

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
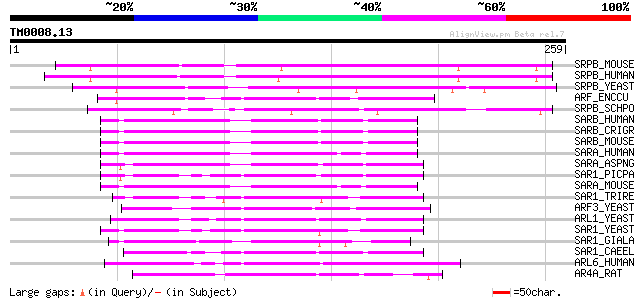
Query= TM0008.13
(259 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SRPB_MOUSE (P47758) Signal recognition particle receptor beta su... 109 8e-24
SRPB_HUMAN (Q9Y5M8) Signal recognition particle receptor beta su... 107 2e-23
SRPB_YEAST (P36057) Signal recognition particle receptor beta su... 72 1e-12
ARF_ENCCU (Q8SQH8) ADP-ribosylation factor 56 8e-08
SRPB_SCHPO (O13950) Putative signal recognition particle recepto... 56 1e-07
SARB_HUMAN (Q9Y6B6) GTP-binding protein SAR1b (GTBPB) 55 1e-07
SARB_CRIGR (Q9QVY3) GTP-binding protein SAR1b (Sar1) (GTBPB) 55 1e-07
SARB_MOUSE (Q9CQC9) GTP-binding protein SAR1b 54 4e-07
SARA_HUMAN (Q9NR31) GTP-binding protein SAR1a (COPII-associated ... 54 5e-07
SARA_ASPNG (P52886) GTP-binding protein sarA 53 8e-07
SAR1_PICPA (Q9P4C8) GTP-binding protein sar1 53 8e-07
SARA_MOUSE (P36536) GTP-binding protein SAR1a 52 1e-06
SAR1_TRIRE (P78976) GTP-binding protein SAR1 50 7e-06
ARF3_YEAST (P40994) ADP-ribosylation factor 3 49 1e-05
ARL1_YEAST (P38116) ADP-ribosylation factor-like protein 1 (Arf-... 48 2e-05
SAR1_YEAST (P20606) GTP-binding protein SAR1 48 3e-05
SAR1_GIALA (Q8MQT8) GTP-binding protein Sar1 48 3e-05
SAR1_CAEEL (Q23445) GTP-binding protein SAR1 48 3e-05
ARL6_HUMAN (Q9H0F7) ADP-ribosylation factor-like protein 6 47 4e-05
AR4A_RAT (P61214) ADP-ribosylation factor-like protein 4A 47 6e-05
>SRPB_MOUSE (P47758) Signal recognition particle receptor beta
subunit (SR-beta)
Length = 269
Score = 109 bits (272), Expect = 8e-24
Identities = 79/244 (32%), Positives = 125/244 (50%), Gaps = 18/244 (7%)
Query: 22 LRQVPPEQLYIAAAI-AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGST 80
L+Q P L +A A+ AV TL+ +++ V+ GL SGKT+LF +L G
Sbjct: 28 LQQRDPTLLSVAVALLAVLLTLVFWKFIWSRKSSQRAVLFVGLCDSGKTLLFVRLLTGQ- 86
Query: 81 HQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKL-DEYLPQAAGVVFVV 139
++ T TS+ + + +++ + + ++D+PGH LR +L D + A VVFVV
Sbjct: 87 YRDTQTSITDSSAIYKVNNN-----RGNSLTLIDLPGHESLRFQLLDRFKSSARAVVFVV 141
Query: 140 DAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDK 199
D+ F + +E+LY +L +K LLI CNK D A + + I++QLEKE++
Sbjct: 142 DSAAFQREVKDVAEFLYQVLIDSMALKNSPSLLIACNKQDIAMAKSAKLIQQQLEKELNT 201
Query: 200 LRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTG-------EISQL 249
LR +RSA D+ T LG G+ F F+Q KV + S G +I L
Sbjct: 202 LRVTRSAAPSTLDSSSTAPAQLGKKGKEFEFSQLPLKVEFLECSAKGGRGDTGSADIQDL 261
Query: 250 EEFI 253
E+++
Sbjct: 262 EKWL 265
>SRPB_HUMAN (Q9Y5M8) Signal recognition particle receptor beta
subunit (SR-beta) (Protein APMCF1)
Length = 271
Score = 107 bits (268), Expect = 2e-23
Identities = 79/249 (31%), Positives = 126/249 (49%), Gaps = 18/249 (7%)
Query: 17 TASDYLRQVPPEQLYIAAAI-AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQL 75
T L+Q P L + A+ AV TL+ L +R+ V+L GL SGKT+LF +L
Sbjct: 25 TLRQELQQTDPTLLSVVVAVLAVLLTLVFWKLIRSRRSSQRAVLLVGLCDSGKTLLFVRL 84
Query: 76 RDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPK-LDEYLPQAAG 134
G ++ T TS+ + + +++ + + ++D+PGH LR + L+ + A
Sbjct: 85 LTG-LYRDTQTSITDSCAVYRVNNN-----RGNSLTLIDLPGHESLRLQFLERFKSSARA 138
Query: 135 VVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLE 194
+VFVVD+ F + +E+LY +L +K LI CNK D A + + I++QLE
Sbjct: 139 IVFVVDSAAFQREVKDVAEFLYQVLIDSMGLKNTPSFLIACNKQDIAMAKSAKLIQQQLE 198
Query: 195 KEIDKLRASRSAVS---DADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTG------- 244
KE++ LR +RSA D+ T LG G+ F F+Q KV + S G
Sbjct: 199 KELNTLRVTRSAAPSTLDSSSTAPAQLGKKGKEFEFSQLPLKVEFLECSAKGGRGDVGSA 258
Query: 245 EISQLEEFI 253
+I LE+++
Sbjct: 259 DIQDLEKWL 267
>SRPB_YEAST (P36057) Signal recognition particle receptor beta
subunit (SR-beta)
Length = 244
Score = 72.0 bits (175), Expect = 1e-12
Identities = 73/248 (29%), Positives = 111/248 (44%), Gaps = 33/248 (13%)
Query: 30 LYIAAAIAVFTTLLLLSLR-------LFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQ 82
L IA + + TT+ L++++ + +++ ++++ G SGKT L L S
Sbjct: 6 LIIACLLVIGTTIALIAVQKASSKTGIKQKSYQPSIIIAGPQNSGKTSLLTLLTTDSVRP 65
Query: 83 GTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAA----GVVFV 138
TV S EP S T +VD PGH +LR KL +YL A G++F+
Sbjct: 66 -TVVSQEPLSAADYDGSGVT---------LVDFPGHVKLRYKLSDYLKTRAKFVKGLIFM 115
Query: 139 VDAVDFLPNCRAASEYLYDILT-KGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEI 197
VD+ +E+L DIL+ S + I +LI CNK++ TA I+ LE EI
Sbjct: 116 VDSTVDPKKLTTTAEFLVDILSITESSCENGIDILIACNKSELFTARPPSKIKDALESEI 175
Query: 198 DKLRASRS--------AVSDADVTNEFTLGV--PGEAFSFTQCCNKVTTADASGLTGEIS 247
K+ R +++ D E TL V + F F V + S +IS
Sbjct: 176 QKVIERRKKSLNEVERKINEEDYA-ENTLDVLQSTDGFKFANLEASVVAFEGSINKRKIS 234
Query: 248 QLEEFIRE 255
Q E+I E
Sbjct: 235 QWREWIDE 242
>ARF_ENCCU (Q8SQH8) ADP-ribosylation factor
Length = 206
Score = 56.2 bits (134), Expect = 8e-08
Identities = 43/158 (27%), Positives = 73/158 (45%), Gaps = 14/158 (8%)
Query: 42 LLLLSLR-LFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
LL LR LF ++ + GL G+GKT L L+ G HQ TV ++ N E
Sbjct: 10 LLYTKLRGLFSGQSERSITMIGLDGAGKTTLLLYLQTGEVHQ-TVPTLGFN-------CE 61
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
G +K + D+ G + +Y+ + G++++VD D + E L+ IL
Sbjct: 62 NVTLGSMK-FQVWDIGGQNSFMRFWHQYINEGCGIIYMVDCAD-PQRFGKSGEELWRIL- 118
Query: 161 KGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEID 198
+++ PLL+L NK D + H + + + + E +
Sbjct: 119 --NILNSPRPLLVLANKIDLIREHERSEVVKSIRNEFN 154
>SRPB_SCHPO (O13950) Putative signal recognition particle receptor
beta subunit (SR-beta)
Length = 227
Score = 55.8 bits (133), Expect = 1e-07
Identities = 62/226 (27%), Positives = 100/226 (43%), Gaps = 34/226 (15%)
Query: 37 AVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQL--RDGSTHQGTVTSMEPNEGT 94
A+ +TL + R + K V L G + SGKT LF +L ++ T TV S+EPNE
Sbjct: 19 AIISTLGIFFTRKTIQKKLPAVFLIGPSDSGKTSLFCELIYKEKKT---TVPSIEPNEAV 75
Query: 95 FILHSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLP---QAAGVVFVVDAVDFLPNCRAA 151
+ K G +VD+PGH R + + VVFV+++ +
Sbjct: 76 W-------KYG----AWLVDLPGHPRAKRWITTKFSGNYNVKAVVFVLNSATIDRDVHEV 124
Query: 152 SEYLYDILTKGSVVKKKIP-LLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRSAVSDA 210
L+D + K K +P LLI CNK D TA E I++ L+ E+ + ++ ++
Sbjct: 125 GLMLFDTILKCR--KHHVPHLLIACNKFDLFTAQPAEKIQQLLKAELHNILEEKNLQLES 182
Query: 211 DVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEI---SQLEEFI 253
V+ E + +++ D L G + S +EE+I
Sbjct: 183 IVS---------EDVDWESVADQIEDTDLKFLPGSVAKMSNIEEWI 219
>SARB_HUMAN (Q9Y6B6) GTP-binding protein SAR1b (GTBPB)
Length = 198
Score = 55.5 bits (132), Expect = 1e-07
Identities = 46/148 (31%), Positives = 68/148 (45%), Gaps = 14/148 (9%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L L L+K KT +V GL +GKT L + L+D Q T +E I T
Sbjct: 15 VLQFLGLYK--KTGKLVFLGLDNAGKTTLLHMLKDDRLGQHVPTLHPTSEELTIAGMTFT 72
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
D+ GH + R YLP G+VF+VD D + E L ++T
Sbjct: 73 T---------FDLGGHVQARRVWKNYLPAINGIVFLVDCADH-ERLLESKEELDSLMTDE 122
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIR 190
++ +P+LIL NK D+ A ++E +R
Sbjct: 123 TIA--NVPILILGNKIDRPEAISEERLR 148
>SARB_CRIGR (Q9QVY3) GTP-binding protein SAR1b (Sar1) (GTBPB)
Length = 198
Score = 55.5 bits (132), Expect = 1e-07
Identities = 46/148 (31%), Positives = 68/148 (45%), Gaps = 14/148 (9%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L L L+K KT +V GL +GKT L + L+D Q T +E I T
Sbjct: 15 VLQFLGLYK--KTGKLVFLGLDNAGKTTLLHMLKDDRLGQHVPTLHPTSEELTIAGMTFT 72
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
D+ GH + R YLP G+VF+VD D + E L ++T
Sbjct: 73 T---------FDLGGHIQARRVWKNYLPAINGIVFLVDCADH-ERLLESKEELDSLMTDE 122
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIR 190
++ +P+LIL NK D+ A ++E +R
Sbjct: 123 TIA--NVPILILGNKIDRPEAISEERLR 148
>SARB_MOUSE (Q9CQC9) GTP-binding protein SAR1b
Length = 198
Score = 53.9 bits (128), Expect = 4e-07
Identities = 45/148 (30%), Positives = 68/148 (45%), Gaps = 14/148 (9%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L L L+K K+ +V GL +GKT L + L+D Q T +E I T
Sbjct: 15 VLQFLGLYK--KSGKLVFLGLDNAGKTTLLHMLKDDRLGQHVPTLHPTSEELTIAGMTFT 72
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
D+ GH + R YLP G+VF+VD D + E L ++T
Sbjct: 73 T---------FDLGGHVQARRVWKNYLPAINGIVFLVDCADH-ERLLESKEELDSLMTDE 122
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIR 190
++ +P+LIL NK D+ A ++E +R
Sbjct: 123 TIA--NVPILILGNKIDRPEAISEERLR 148
>SARA_HUMAN (Q9NR31) GTP-binding protein SAR1a (COPII-associated
small GTPase)
Length = 198
Score = 53.5 bits (127), Expect = 5e-07
Identities = 45/148 (30%), Positives = 68/148 (45%), Gaps = 14/148 (9%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L L L+K K+ +V GL +GKT L + L+D Q T +E I T
Sbjct: 15 VLQFLGLYK--KSGKLVFLGLDNAGKTTLLHMLKDDRLGQHVPTLHPTSEELTIAGMTFT 72
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
D+ GH + R YLP G+VF+VD D + E L ++T
Sbjct: 73 T---------FDLGGHEQARRVWKNYLPAINGIVFLVDCADHSRLVESKVE-LNALMTDE 122
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIR 190
++ +P+LIL NK D+ A ++E +R
Sbjct: 123 TI--SNVPILILGNKIDRTDAISEEKLR 148
>SARA_ASPNG (P52886) GTP-binding protein sarA
Length = 189
Score = 52.8 bits (125), Expect = 8e-07
Identities = 48/152 (31%), Positives = 72/152 (46%), Gaps = 16/152 (10%)
Query: 43 LLLSLRLF-KRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSET 101
LL SL L K AK ++ GL +GKT L + L++ T+ +E I ++
Sbjct: 10 LLASLGLLNKHAK---LLFLGLDNAGKTTLLHMLKNDRVAILQPTAHPTSEELAIGNNRF 66
Query: 102 TKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTK 161
T D+ GH + R +Y P+ +G+VF+VDA D C S+ D L
Sbjct: 67 TT---------FDLGGHQQARRLWKDYFPEVSGIVFLVDAKDH--ECFPESKAELDALLA 115
Query: 162 GSVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
+ K+P LIL NK D A +++ +R QL
Sbjct: 116 MEEL-AKVPFLILGNKIDHPDAVSEDDVRHQL 146
>SAR1_PICPA (Q9P4C8) GTP-binding protein sar1
Length = 190
Score = 52.8 bits (125), Expect = 8e-07
Identities = 48/152 (31%), Positives = 76/152 (49%), Gaps = 16/152 (10%)
Query: 43 LLLSLRLF-KRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSET 101
+L SL L+ K AK ++ GL +GKT L + L++ + +++P T+ SE
Sbjct: 10 VLASLGLWNKHAK---LLFLGLDNAGKTTLLHMLKNDR-----LATLQP---TWHPTSEE 58
Query: 102 TKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTK 161
G V+ D+ GH + R +Y P+ G+V++VD D P S D L K
Sbjct: 59 LSIGNVR-FTTFDLGGHEQARRVWKDYFPEVDGIVYLVDIAD--PERFEESRVELDALLK 115
Query: 162 GSVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
+ K+P+L+L NK DK TA ++ +R L
Sbjct: 116 IEEL-SKVPVLVLGNKIDKSTAVSENELRHAL 146
>SARA_MOUSE (P36536) GTP-binding protein SAR1a
Length = 198
Score = 52.4 bits (124), Expect = 1e-06
Identities = 45/148 (30%), Positives = 67/148 (44%), Gaps = 14/148 (9%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L L L+K K+ +V GL +GKT L L+D Q T +E I T
Sbjct: 15 VLQFLGLYK--KSGKLVFLGLDNAGKTTLLQMLKDDRLGQHVPTLHPTSEELTIAGMTFT 72
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
D+ GH + R YLP G+VF+VD D + E L ++T
Sbjct: 73 T---------FDLGGHEQARRVWKNYLPAINGIVFLVDCADHSRLMESKVE-LNALMTDE 122
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIR 190
++ +P+LIL NK D+ A ++E +R
Sbjct: 123 TI--SNVPILILGNKIDRTDAISEEKLR 148
>SAR1_TRIRE (P78976) GTP-binding protein SAR1
Length = 189
Score = 49.7 bits (117), Expect = 7e-06
Identities = 47/149 (31%), Positives = 70/149 (46%), Gaps = 23/149 (15%)
Query: 49 LFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILH--SETTKKGK 106
L K AK ++ GL +GKT L + L++ V ++P LH SE G
Sbjct: 17 LNKHAK---LLFLGLDNAGKTTLLHMLKNDR-----VAILQPT-----LHPTSEELAIGN 63
Query: 107 VKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDF--LPNCRAASEYLYDILTKGSV 164
V+ + D+ GH + R +Y P GVVF+VDA D P +A + L +
Sbjct: 64 VR-FNTFDLGGHQQARRIWRDYFPDVNGVVFLVDAKDHERFPEAKAELDALLSM-----E 117
Query: 165 VKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
K+P +IL NK D A +++ +R QL
Sbjct: 118 ELSKVPFVILGNKIDHPDAVSEDELRHQL 146
>ARF3_YEAST (P40994) ADP-ribosylation factor 3
Length = 182
Score = 48.9 bits (115), Expect = 1e-05
Identities = 40/144 (27%), Positives = 65/144 (44%), Gaps = 12/144 (8%)
Query: 53 AKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHI 112
+K +++ GL +GKT + Y+L+ + ++ + T + ET VK ++
Sbjct: 14 SKEMKILMLGLDKAGKTTILYKLK--------LNKIKTSTPTVGFNVETVTYKNVK-FNM 64
Query: 113 VDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLL 172
DV G RLRP Y P ++FV+D+ A E LY I+ G + + LL
Sbjct: 65 WDVGGQQRLRPLWRHYFPATTALIFVIDS-SARNRMEEAKEELYSII--GEKEMENVVLL 121
Query: 173 ILCNKTDKVTAHTKEFIRRQLEKE 196
+ NK D A + + LE E
Sbjct: 122 VWANKQDLKDAMKPQEVSDFLELE 145
>ARL1_YEAST (P38116) ADP-ribosylation factor-like protein 1
(Arf-like GTPase 1)
Length = 182
Score = 48.1 bits (113), Expect = 2e-05
Identities = 40/146 (27%), Positives = 73/146 (49%), Gaps = 12/146 (8%)
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
+L+ K +++ GL G+GKT + Y+L+ G V + +P G + ET +
Sbjct: 10 KLWGSNKELRILILGLDGAGKTTILYRLQIGE-----VVTTKPTIG---FNVETLSYKNL 61
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
K +++ D+ G + +RP Y A V+FVVD+ D AS+ L+ +L + + +
Sbjct: 62 K-LNVWDLGGQTSIRPYWRCYYADTAAVIFVVDSTD-KDRMSTASKELHLMLQEEEL--Q 117
Query: 168 KIPLLILCNKTDKVTAHTKEFIRRQL 193
LL+ NK D+ A + + ++L
Sbjct: 118 DAALLVFANKQDQPGALSASEVSKEL 143
>SAR1_YEAST (P20606) GTP-binding protein SAR1
Length = 190
Score = 47.8 bits (112), Expect = 3e-05
Identities = 44/153 (28%), Positives = 72/153 (46%), Gaps = 18/153 (11%)
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L+ K ++ GL +GKT L + L++ + +++P T+ SE
Sbjct: 13 VLASLGLWN--KHGKLLFLGLDNAGKTTLLHMLKNDR-----LATLQP---TWHPTSEEL 62
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVD--FLPNCRAASEYLYDILT 160
G +K D+ GH + R +Y P+ G+VF+VDA D R + L++I
Sbjct: 63 AIGNIK-FTTFDLGGHIQARRLWKDYFPEVNGIVFLVDAADPERFDEARVELDALFNI-- 119
Query: 161 KGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
K +P +IL NK D A ++ +R L
Sbjct: 120 ---AELKDVPFVILGNKIDAPNAVSEAELRSAL 149
>SAR1_GIALA (Q8MQT8) GTP-binding protein Sar1
Length = 191
Score = 47.8 bits (112), Expect = 3e-05
Identities = 44/146 (30%), Positives = 67/146 (45%), Gaps = 24/146 (16%)
Query: 47 LRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGK 106
L L+K K T+V GL +GK+ L L++ +T T +P ++ S K
Sbjct: 14 LGLYK--KKATIVFVGLDNAGKSTLLAMLKNSATTTVAPTQ-QPTSQELVMGSIRFKT-- 68
Query: 107 VKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVD---FLPNCRAASEYL--YDILTK 161
D+ GH R ++Y+ + G+VF+VD+ D F + R E L +D+ T
Sbjct: 69 ------FDLGGHEVARQLWEQYVTNSDGIVFLVDSADPSRFEESRRTLQELLDNHDLAT- 121
Query: 162 GSVVKKKIPLLILCNKTDKVTAHTKE 187
P+LIL NK D TA + E
Sbjct: 122 -------TPILILSNKVDIQTAVSME 140
>SAR1_CAEEL (Q23445) GTP-binding protein SAR1
Length = 193
Score = 47.8 bits (112), Expect = 3e-05
Identities = 40/140 (28%), Positives = 64/140 (45%), Gaps = 12/140 (8%)
Query: 54 KTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIV 113
K +V GL +GKT L + L+D Q V ++ P SE G +
Sbjct: 20 KKGKLVFLGLDNAGKTTLLHMLKDDRIAQH-VPTLHPT-------SEQMSLGGIS-FTTY 70
Query: 114 DVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLI 173
D+ GH++ R +Y P VVF++D D + + L +L + +P+LI
Sbjct: 71 DLGGHAQARRVWKDYFPAVDAVVFLIDVAD-AERMQESRVELESLLQDEQIA--SVPVLI 127
Query: 174 LCNKTDKVTAHTKEFIRRQL 193
L NK DK A +++ ++ QL
Sbjct: 128 LGNKIDKPGALSEDQLKWQL 147
>ARL6_HUMAN (Q9H0F7) ADP-ribosylation factor-like protein 6
Length = 186
Score = 47.4 bits (111), Expect = 4e-05
Identities = 45/166 (27%), Positives = 70/166 (42%), Gaps = 8/166 (4%)
Query: 45 LSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKK 104
LS+ L + K V+ GL SGKT + +L+ + + P G I E K
Sbjct: 7 LSVLLGLKKKEVHVLCLGLDNSGKTTIINKLKPSNAQSQNIL---PTIGFSI---EKFKS 60
Query: 105 GKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSV 164
+ + D+ G R R + Y + ++FV+D+ D L A E L +L +
Sbjct: 61 SSLS-FTVFDMSGQGRYRNLWEHYYKEGQAIIFVIDSSDRL-RMVVAKEELDTLLNHPDI 118
Query: 165 VKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRSAVSDA 210
++IP+L NK D A T + + L E K + SDA
Sbjct: 119 KHRRIPILFFANKMDLRDAVTSVKVSQLLCLENIKDKPWHICASDA 164
>AR4A_RAT (P61214) ADP-ribosylation factor-like protein 4A
Length = 200
Score = 46.6 bits (109), Expect = 6e-05
Identities = 44/146 (30%), Positives = 62/146 (42%), Gaps = 17/146 (11%)
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPG 117
+V+ GL +GKT + Y+L+ T E + T K H DV G
Sbjct: 23 IVILGLDCAGKTTVLYRLQFNEFVNTVPTKGFNTEKIKV----TLGNSKTVTFHFWDVGG 78
Query: 118 HSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNK 177
+LRP Y G+VFVVD+VD + A L+ I + +P+LI+ NK
Sbjct: 79 QEKLRPLWKSYTRCTDGIVFVVDSVD-VERMEEAKTELHKITRISE--NQGVPVLIVANK 135
Query: 178 TDKVTAHTKEFIRRQLE-KEIDKLRA 202
D +R L EI+KL A
Sbjct: 136 QD---------LRNSLSLSEIEKLLA 152
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,733,308
Number of Sequences: 164201
Number of extensions: 1128055
Number of successful extensions: 5100
Number of sequences better than 10.0: 457
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 348
Number of HSP's that attempted gapping in prelim test: 4851
Number of HSP's gapped (non-prelim): 462
length of query: 259
length of database: 59,974,054
effective HSP length: 108
effective length of query: 151
effective length of database: 42,240,346
effective search space: 6378292246
effective search space used: 6378292246
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0008.13


