAKRKKEQAEEAARQAAAQAEQARLEAERLE 876
+E K+ EL+++ +L ++ A+++++Q ++ + A QA
Sbjct: 2978 -LEHEKK---------ELQKKQQLSAQLQPAQQQQQQQQQHSLLPAPGPAQAMSLPHEGS 3027
Query: 877 AEAEALRCQALAPVVFTPAASASIPDPQAAQNVPSSSTQTSSSRLDIMEQRLDTHESMLI 936
+ + A Q L+ + A +P P P+ + ++QRL +M+
Sbjct: 3028 SPSLAGSQQQLS-LGLAVARQPGLPQPLMPTQPPAHA----------LQQRLAPSMAMVS 3076
Query: 937 EMKHMM 942
HM+
Sbjct: 3077 NQGHML 3082
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 54.3 bits (129), Expect = 2e-06
Identities = 94/426 (22%), Positives = 140/426 (32%), Gaps = 57/426 (13%)
Query: 369 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRI 428
PAP P+PP + S P + P G + PP
Sbjct: 43 PAPPSPAPPS-----------PAPPSPAP---PSPAPPSPGPPSPAPPSPPSPAPPSPA- 87
Query: 429 VVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPL 488
P P + PP+ S A S PA S P + +P
Sbjct: 88 ---PPSPAPPSPAPPSPAPPSPAPPSPAPPS-PAPPSPPSPAPPSPSPPAPPSPSPPSPA 143
Query: 489 TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSS 548
P+P S PA + SPP P PSP P P P+ +P P
Sbjct: 144 PPLPPS-PAPPSPSPPVPP-----SPSPPVPP-----SPAPPSPTPPSPSPPVPPSPAPP 192
Query: 549 EPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSN 608
P P P P P AP + PA + + + + +P + P + V SP+
Sbjct: 193 SPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPA-PPSPVPPSPAP 251
Query: 609 NSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQ 668
S + K A P P P P P + P+ +P P
Sbjct: 252 PSPA------PPSPKPPAPP------PPPSPPPPPPPRPPFPANTPMPPSP---PSPPPS 296
Query: 669 QAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGA 728
A P P P P P P V + P P + P S P +P SP +
Sbjct: 297 PAPPTP----PTPPSPSPPSPVPPSPAPVPPSP----APPSPAPSPPPSPAPPTPSPSPS 348
Query: 729 SEAHSSNHPA---SPAPNLSIIPYTHLQPTSLSECINIFYHEASLRLRNVHGQTDLSENA 785
S P+ SP+P+ S IP +P+ + + + + ++ +++G + +A
Sbjct: 349 PSPSPSPSPSPSPSPSPSPSPIPSPSPKPSPSPVAVKLVWADDAIAFDDLNGTSTRPGSA 408
Query: 786 ENVADE 791
+ E
Sbjct: 409 SRMVGE 414
Score = 45.4 bits (106), Expect = 7e-04
Identities = 56/239 (23%), Positives = 71/239 (29%), Gaps = 19/239 (7%)
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGP-------PQ 420
PPAP PSPP S VP P + P P
Sbjct: 131 PPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPA 190
Query: 421 KKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISAL 480
+ P PA V PP+ + + PA S +P S
Sbjct: 191 PPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSP- 249
Query: 481 LQHSSNPLTPIPESQPAEQTTSPPHSPRSSF--------FQPSPTEAPLWNLLQNQPSRS 532
S P +P P + P + PP PR F PSP +P PS S
Sbjct: 250 APPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAPPTPPTPPSPS 309
Query: 533 DE---PTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDS 588
P S +P P P P P P P S +P S P+ + + S
Sbjct: 310 PPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSPSPSPSPSPS 368
>SON_MOUSE (Q9QX47) SON protein
Length = 2404
Score = 53.5 bits (127), Expect = 3e-06
Identities = 84/409 (20%), Positives = 150/409 (36%), Gaps = 58/409 (14%)
Query: 369 PAPEFPSPPKRKAQR-KMVLDESSEESDVPLVKKTKRKPDDGDDD---------DSEDGP 418
P E P+ +K ++ K + +E + KK KR+P++ + +S+
Sbjct: 99 PTDEVPTKKSKKHKKHKNKKKKKKKEKE----KKYKRQPEESESKLKSHHDGNLESDSFL 154
Query: 419 PQKKKKKVRIVVKPTR---VEPAAEVVRRTEPPA-----------RVTRSSAHSSKPALT 464
+ + P R + A+E EPP + + +++ ++
Sbjct: 155 KFDSEPSAAALEHPVRAFGLSEASETALVLEPPVVSMEVQESHVLETLKPATKAAELSVV 214
Query: 465 SDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNL 524
S ++ P+ +L+ S + + P +T+ SP + +
Sbjct: 215 STSVISEQSEQPMPGMLEPSMTKILDSFTAAPVPMSTAALKSPEPVVTMSVEYQKSVLKS 274
Query: 525 LQNQPSRSDEPTSLLTIPY----DPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPA 580
L+ P + + T+L+ +P +P + I+ E P EP P T D P
Sbjct: 275 LETMPPETSK-TTLVELPIAKVVEPSETLTIVSETPTEVHPEPSPSTMDF--------PE 325
Query: 581 ARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRY 640
+ TTD PV P + DSS + ES+ K +A+E T S
Sbjct: 326 SSTTDVQRLPE-QPVEAPSEIADSSMTRPQESL------ELPKTTAVELQESTVASALEL 378
Query: 641 PGPRPERLVD----PDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSV 696
PGP +++ P P+ P A P+ + + P+ P+ P P +V +
Sbjct: 379 PGPPATSILELQGPPVTPVPELPGPSATPVPELSGPL---STPVPELPGPPATVVPELPG 435
Query: 697 RSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLS 745
S P+ + S G P AP + + PQ E S P +S
Sbjct: 436 PSVTPVPQLSQELPG---PPAPSMGLEPPQEVPEPPVMAQELSGVPAVS 481
Score = 35.0 bits (79), Expect = 0.95
Identities = 68/316 (21%), Positives = 106/316 (33%), Gaps = 64/316 (20%)
Query: 440 EVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQ 499
E + + EPP + S + PA S + +++L + P P +P
Sbjct: 1159 ESLSQPEPPVSQSEISEPMAVPANYSMSESE-------TSMLASEAVMTVPEPAREPESS 1211
Query: 500 TTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLS--SEPIIHEQP- 556
TS P E P+ ++ ++ + S EP ++LT +S SE +P
Sbjct: 1212 VTSAPVESAVVAEHEMVPERPMTYMV-SETTMSVEP-AVLTSEASVISETSETYDSMRPS 1269
Query: 557 ------------EPNQTEPQPRTSDHSAPRASARPAARTTDTDSS----TAFTPVSFPIN 600
EP T QP P + + T T+S T PV+ P
Sbjct: 1270 GHAISEVTMSLLEPAVTISQPAEDSLELPSMTVPAPSTMTTTESPVVAVTEIPPVAVPEP 1329
Query: 601 VVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPR---------------- 644
+ + P + M V K A+ E + P P P
Sbjct: 1330 PIMAVPELPT------MAVVKTPAVAVPEPLVAAPEPPTMATPELCSLSVSEPPVAVSEL 1383
Query: 645 -----PERLV---------DPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSV 690
PE + +P PIL + P++ + P QEP EP V
Sbjct: 1384 PALADPEHAITAVSGVSSLEPSVPILEPAVSVLQPVMIVSEPSVPVQEPTVAVSEPAVIV 1443
Query: 691 SNQSSVRSPHPLVETS 706
S + + SP VE+S
Sbjct: 1444 SEHTQITSPEMAVESS 1459
Score = 34.3 bits (77), Expect = 1.6
Identities = 37/155 (23%), Positives = 59/155 (37%), Gaps = 20/155 (12%)
Query: 530 SRSDEPTSLLTIPYDPLSSEPIIHEQP--EPNQTEPQPRTSDHSAPRASARPAARTTDTD 587
S +D T +P P P + P EP T P P P S +A T D
Sbjct: 1089 SYTDSYTEAYMVPPLPPEEPPTMPPLPPEEPPMTPPLPPEEPPEGPALSTEQSALTADNT 1148
Query: 588 SSTAFT-----PVSFPINVVDSSPSNNSESIRKFMEVRKEKVSAL-EEYYLTCPSPRRYP 641
ST T +S P V S + ++ + + + S L E +T P P R
Sbjct: 1149 WSTEVTLSTGESLSQPEPPVSQSEISEPMAVPANYSMSESETSMLASEAVMTVPEPAR-- 1206
Query: 642 GPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQ 676
+P+ + + P++ A +V + VP++
Sbjct: 1207 --------EPESSVTSAPVESA--VVAEHEMVPER 1231
>PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment)
Length = 5120
Score = 53.1 bits (126), Expect = 3e-06
Identities = 59/270 (21%), Positives = 100/270 (36%), Gaps = 30/270 (11%)
Query: 362 DFVSRLPPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQK 421
D S P P+ P PPK+K + S + K KPD DS+ PQK
Sbjct: 428 DLASGHGPGPQLP-PPKQKTPTPASTAKPSPQLQPGQKKDASPKPDPSQQADSKKPVPQK 486
Query: 422 KKKKV----RIVVKPTRVEPAAEVVRRTEPP----ARVTRSSAHSSKPALTSDDDLNLFD 473
K+ + + K T EP+ + P + T++ ++P++ D
Sbjct: 487 KQPSMPGSPPVKSKQTHAEPSDTGQQIDSTPKSDQVKPTQAEEKQNQPSIQKP----TMD 542
Query: 474 ALPISA---LLQHSSNPLTPIPESQ----PAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQ 526
+P SA + Q ++P +P + + P +TT PP + +P P + +
Sbjct: 543 TVPTSAAPGVKQDLADPQSPSTQQKVTDSPMPETTKPPADTHPAGDKPDSKPLPQVSRQK 602
Query: 527 NQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDT 586
+ P + + + S + + P+ QT+P P+ AP+ P A T
Sbjct: 603 SDPKLASQSGAKSDAKTQKPSEPAPVKDDPKKLQTKPAPKPDTKPAPKG---PQAGTGPR 659
Query: 587 DSSTAFTPVSFPINVVDSSPSNNSESIRKF 616
+S P P E R+F
Sbjct: 660 PTSAQPAP-------QPQQPQKTPEQSRRF 682
Score = 40.0 bits (92), Expect = 0.030
Identities = 84/419 (20%), Positives = 148/419 (35%), Gaps = 56/419 (13%)
Query: 365 SRLPPAPEFPSPPKR--KAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKK 422
S+ P + P PK+ K D +E+ P + +K + ++ G Q
Sbjct: 148 SKPVPQQQQPGEPKQGQKPGPSHPGDSKAEQVKQPPQPRGPQKSQLQQSEPTKPGQQQTS 207
Query: 423 KKKVRIVVKPTRVEP-AAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALL 481
K KP +P +A+ + PP + + + S K P+
Sbjct: 208 AKTSAGPTKPLPQQPDSAKTSSQAPPPTKPSLQQSGSVK----QPSQQPARQGGPVKPSA 263
Query: 482 QHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNL--LQNQPSRSD--EPTS 537
Q + P +P Q T P P QP P + PL ++ P ++ +P+S
Sbjct: 264 QQAGPPKQQPGSEKPTAQQTGPAKQPP----QPGPGKTPLQQTGPVKQVPPQAGPTKPSS 319
Query: 538 LLTIPYDPLSSEPIIH----EQPEPNQTEPQPRTSD----HSAPRASARPAARTTD---- 585
L+ +P + +QP P + Q + S S P+ + P TT+
Sbjct: 320 QTAGAAKSLAQQPGLTKPPGQQPGPEKPLQQKQASTTQPVESTPKKTFCPLCTTTELLLH 379
Query: 586 TDSSTAFTPVSFPINVVDS----SPSNNSESIRKFMEVRKEKVSALEEYYLTCPSP---- 637
T + + VV S +P+ + I++++ + + AL + P
Sbjct: 380 TPEKANYNTCTQCHTVVCSLCGFNPNPHITEIKEWLCLNCQMQRALGGDLASGHGPGPQL 439
Query: 638 ----RRYPGP----RPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQS 689
++ P P +P + P + A+P + PVP +++P P P
Sbjct: 440 PPPKQKTPTPASTAKPSPQLQPGQKKDASPKPDPSQQADSKKPVPQKKQPSMPGSPP--- 496
Query: 690 VSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIP 748
V+S E SD G + P + P A E N P+ P + +P
Sbjct: 497 ------VKSKQTHAEPSD--TGQQIDSTPKSDQVKPTQAEE--KQNQPSIQKPTMDTVP 545
Score = 38.9 bits (89), Expect = 0.066
Identities = 61/258 (23%), Positives = 89/258 (33%), Gaps = 70/258 (27%)
Query: 490 PIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSE 549
P P+ P +Q T P +S +PSP Q QP + + + P S+
Sbjct: 435 PGPQLPPPKQKTPTP----ASTAKPSP---------QLQPGQKKDASP------KPDPSQ 475
Query: 550 PIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNN 609
++P P + +P + S P S + A +DT TP S + + N
Sbjct: 476 QADSKKPVPQKKQP---SMPGSPPVKSKQTHAEPSDTGQQIDSTPKSDQVKPTQAEEKQN 532
Query: 610 SESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEP-----ILANPIQEAD 664
SI+K T P+ PG + + L DP P + +P+ E
Sbjct: 533 QPSIQK-------------PTMDTVPTSAA-PGVKQD-LADPQSPSTQQKVTDSPMPETT 577
Query: 665 PLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGS 724
HP D+ PD +P VS Q SDP L S
Sbjct: 578 KPPADTHPAGDK-----PDSKPLPQVSRQK-----------SDPKL------------AS 609
Query: 725 PQGASEAHSSNHPASPAP 742
GA + P+ PAP
Sbjct: 610 QSGAKSDAKTQKPSEPAP 627
Score = 37.4 bits (85), Expect = 0.19
Identities = 104/552 (18%), Positives = 195/552 (34%), Gaps = 80/552 (14%)
Query: 367 LPPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKV 426
LP S PK +Q D +++ P K K D P K +
Sbjct: 595 LPQVSRQKSDPKLASQSGAKSDAKTQKPSEPAPVKDDPKKLQTKPAPKPDTKPAPKGPQA 654
Query: 427 RIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSN 486
+PT +PA + + + P + R S LNL
Sbjct: 655 GTGPRPTSAQPAPQPQQPQKTPEQSRRFS-------------LNL--------------G 687
Query: 487 PLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPL 546
+T P+ QP T+P + F A +++ + S + +P S + P P
Sbjct: 688 GITDAPKPQP----TTPQETVTGKLF---GFGASIFSQASSLISTAGQPGSQTSGPAPPA 740
Query: 547 SSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINV----- 601
+ +P QP +Q P+ P+A+ P + T +S P++ +
Sbjct: 741 TKQPQPPSQPPASQAPPKEAAQAQPPPKAA--PTKKETKPLASEKLGPMASDSTLTTKGS 798
Query: 602 -VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPI 660
++ PS +S K +K + L E P + + +P N
Sbjct: 799 DLEKKPSLAKDS--KHQTAEAKKPAELSEQEKASQPKVSCPLCKTGLNIGSKDPPNFNTC 856
Query: 661 QEADPLVQQA---HPVP---DQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSE 714
E +V +P+P + QE + + + ++++S Q PL P LG S+
Sbjct: 857 TECKKVVCNLCGFNPMPHIVEVQEWLCLNCQTQRAMSGQLGDMGKVPL-----PKLGPSQ 911
Query: 715 P-NAPMINIGSPQGASEAHSSNHPASPAPNLSIIP---------YTHLQPTSLSECINIF 764
P + P + +HS ++P + P LQ L + ++
Sbjct: 912 PVSKPPATPQKQPVPAVSHSPQKSSTPPTPAATKPKEEPSVPKEVPKLQQGKLEKTLSAD 971
Query: 765 YHEASLRLRNVHG------QTDLSENAENVAD-EWNNLSTWLVAQVPIMMQLLHAEGSQS 817
+ ++ + +T ++ + V+ E + L + + P ++LH +
Sbjct: 972 KIQQGIQKEDAKSKQGKLFKTPSADKIQRVSQKEDSRLQQTKLTKTPSSDKILHGVQKED 1031
Query: 818 IEAAKQRFARRVA----LHELEQRNKLLEAIEEAKRKKEQAEEAARQAAAQAEQARLEAE 873
I+ + + A+ + LH L++ + L+ ++ AK Q AQ E +L+
Sbjct: 1032 IKFQEAKLAKIPSADKILHRLQKEDPKLQQMKMAKALSAD----KIQPEAQKEDVQLQEV 1087
Query: 874 RLEAEAEALRCQ 885
RL A + Q
Sbjct: 1088 RLSKAVSADKIQ 1099
Score = 36.6 bits (83), Expect = 0.33
Identities = 40/151 (26%), Positives = 60/151 (39%), Gaps = 22/151 (14%)
Query: 598 PINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTC-PSPRRYPGPRPERLVDPDEPIL 656
P +++ S + + E+ RK +KE+ E+ L PS ++ P +LV P+
Sbjct: 85 PFDLISDSDAAHEEAGRKQKVTQKEQGKPEEQRGLAKHPSQQQSP-----KLVQQQGPVK 139
Query: 657 ANPIQEADPLVQQAHPVPDQQEPIQP----DPEPEQ-SVSNQSSVRSPHPLVETSDPHLG 711
P Q + + PVP QQ+P +P P P S V+ P L
Sbjct: 140 PTPQQ-----TESSKPVPQQQQPGEPKQGQKPGPSHPGDSKAEQVKQPPQPRGPQKSQLQ 194
Query: 712 TSEPNAPMINIGSPQGASEAHSSNHPASPAP 742
SEP P Q + A +S P P P
Sbjct: 195 QSEPTKP------GQQQTSAKTSAGPTKPLP 219
Score = 33.1 bits (74), Expect = 3.6
Identities = 26/92 (28%), Positives = 44/92 (47%), Gaps = 13/92 (14%)
Query: 671 HPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPM---------IN 721
+P+ +Q E I + +S S Q+S +SP P V S H +P+ M +
Sbjct: 4742 YPLKEQSENI----DHGKSHSGQNSQQSPKPSVIKSRSHGIFPDPSKDMQVPTIEKSHSS 4797
Query: 722 IGSPQGASEAHSSNHPASPAPNLSIIPYTHLQ 753
GS + +SE H +H S + + + + THL+
Sbjct: 4798 PGSSKSSSEGHLRSHGPSRSQSKTSVTQTHLE 4829
>NPBL_COPCI (Q00333) Rad9 protein (SCC2 homolog)
Length = 2157
Score = 52.0 bits (123), Expect = 8e-06
Identities = 71/277 (25%), Positives = 106/277 (37%), Gaps = 26/277 (9%)
Query: 492 PESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPI 551
P +P + + P S +S+ F S + N P TS ++ + L P
Sbjct: 154 PYQEPQQPSNWPASSYQSTPFAQSVFQRTTPNSAYPTPPPPGISTSSSSLAF-ALGGPP- 211
Query: 552 IHEQPEPNQ-TEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNS 610
QP P Q T QP AP PA + +T F P S P S P+ N+
Sbjct: 212 ---QPPPRQQTVTQPLFHPTQAPLPQPAPAPQPAPVPQATVFPPPSNPPAAQASQPTYNA 268
Query: 611 ES----IRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPL 666
+ F+E + +++A + T + +P P P +L P P + P
Sbjct: 269 DDSASFFNSFLERKTREMNATMQQ--TTSAKSIFP-PAPAKLPLPVPVASKPPARMKTPP 325
Query: 667 VQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPL-----VETSDPHLGTSEPNAPMIN 721
++ VPD PDP S+S+ ++ S PL VE P + PM
Sbjct: 326 PREQKAVPDS----SPDP---LSLSSNMALPSSTPLKRKCVVEIHSPPPKRIQSYKPMTL 378
Query: 722 IGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTSLS 758
I P S + P S A N S +P T TS++
Sbjct: 379 ISQPLTPSRG-APLTPTSRASNASSVPTTASSSTSIT 414
Score = 34.3 bits (77), Expect = 1.6
Identities = 51/211 (24%), Positives = 84/211 (39%), Gaps = 38/211 (18%)
Query: 418 PPQKKKKKVRIVVKPTRV---EPA-----AEVVRRTEPPARVTRSSAHSSKPALTSDDDL 469
PP +++ + + PT+ +PA A V + T P +A +S+P +DD
Sbjct: 213 PPPRQQTVTQPLFHPTQAPLPQPAPAPQPAPVPQATVFPPPSNPPAAQASQPTYNADDSA 272
Query: 470 NLFDAL------PISALLQHSSN------------PL-TPIPESQPAEQTTSPPHSPRSS 510
+ F++ ++A +Q +++ PL P+ PA T PP ++
Sbjct: 273 SFFNSFLERKTREMNATMQQTTSAKSIFPPAPAKLPLPVPVASKPPARMKTPPPREQKA- 331
Query: 511 FFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDH 570
P + PL +L N S P + + S P + +P QP T
Sbjct: 332 --VPDSSPDPL-SLSSNMALPSSTPLKRKCV-VEIHSPPPKRIQSYKPMTLISQPLTPSR 387
Query: 571 SAP-----RASARPAARTTDTDSSTAFTPVS 596
AP RAS + TT SST+ T +S
Sbjct: 388 GAPLTPTSRASNASSVPTT-ASSSTSITSMS 417
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 51.6 bits (122), Expect = 1e-05
Identities = 106/543 (19%), Positives = 191/543 (34%), Gaps = 75/543 (13%)
Query: 368 PPAPEFPSPPKR------KAQRKMVLDESSEESD-----------------VPLVKKTKR 404
PPA + P KR KA K SSEES P K K
Sbjct: 100 PPAKKAAVPAKRVGLPPGKAAAKASESSSSEESRDDDDEEDQKKQPVQKGVKPQAKAAKA 159
Query: 405 KP------DDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHS 458
P D D SED PP+ +K K+ V + + + R A +S+ S
Sbjct: 160 PPKKAKSSDSDSDSSSEDEPPKNQKPKITPVTVKAQTKAPPKPARAAPKIANGKAASSSS 219
Query: 459 SKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQ-----PAEQTTSPPHSPRSSFFQ 513
S + +S DD S + ++ P +P+ Q P + T+P SS
Sbjct: 220 SSSSSSSSDD---------SEEEKAAATPKKTVPKKQVVAKAPVKAATTPTRKSSSSEDS 270
Query: 514 PSPTEAPLWNLLQNQPS-RSDEPTSLLTIPYDPLSSEP--IIHEQPEPNQTEPQPRTSDH 570
S E ++N+P S P P L ++P E+ +P ++
Sbjct: 271 SSDEEEEQKKPMKNKPGPYSYAPPPSAPPPKKSLGTQPPKKAVEKQQPVESSEDSSDESD 330
Query: 571 SAPRASARPAARTTDTDSSTAFTPVSFPI-NVVDSSPSNNSESIRKFMEVRKEKVSALEE 629
S+ +P + + ++T P + DSS S++SE + ++ +
Sbjct: 331 SSSEEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSEDDEAPSKPAGTTKNSSNK 390
Query: 630 YYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQS 689
+T SP P P++ V + +L + D +
Sbjct: 391 PAVTTKSPAVKPAAAPKQPVGGGQKLLTR----------------------KADSSSSEE 428
Query: 690 VSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPY 749
S+ S +V T+ P T++ + +PQG+ ++ S + +S
Sbjct: 429 ESSSSEEEKTKKMVATTKPK-ATAKAALSLPAKQAPQGSRDSSSDSDSSSSEEEEEKTSK 487
Query: 750 THLQPTSLSECINIFYHEASLRLRNVHGQTDLSENAENVADEWNNLSTWLVAQVPIMMQL 809
+ ++ + + A + + S N+ + D L + Q
Sbjct: 488 SAVK----KKPQKVAGGAAPSKPASAKKGKAESSNSSSSDDSSEEEEEKLKGKGSPRPQA 543
Query: 810 LHAEGSQSIEAAKQRFARRVALHELEQRNKLLEAIEEAKRKKEQAEEAARQA-AAQAEQA 868
A G+ ++ A + A+ E E++ + + KK + EAA++A QA++
Sbjct: 544 PKANGTSALTAQNGKAAKNSEEEEEEKKKAAVVVSKSGSLKKRKQNEAAKEAETPQAKKI 603
Query: 869 RLE 871
+L+
Sbjct: 604 KLQ 606
Score = 41.6 bits (96), Expect = 0.010
Identities = 73/373 (19%), Positives = 129/373 (34%), Gaps = 48/373 (12%)
Query: 389 ESSEESDVPLVKKTKRKPDDGDDDDSED------GPPQKKKKKVRIVVKPTRVEPAAEVV 442
E +++ P+ KK K+K D +DS + GPP KK A
Sbjct: 65 ERKLQANGPVAKKAKKKASSSDSEDSSEEEEEVQGPPAKK--------------AAVPAK 110
Query: 443 RRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTS 502
R PP + ++ SS + DDD + +Q P ++ P + +S
Sbjct: 111 RVGLPPGKAAAKASESSSSEESRDDD---DEEDQKKQPVQKGVKPQAKAAKAPPKKAKSS 167
Query: 503 PPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPII--HEQPEPNQ 560
S SS +P + P + P T P P + P I + +
Sbjct: 168 DSDSDSSSEDEPPKNQKP-----KITPVTVKAQTK---APPKPARAAPKIANGKAASSSS 219
Query: 561 TEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVR 620
+ +SD S +A +T A PV S S+ S + E +
Sbjct: 220 SSSSSSSSDDSEEEKAAATPKKTVPKKQVVAKAPVKAATTPTRKSSSSEDSSSDEEEEQK 279
Query: 621 KEKVSALEEYYL----TCPSPRRYPGPR-PERLVDPDEPILANPIQEADPLVQQAHPVPD 675
K + Y + P P++ G + P++ V+ +P+ ++ + + D
Sbjct: 280 KPMKNKPGPYSYAPPPSAPPPKKSLGTQPPKKAVEKQQPVESS---------EDSSDESD 330
Query: 676 QQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSN 735
+ P + VS ++ P S S+ + P G ++ +SSN
Sbjct: 331 SSSEEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSEDDEAPSKPAGTTK-NSSN 389
Query: 736 HPASPAPNLSIIP 748
PA + ++ P
Sbjct: 390 KPAVTTKSPAVKP 402
>TEGU_HHV11 (P10220) Large tegument protein (Virion protein UL36)
Length = 3164
Score = 50.4 bits (119), Expect = 2e-05
Identities = 96/431 (22%), Positives = 138/431 (31%), Gaps = 43/431 (9%)
Query: 361 DDFVSRLPPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQ 420
DDF RLP P+PP A R +S S + R+ Q
Sbjct: 2644 DDF-RRLPSPQSSPAPPDATAPRPPASSRASAASSSGSRARRHRRARSLARATQASATTQ 2702
Query: 421 KKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISAL 480
+ P V P + R PP KP + L LP+
Sbjct: 2703 GWRPPAL----PDTVAPVTDFARPPAPP-----------KPPEPAPHALVSGVPLPLGPQ 2747
Query: 481 LQHSSNPLTPI-PESQPAEQTTSPP-----HSPRSSFFQPSPTEAPLWN----LLQNQPS 530
++P PI P P T P P +S P+P P+ L + +
Sbjct: 2748 AAGQASPALPIDPVPPPVATGTVLPGGENRRPPLTSGPAPTPPRVPVGGPQRRLTRPAVA 2807
Query: 531 RSDEPTSLLTIPYDPLS-SEPIIHEQP-EPNQTEPQ------PRTSDHSAPRASARPAAR 582
E L P+DP + P++ P EP + P P + P S P
Sbjct: 2808 SLSESRESLPSPWDPADPTAPVLGRNPAEPTSSSPAGPSPPPPAVQPVAPPPTSGPPPTY 2867
Query: 583 TTDTDSSTAFTPVSF-PINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYP 641
T PVS P + S R + V + + P P+ P
Sbjct: 2868 LTLEGGVAPGGPVSRRPTTRQPVATPTTSARPRGHLTVSRLSAPQPQPQPQPQPQPQPQP 2927
Query: 642 GPRPERLVDPDEPILANPIQEADPLVQ---QAHPVPDQQEPIQPDPEPEQSVSNQSSVRS 698
P+P+ P P + P Q Q P P Q QP P+P+ N
Sbjct: 2928 QPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQPQNGHVAPG 2987
Query: 699 PHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTSLS 758
+P V P S P A + G+S + S+ +S A ++ P P SL
Sbjct: 2988 EYPAVRFRAPQNRPSVP-ASASSTNPRTGSSLSGVSSWASSLALHIDATP----PPVSLL 3042
Query: 759 ECINIFYHEAS 769
+ + + E S
Sbjct: 3043 QTLYVSDDEDS 3053
>TARS_HUMAN (Q7Z7G0) Target of Nesh-SH3 precursor (Tarsh) (Nesh
binding protein) (NeshBP)
Length = 1075
Score = 49.7 bits (117), Expect = 4e-05
Identities = 74/317 (23%), Positives = 122/317 (38%), Gaps = 33/317 (10%)
Query: 418 PPQKKKKKV-RIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALP 476
P + K +V +I +PT V P V R T+P T S+ S+ L S + +
Sbjct: 319 PAESKTPEVEKISARPTTVTPET-VPRSTKP---TTSSALDVSETTLASSEKPWIVPTAK 374
Query: 477 ISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPT 536
IS S L P + + + P S + ++ L ++ ++P
Sbjct: 375 IS----EDSKVLQPQTATYDVFSSPTTSDEPEISDSYTATSDRILDSIPPKTSRTLEQPR 430
Query: 537 SLLT---IPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFT 593
+ L P+ P E + +P PQ TS S P+ RP T + +T+
Sbjct: 431 ATLAPSETPFVPQKLEIFTSPEMQPTTPAPQQTTSIPSTPKRRPRPKPPRTKPERTTSAG 490
Query: 594 PVSFPINVVDSSPSNNSESIRKFMEVR-KEKVSALEEYYLTCPSPRRYPGPRPERLVDPD 652
++ I+ P+ + + K + K K+ E T P+P++ P P+ P
Sbjct: 491 TITPKIS-KSPEPTWTTPAPGKTQFISLKPKIPLSPEVTHTKPAPKQTPRAPPKPKTSP- 548
Query: 653 EPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHP----LVETSDP 708
P + Q PVP + P + +P+ S S + S +P P L P
Sbjct: 549 -----------RPRIPQTQPVP--KVPQRVTAKPKTSPSPEVSYTTPAPKDVLLPHKPYP 595
Query: 709 HLGTSEPNAPMINIGSP 725
+ SEP AP+ G P
Sbjct: 596 EVSQSEP-APLETRGIP 611
Score = 43.5 bits (101), Expect = 0.003
Identities = 94/436 (21%), Positives = 153/436 (34%), Gaps = 73/436 (16%)
Query: 354 QGFEIDLDDFVSRLPPAPE----FPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDG 409
Q EI + PAP+ PS PKR+ + K + + + K +
Sbjct: 443 QKLEIFTSPEMQPTTPAPQQTTSIPSTPKRRPRPKPPRTKPERTTSAGTITPKISKSPEP 502
Query: 410 DDDDSEDGPPQKKKKKVRIVVKP--TRVEPAAEVVRR------TEPPARVTRSSAH---- 457
G Q K +I + P T +PA + R T P R+ ++
Sbjct: 503 TWTTPAPGKTQFISLKPKIPLSPEVTHTKPAPKQTPRAPPKPKTSPRPRIPQTQPVPKVP 562
Query: 458 ---SSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAE-QTTSPPHSPRSSFFQ 513
++KP + +++ P LL H P + +S+PA +T P P S
Sbjct: 563 QRVTAKPKTSPSPEVSYTTPAPKDVLLPHK--PYPEVSQSEPAPLETRGIPFIPMIS--- 617
Query: 514 PSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAP 573
PSP++ L L+ + EP + L+ + P T +PRTSD
Sbjct: 618 PSPSQEELQTTLEETDQSTQEPFTTKIPRTTELAKTT---QAPHRFYTTVRPRTSDKPHI 674
Query: 574 RASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLT 633
R + A R + D + ++V + P+ + R + R
Sbjct: 675 RPGVKQAPRPSGADRN---------VSVDSTHPTKKPGTRRPPLPPRP------------ 713
Query: 634 CPSPRRYPGPRPERLVDPDEP--ILANPIQEADPLVQQAHPVPDQQEPIQPD------PE 685
PRR P P P I + PI PL P E I+ D P
Sbjct: 714 -THPRRKPLPPNNVTGKPGSAGIISSGPI-TTPPLRSTPRPTGTPLERIETDIKQPTVPA 771
Query: 686 PEQSVSN-----QSSVRSPHPLVET--SDPHLG--TSEPNAPMINIGS-----PQGASEA 731
+ + N S R PL + PH+ N+P S + A+E
Sbjct: 772 SGEELENITDFSSSPTRETDPLGKPRFKGPHVRYIQKPDNSPCSITDSVKRFPKEEATEG 831
Query: 732 HSSNHPASPAPNLSII 747
++++ P +P NL+++
Sbjct: 832 NATSPPQNPPTNLTVV 847
>EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein)
Length = 620
Score = 48.9 bits (115), Expect = 6e-05
Identities = 66/312 (21%), Positives = 91/312 (29%), Gaps = 52/312 (16%)
Query: 483 HSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIP 542
+S P +PI P + SPP +P +F P P +P P+ S P + L +P
Sbjct: 289 YSPPPPSPIYSPPPPAYSPSPPPTPTPTFSPPPPAYSP-------PPTYSPPPPTYLPLP 341
Query: 543 YDPLSSEP--IIHEQPEPNQTEPQPR--------------------TSDHSAPRASA--- 577
P+ S P + P P+ + P P T + S P A
Sbjct: 342 SSPIYSPPPPVYSPPPPPSYSPPPPTYLPPPPPSSPPPPSFSPPPPTYEQSPPPPPAYSP 401
Query: 578 --------RPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEE 629
P T T P P P+ + +
Sbjct: 402 PLPAPPTYSPPPPTYSPPPPTYAQPPPLPPTYSPPPPAYSPPPPPTYSPPPPTYSPPPPA 461
Query: 630 YYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPE---- 685
Y P P Y P P P PI + P + PL P P ++ + P P
Sbjct: 462 YAQPPPPPPTYSPPPPAYSPPPPSPIYSPPPPQVQPLPPTFSPPPPRRIHLPPPPHRQPR 521
Query: 686 ---PEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAP 742
P SP P + P +P P G P A P
Sbjct: 522 PPTPTYGQPPSPPTFSPPPPRQIHSPPPPHWQPRTPTPTYGQPPS-----PPTFSAPPPR 576
Query: 743 NLSIIPYTHLQP 754
+ P H QP
Sbjct: 577 QIHSPPPPHRQP 588
Score = 47.4 bits (111), Expect = 2e-04
Identities = 70/281 (24%), Positives = 90/281 (31%), Gaps = 51/281 (18%)
Query: 483 HSSNPLT---PIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLL 539
H +PL P P QP T SPP + QPSPT +P P+ S P S +
Sbjct: 244 HQPSPLRHLPPSPRRQPQPPTYSPPPPAYAQSPQPSPTYSP------PPPTYSPPPPSPI 297
Query: 540 TIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPI 599
P P S P P P + P P +S P + P SS ++P P+
Sbjct: 298 YSPPPPAYS-PSPPPTPTPTFSPPPPA---YSPPPTYSPPPPTYLPLPSSPIYSPPP-PV 352
Query: 600 NVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANP 659
PS + P P P P P P P + P
Sbjct: 353 YSPPPPPSYSP------------------------PPPTYLPPPPPS---SPPPPSFSPP 385
Query: 660 IQEADPLVQQAHPVPDQQEPIQP-----DPEPEQSVSNQSSVRSPHPLVET-SDPHLGTS 713
P +Q+ P P P P P P + P PL T S P S
Sbjct: 386 ----PPTYEQSPPPPPAYSPPLPAPPTYSPPPPTYSPPPPTYAQPPPLPPTYSPPPPAYS 441
Query: 714 EPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQP 754
P P + P + + P P P S P + P
Sbjct: 442 PPPPPTYSPPPPTYSPPPPAYAQPPPPPPTYSPPPPAYSPP 482
>NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 kDa
protein) (140 kDa nucleolar phosphoprotein) (Nopp140)
(Nucleolar and coiled-body phosphoprotein 1)
Length = 704
Score = 48.5 bits (114), Expect = 8e-05
Identities = 80/377 (21%), Positives = 130/377 (34%), Gaps = 38/377 (10%)
Query: 378 KRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEP 437
K+ Q+K V ++ P K + D SED PQ +K K +
Sbjct: 145 KKPVQQKAVKPQAKAVRPPP---KKAESSESESDSSSEDEAPQTQKPKAAATAAKAPTKA 201
Query: 438 AAEVVRRTEPPARV--------TRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLT 489
+ + PPA+ SS+ SS + + D + A P+ + P
Sbjct: 202 QTKAPAKPGPPAKAQPKAANGKAGSSSSSSSSSSSDDSEEEKKAAAPLK-----KTAPKK 256
Query: 490 PIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDE-PTSLLTIPYDPLSS 548
+ P + T +P SS S E Q +P + P S + P LS
Sbjct: 257 QVVAKAPVKVTAAPTQKSSSSEDSSSEEEEE-----QKKPMKKKAGPYSSVPPPSVSLSK 311
Query: 549 EPIIHEQPE--PNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVS-FPINVVDSS 605
+ + + P+ QT+P ++D S S+ + T + + TP P+ S
Sbjct: 312 KSVGAQSPKKAAAQTQPADSSADSSEESDSSSEEEKKTPAKTVVSKTPAKPAPVKKKAES 371
Query: 606 PSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADP 665
S++S+S E + VSA + SP P P+ P +A P Q A
Sbjct: 372 SSDSSDSDSSEDEAPAKPVSATK-------SPLSKPAVTPK---PPAAKAVATPKQPAGS 421
Query: 666 LVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSP 725
+ D + E+ + +S + + P L + AP G
Sbjct: 422 GQKPQSRKADSSSSEEESSSSEEEATKKSVTTPKARVTAKAAPSLPAKQ--APRAG-GDS 478
Query: 726 QGASEAHSSNHPASPAP 742
SE+ SS P
Sbjct: 479 SSDSESSSSEEEKKTPP 495
Score = 46.6 bits (109), Expect = 3e-04
Identities = 106/553 (19%), Positives = 199/553 (35%), Gaps = 106/553 (19%)
Query: 393 ESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVT 452
+S+ P+ KK K+ + D SED ++ K +V PT + AA +R P
Sbjct: 69 QSNGPVAKKAKK--ETSSSDSSEDSSEEEDKAQV-----PT--QKAAAPAKRASLPQHAG 119
Query: 453 RSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFF 512
+++A +S+ + + + S + +P +Q P++
Sbjct: 120 KAAAKASESSSSEES----------------SEEEEEKDKKKKPVQQKAV---KPQAKAV 160
Query: 513 QPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSA 572
+P P +A ++ S SD + P + +P T+ Q +
Sbjct: 161 RPPPKKA------ESSESESDSSSE----DEAPQTQKPKAAATAAKAPTKAQTKAPAKPG 210
Query: 573 PRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYL 632
P A A+P A SS++ + SS S++SE +K
Sbjct: 211 PPAKAQPKAANGKAGSSSSSS---------SSSSSDDSEEEKK----------------- 244
Query: 633 TCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQE-PIQPDPEPEQSVS 691
+P + P+ + + + A P Q++ + ++Q+ P++ P SV
Sbjct: 245 -AAAPLKKTAPKKQVVAKAPVKVTAAPTQKSSSSEDSSSEEEEEQKKPMKKKAGPYSSV- 302
Query: 692 NQSSVRSPHPLVETSDPHLGTSEPNAPMINI----GSPQGASEAHSSNHPASPAPNLSII 747
P P V S +G P S + E+ SS+ P +++
Sbjct: 303 -------PPPSVSLSKKSVGAQSPKKAAAQTQPADSSADSSEESDSSSEEEKKTPAKTVV 355
Query: 748 PYTHLQPTSLSECINIFYHEASLRLRNVHGQTDLSEN---AENVADEWNNLSTWLVAQVP 804
T +P + + + +D SE+ A+ V+ + LS V P
Sbjct: 356 SKTPAKPAPVKK---------KAESSSDSSDSDSSEDEAPAKPVSATKSPLSKPAVTPKP 406
Query: 805 IMMQLLHAEGSQSIEAAKQRFARRVALHELEQRNKLLEAIEEAKRKKEQAEEA------- 857
+ + A Q + ++ +R+ E+ + E EEA +K +A
Sbjct: 407 PAAKAV-ATPKQPAGSGQKPQSRKADSSSSEEESSSSE--EEATKKSVTTPKARVTAKAA 463
Query: 858 ----ARQAAAQAEQARLEAERLEAEAEALRCQALAPVVFTPAASASIPDPQAAQNVPSSS 913
A+QA + ++E +E E + P AA A++P P + + S
Sbjct: 464 PSLPAKQAPRAGGDSSSDSESSSSEEE--KKTPPKPPAKKKAAGAAVPKPTPVKKAAAES 521
Query: 914 TQTSSSRLDIMEQ 926
+ +SSS D E+
Sbjct: 522 SSSSSSSEDSSEE 534
Score = 34.7 bits (78), Expect = 1.2
Identities = 47/220 (21%), Positives = 90/220 (40%), Gaps = 15/220 (6%)
Query: 375 SPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRI-VVKP- 432
S ++K K V+ ++ + P+ KK + D D D SED P K + + KP
Sbjct: 343 SEEEKKTPAKTVVSKTPAKP-APVKKKAESSSDSSDSDSSEDEAPAKPVSATKSPLSKPA 401
Query: 433 -TRVEPAAEVVRRTEPPARVTR-------SSAHSSKPALTSDDDLNLFDALPISALLQHS 484
T PAA+ V + PA + S+ S + + +S+++ A +
Sbjct: 402 VTPKPPAAKAVATPKQPAGSGQKPQSRKADSSSSEEESSSSEEEATKKSVTTPKARVTAK 461
Query: 485 SNPLTPIPESQPA-EQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPY 543
+ P P ++ A ++S S S + +P + P + + +PT +
Sbjct: 462 AAPSLPAKQAPRAGGDSSSDSESSSSEEEKKTPPKPPAKK--KAAGAAVPKPTPVKKAAA 519
Query: 544 DPLSSEPIIHEQPEPNQTEPQPR-TSDHSAPRASARPAAR 582
+ SS + E + +P+ + T A +A+ PA++
Sbjct: 520 ESSSSSSSSEDSSEEEKKKPKSKATPKPQAGKANGVPASQ 559
>SPKC_SYNY3 (P74745) Serine/threonine-protein kinase C (EC 2.7.1.37)
Length = 535
Score = 48.1 bits (113), Expect = 1e-04
Identities = 47/200 (23%), Positives = 67/200 (33%), Gaps = 48/200 (24%)
Query: 527 NQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDT 586
N P +EPT IP L P + E P P T P P S P+ TT +
Sbjct: 372 NPPPAVEEPTEETPIPLPSLEPRPNLFETPSPIPTPATPSPE----PTPSPSPSPETTSS 427
Query: 587 DSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPE 646
+ TP+ E +L+E P+P P P P
Sbjct: 428 PTEDTITPM--------------------------EPEPSLDE-----PAPIPEPKPSPS 456
Query: 647 RLVDPD-EPILANPIQEADPLVQQAHPVPDQQEP--IQPDPEPEQSVSNQSSVRSPHPLV 703
+ P P ++ P+ A P P P I P P+P +V P++
Sbjct: 457 PTISPQPSPTISIPVTPAPVPKPSPSPTPKPTVPPQISPTPQPSNTV----------PVI 506
Query: 704 ETSDPHLGTSEPNAPMINIG 723
+ +EPN P +G
Sbjct: 507 PPPENPSAETEPNLPAPPVG 526
Score = 40.4 bits (93), Expect = 0.023
Identities = 43/174 (24%), Positives = 62/174 (34%), Gaps = 30/174 (17%)
Query: 443 RRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTS 502
RR PP V + + P + + NLF+ P + +P P P P+ +TTS
Sbjct: 369 RRNNPPPAVEEPTEETPIPLPSLEPRPNLFET-PSPIPTPATPSP-EPTPSPSPSPETTS 426
Query: 503 PPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPN--- 559
P + +P P+ DEP IP S P I QP P
Sbjct: 427 SPTEDTITPMEPEPS--------------LDEPA---PIPEPKPSPSPTISPQPSPTISI 469
Query: 560 --QTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSE 611
P P+ S P+ + P T S+T P+ +PS +E
Sbjct: 470 PVTPAPVPKPSPSPTPKPTVPPQISPTPQPSNTV------PVIPPPENPSAETE 517
>PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix
protein) (Aczonin) (Brain-derived HLMN protein)
Length = 5038
Score = 48.1 bits (113), Expect = 1e-04
Identities = 77/366 (21%), Positives = 125/366 (34%), Gaps = 69/366 (18%)
Query: 393 ESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVT 452
+S+ + TK++ D +G + ++ + P + P EV+ + P V+
Sbjct: 178 DSEAVQEETTKKQKVAQKDQGKSEGITKPSLQQPSPKLIPKQQGPGKEVIPQDIPSKSVS 237
Query: 453 RSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFF 512
A +KP P +A S TP +++P Q P +++
Sbjct: 238 SQQAEKTKP------------QAPGTAKPSQQSPAQTPAQQAKPVAQQPGP---AKATVQ 282
Query: 513 QPSPTEAPLWNLLQNQPSRSD----EPTSLLTIPYDPLSSEPIIHEQPEP---------N 559
QP P ++P QP+ + +P P E +QP P
Sbjct: 283 QPGPAKSPA------QPAGTGKSPAQPPVTAKPPAQQAGLEKTSLQQPGPKSLAQTPGQG 336
Query: 560 QTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEV 619
+ P P S P + PA + +S P P + S P
Sbjct: 337 KVPPGPAKSPAQQPGTAKLPAQQPGPQTASKVPGPTKTPAQL--SGPGK----------- 383
Query: 620 RKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILA--NPIQEADPLVQQAHPVPDQQ 677
+ ++ T PSP++ +P+ P +P+ P Q A Q HP P +
Sbjct: 384 -----TPAQQPGPTKPSPQQPIPAKPQ----PQQPVATKPQPQQPAPAKPQPQHPTPAKP 434
Query: 678 EPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINI-----GSPQGASEAH 732
+P QP P Q Q + P P P LG P +I G P A
Sbjct: 435 QPQQPTPAKPQ--PQQPTPAKPQP----QHPGLGKPSAQQPSKSISQTVTGRPLQAPPTS 488
Query: 733 SSNHPA 738
++ PA
Sbjct: 489 AAQAPA 494
Score = 38.9 bits (89), Expect = 0.066
Identities = 83/411 (20%), Positives = 147/411 (35%), Gaps = 47/411 (11%)
Query: 374 PSPPKRKAQRKMVLDESSEESDVPLV-----KKTKRKPDDGDDDDSEDGPPQKKKKKVRI 428
PS P+ + V ++ +S VP K+ K D +S+ PP K+ +
Sbjct: 567 PSSPQPTPKAASVQPATASKSPVPSQQASPKKELPSKQDSPKAPESKKPPPLVKQPTLHG 626
Query: 429 VVKPTRVEP-AAEVVRRTEPPARVTRSSAHSSKPALTSDDDLN------LFDALPISALL 481
T +P AE + + PP + + + +K + + L D+ P SA
Sbjct: 627 PTPATAPQPPVAEALPKPAPPKKPSAALPEQAKAPVADVEPKQPKTTETLTDS-PSSAAA 685
Query: 482 QHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPS--PTEAPLWNLLQNQPSRSDEPTSLL 539
L+ ++Q A+ TT+PP S+ S PT + L R + ++
Sbjct: 686 TSKPAILSSQVQAQ-AQVTTAPPLKTDSAKTSQSFPPTGDTITPLDSKAMPRPASDSKIV 744
Query: 540 TIPYDPLSSEPIIHEQPEPN--QTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSF 597
+ P S+ + ++ EP QT+ P+ P+ S P+ T + TP S
Sbjct: 745 SHPGPTSESKDPVQKKEEPKKAQTKVTPKPDTKPVPKGSPTPSGTRPTTGQA---TPQS- 800
Query: 598 PINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILA 657
P E R+F + + + +P+ P E + A
Sbjct: 801 ------QQPPKPPEQSRRF----SLNLGGIAD------APKSQPTTPQETVTGKLFGFGA 844
Query: 658 NPIQEADPLV-----QQAHPVPDQQEPIQPDPEPEQSVSNQSSVRS----PHPLVETSDP 708
+ +A L+ Q HP P + P P Q+++ Q +S P +T+
Sbjct: 845 SIFSQASNLISTAGQQAPHPQTGPAAPSKQAPPPSQTLAAQGPPKSTGQHPSAPAKTTAV 904
Query: 709 HLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTSLSE 759
T P A + Q + + P +S P T + L++
Sbjct: 905 KKETKGPAAENLEAKPAQAPTVKKAEKDKKHPPGKVSKPPPTEPEKAVLAQ 955
Score = 35.0 bits (79), Expect = 0.95
Identities = 57/244 (23%), Positives = 83/244 (33%), Gaps = 18/244 (7%)
Query: 488 LTPIPESQPAEQTTSPPHSPRSSFFQPSPT-EAPLWNL-LQNQPSRSDE--PTSLLTIPY 543
L P P SQ EQ SP Q PT +P L + PS+S +S L I
Sbjct: 2244 LAPTPSSQTKEQPGSPHSVSGEISGQEKPTYRSPSGGLPVSTHPSKSHPFFRSSSLDISA 2303
Query: 544 DPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVD 603
P P P P P P ++P+ P + ++ P + D
Sbjct: 2304 QPPPPPPPPPPPPPPPPPPPPPPLPPATSPKPPTYPKRKLA---AAAPVAPTAIVTAHAD 2360
Query: 604 SSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEA 663
+ P+ + + R+ + K+ A P P P LV P + P
Sbjct: 2361 AIPTVEATAARRSNGLPATKICAAAP-----PPVPPKPSSIPTGLVFTHRPEASKPPIAP 2415
Query: 664 DPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHL-GTSEPNAPMINI 722
P V + + P P SN S LV ++D L + P +P N
Sbjct: 2416 KPAVPEIPVTTQKTTDTCPKPTGLPLTSNMSL-----NLVTSADYKLPSPTSPLSPHSNK 2470
Query: 723 GSPQ 726
SP+
Sbjct: 2471 SSPR 2474
Score = 35.0 bits (79), Expect = 0.95
Identities = 77/377 (20%), Positives = 136/377 (35%), Gaps = 40/377 (10%)
Query: 366 RLPPAPEFPSPPKRKAQRKMVLDESSEE--SDVPLVKKTKRKPDDGDDDDSEDGPPQKKK 423
++PP P SP ++ K+ + + S VP KT + ++ P K
Sbjct: 337 KVPPGPA-KSPAQQPGTAKLPAQQPGPQTASKVPGPTKTPAQLSGPGKTPAQQPGPTKPS 395
Query: 424 KKVRIVVKPTRVEPAAEVVRRTEP-PARVTRSSAHSSKPALTSDDDLNLFDALPISALLQ 482
+ I KP +P A + +P PA+ +KP P A Q
Sbjct: 396 PQQPIPAKPQPQQPVATKPQPQQPAPAKPQPQHPTPAKPQPQQPTPAKPQPQQPTPAKPQ 455
Query: 483 HSSNPLTPIPESQPAEQ-----TTSPPHSPRSSFFQP-----SPTEAPLWN---LLQNQP 529
L QP++ T P +P +S Q S T PL N LL + P
Sbjct: 456 PQHPGLGKPSAQQPSKSISQTVTGRPLQAPPTSAAQAPAQGLSKTICPLCNTTELLLHTP 515
Query: 530 SRSD-------EPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAAR 582
+++ + T ++P I E N + + +A +S +P +
Sbjct: 516 EKANFNTCTECQSTVCSLCGFNPNPHLTEIKEWLCLNCQMQRALGGELAAIPSSPQPTPK 575
Query: 583 TTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSA--LEEYYLTCPSPRRY 640
+TA P+ +SP S + + + K +++ L P+P
Sbjct: 576 AASVQPATASKS---PVPSQQASPKKELPSKQDSPKAPESKKPPPLVKQPTLHGPTPATA 632
Query: 641 PGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPH 700
P +P +A + + P + + +P+Q + D EP+Q + ++ SP
Sbjct: 633 P-----------QPPVAEALPKPAPPKKPSAALPEQAKAPVADVEPKQPKTTETLTDSPS 681
Query: 701 PLVETSDPHLGTSEPNA 717
TS P + +S+ A
Sbjct: 682 SAAATSKPAILSSQVQA 698
Score = 34.7 bits (78), Expect = 1.2
Identities = 27/92 (29%), Positives = 45/92 (48%), Gaps = 13/92 (14%)
Query: 671 HPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPM---------IN 721
+P+ +Q E I E +S S+Q+S +SP P V S H +P+ M +
Sbjct: 4721 YPLKEQTESI----EHGKSHSSQNSQQSPKPSVIKSRSHGIFPDPSKDMQVPTIEKSHSS 4776
Query: 722 IGSPQGASEAHSSNHPASPAPNLSIIPYTHLQ 753
GS + +SE H +H S + + + + THL+
Sbjct: 4777 PGSSKSSSEGHLRSHGPSRSQSKTSVAQTHLE 4808
>YDC9_SCHPO (Q10172) Hypothetical protein C25G10.09c in chromosome I
Length = 1794
Score = 47.0 bits (110), Expect = 2e-04
Identities = 87/425 (20%), Positives = 146/425 (33%), Gaps = 64/425 (15%)
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSE------DGPPQK 421
P AP+ +PP ++ ES+ + P VK T P + + + +
Sbjct: 1184 PAAPQTLNPPSVSTVQQSKPIESNTHT--PEVKATSESPSASSNLEDRAARIKAEAQRRM 1241
Query: 422 KKKKVRIVVKPTRV---EPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPIS 478
++ + +KP + PA + P A T K A ++ +S
Sbjct: 1242 NERLAALGIKPRQKGTPSPAPVNSATSTPVAAPTAQQIQPGKQASAVSSNVPA-----VS 1296
Query: 479 ALLQHSSNPLTPIPESQPAEQ----TTSPPHSPRSSF-FQPSPTEAPLWNLLQNQPSRSD 533
A + + + QP +Q P + +SF P P +APL N
Sbjct: 1297 ASISTPPAVVPTVQHPQPTKQIPTAAVKDPSTTSTSFNTAPIPQQAPLENQFSKMSLEPP 1356
Query: 534 EPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAA------------ 581
++ T P + +H P P +P H+A +ARP+A
Sbjct: 1357 VRPAVPTSPKPQIPDSSNVHAPPPP--VQPMNAMPSHNA--VNARPSAPERRDSFGSVSS 1412
Query: 582 ----RTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSP 637
+ + ++ST S P N +PSN++ +V + P
Sbjct: 1413 GSNVSSIEDETSTMPLKASQPTN--PGAPSNHA-----------PQVVPPAPMHAVAPVQ 1459
Query: 638 RRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVR 697
+ PG P P+ + P V PVP + +P SV+ ++
Sbjct: 1460 PKAPGMVTNAPAPSSAPAPPAPVSQLPPAVPNV-PVPSMIPSVA--QQPPSSVAPATAPS 1516
Query: 698 SPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQPTSL 757
S P ++S H+ + P A PQ S A S+ PA + PY +P
Sbjct: 1517 STLPPSQSSFAHVPSPAPPA-------PQHPSAAALSSAPADNSMPHRSSPYAPQEPVQK 1569
Query: 758 SECIN 762
+ IN
Sbjct: 1570 PQAIN 1574
>MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia
protein 4 (Trithorax homolog 2)
Length = 2715
Score = 47.0 bits (110), Expect = 2e-04
Identities = 83/379 (21%), Positives = 125/379 (32%), Gaps = 58/379 (15%)
Query: 391 SEESDVPLVKKTKRKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPAR 450
S + P KK ++K DD +++ E+ K+ ++ + PAAE PP
Sbjct: 346 SGQGGSPCWKKQEQKLDDEEEEKKEEEEKDKEGEEKEERAVAEEMMPAAEKEEAKLPPPP 405
Query: 451 VTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSS 510
+T PA S P P P + P PP P S
Sbjct: 406 LT-------PPA--------------------PSPPPPLPPPSTSPPPPLCPPPPPPVSP 438
Query: 511 FFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQ-----PE-PNQTEPQ 564
PSP P + P T PL+ + PE + P
Sbjct: 439 PPLPSPPPPPAQEEQEESPPPVVPATCSRKRGRPPLTPSQRAEREAARAGPEGTSPPTPT 498
Query: 565 PRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKV 624
P T+ P S A ++T + F + VV + S ++ R+FM+ K
Sbjct: 499 PSTATGGPPEDSPTVAPKSTTFLKNIR----QFIMPVVSARSSRVIKTPRRFMDEDPPKP 554
Query: 625 SALE----------------EYYLTCPSPRRYPGP--RPERLVDPDEPILANPIQEADPL 666
+E + PSP R P P P L + IL P L
Sbjct: 555 PKVEVSPVLRPPITTSPPVPQEPAPVPSPPRAPTPPSTPVPLPEKRRSILREPTFRWTSL 614
Query: 667 VQQAHPVPD-QQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSP 725
++ P P P P P P + S++ + P S+ HL E +G+P
Sbjct: 615 TRELPPPPPAPPPPPAPSPPPAPATSSRRPLLLRAPQFTPSEAHLKIYESVLTPPPLGAP 674
Query: 726 QG--ASEAHSSNHPASPAP 742
+ + + PA P P
Sbjct: 675 EAPEPEPPPADDSPAEPEP 693
>ENL2_ARATH (Q9T076) Early nodulin-like protein 2 precursor
(Phytocyanin-like protein)
Length = 349
Score = 47.0 bits (110), Expect = 2e-04
Identities = 61/242 (25%), Positives = 94/242 (38%), Gaps = 51/242 (21%)
Query: 386 VLDESSEESDVPLVKKTKRKPDDGDDDDSED--GPPQ---------KKKKKVRIVVKPTR 434
VL+ + + D K ++ DDGD + S D GP KK +K+ +VV R
Sbjct: 73 VLEVNKADYDACNTKNPIKRVDDGDSEISLDRYGPFYFISGNEDNCKKGQKLNVVVISAR 132
Query: 435 VEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPES 494
+ A+ S H++ P ++ + P A SS+P++P +
Sbjct: 133 IPSTAQ--------------SPHAAAPGSSTPGSMTP----PGGAHSPKSSSPVSPT--T 172
Query: 495 QPAEQTTSP--PHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPII 552
P TT P HSP+SS T P + +S P S T P P
Sbjct: 173 SPPGSTTPPGGAHSPKSSSAVSPATSPP-----GSMAPKSGSPVSPTTSPPAP------- 220
Query: 553 HEQPEPNQTEP-QPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSE 611
P T P P ++ ++P A P + +T SS T + SSP +NS
Sbjct: 221 -----PKSTSPVSPSSAPMTSPPAPMAPKSSSTIPPSSAPMTSPPGSMAPKSSSPVSNSP 275
Query: 612 SI 613
++
Sbjct: 276 TV 277
>ZAN_HUMAN (Q9Y493) Zonadhesin precursor
Length = 2812
Score = 46.6 bits (109), Expect = 3e-04
Identities = 62/284 (21%), Positives = 110/284 (37%), Gaps = 34/284 (11%)
Query: 447 PPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQPAEQTTSPPHS 506
PP S+ S LT + ++ P ++ + S P + P E+ T P
Sbjct: 543 PPVSPVSSTGPSETTGLTENPTIST--KKPTVSIEKPSVTTEKP---TVPKEKPTIPTEK 597
Query: 507 PRSSFFQPS-PTEAPLWNLLQNQPS-RSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQ 564
P S +P+ P+E P N+ +P+ S++PT L P P I E+P + +P
Sbjct: 598 PTISTEKPTIPSEKP--NMPSEKPTIPSEKPTILTEKPTIPSEKPTIPSEKPTISTEKPT 655
Query: 565 PRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKV 624
T + + P T T P P ++ PS +E ME E +
Sbjct: 656 VPTEEPTTP-------TEETTTSMEEPVIPTEKP-SIPTEKPSIPTEKPTISME---ETI 704
Query: 625 SALEEYYLTCPSP----RRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPI 680
+ E+ ++ P + P + + P++P P ++ P + P ++P
Sbjct: 705 ISTEKPTISPEKPTIPTEKPTIPTEKSTISPEKP--TTPTEK--PTIPTEKPTISPEKPT 760
Query: 681 QPDPEPEQSV------SNQSSVRSPHPLVETSDPHLGTSEPNAP 718
P +P S + + ++ + P + T P + T EP P
Sbjct: 761 TPTEKPTISPEKLTIPTEKPTIPTEKPTIPTEKPTISTEEPTTP 804
Score = 35.8 bits (81), Expect = 0.56
Identities = 71/357 (19%), Positives = 124/357 (33%), Gaps = 84/357 (23%)
Query: 391 SEESDVPLVKKT--KRKPDDGDDDDS--EDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTE 446
+E+ +P K T KP + + + P +K KPT + P + +
Sbjct: 721 TEKPTIPTEKSTISPEKPTTPTEKPTIPTEKPTISPEKPTTPTEKPT-ISPEKLTIPTEK 779
Query: 447 PPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPE-SQPAEQTTS--- 502
P + + + KP +++++ + IS + P P+ + + P E+TT+
Sbjct: 780 PTIPTEKPTIPTEKPTISTEEPTTPTEETTIS-----TEKPSIPMEKPTLPTEETTTSVE 834
Query: 503 -----------PPHSPRSSFFQPS-PTEAPLWNLLQNQPSRSDEPTSLLTIPYDP----- 545
P P S +P+ PTE P + P + PT LTIP +
Sbjct: 835 ETTISTEKLTIPMEKPTISTEKPTIPTEKPTIS-----PEKLTIPTEKLTIPTEKPTIPI 889
Query: 546 ----LSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINV 601
+S+E + +P + +P S + +P T +T ST
Sbjct: 890 EETTISTEKLTIPTEKPTISPEKPTISTEKPTIPTEKPTIPTEETTIST----------- 938
Query: 602 VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQ 661
EK LT P+ + P PE+L P E + +
Sbjct: 939 --------------------EK--------LTIPTEK--PTISPEKLTIPTEKPTISTEK 968
Query: 662 EADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAP 718
P + P ++P P +P +++R PHP + P+AP
Sbjct: 969 PTIPTEKLTIPT---EKPTIPTEKPTIPTEKLTALRPPHPSPTATGLAALVMSPHAP 1022
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 114,776,027
Number of Sequences: 164201
Number of extensions: 5342093
Number of successful extensions: 27335
Number of sequences better than 10.0: 793
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 731
Number of HSP's that attempted gapping in prelim test: 22971
Number of HSP's gapped (non-prelim): 3187
length of query: 952
length of database: 59,974,054
effective HSP length: 120
effective length of query: 832
effective length of database: 40,269,934
effective search space: 33504585088
effective search space used: 33504585088
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0012a.3


