

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004b.4
(929 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
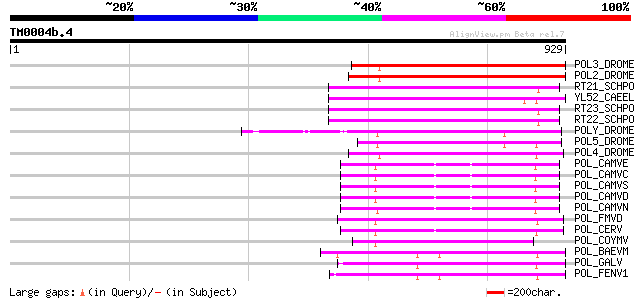
Score E
Sequences producing significant alignments: (bits) Value
POL3_DROME (P04323) Retrovirus-related Pol polyprotein from tran... 284 7e-76
POL2_DROME (P20825) Retrovirus-related Pol polyprotein from tran... 281 7e-75
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 259 2e-68
YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III 257 1e-67
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 257 1e-67
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 257 1e-67
POLY_DROME (P10401) Retrovirus-related Pol polyprotein from tran... 256 3e-67
POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from tran... 254 1e-66
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 238 4e-62
POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic pro... 184 1e-45
POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic pro... 184 1e-45
POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic pro... 182 4e-45
POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic pro... 179 2e-44
POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic pro... 177 9e-44
POL_FMVD (P09523) Enzymatic polyprotein [Contains: Aspartic prot... 168 7e-41
POL_CERV (P05400) Enzymatic polyprotein [Contains: Aspartic prot... 162 5e-39
POL_COYMV (P19199) Putative polyprotein [Contains: Coat protein;... 153 2e-36
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 152 5e-36
POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC 3.4.23... 151 7e-36
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 149 3e-35
>POL3_DROME (P04323) Retrovirus-related Pol polyprotein from
transposon 17.6 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1058
Score = 284 bits (727), Expect = 7e-76
Identities = 146/363 (40%), Positives = 223/363 (61%), Gaps = 6/363 (1%)
Query: 573 YRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRS-----RLCVDYRQLN 627
Y A E++SQ++D+ +G IR S SP+ +P+ +V KK S R+ +DYR+LN
Sbjct: 213 YSYPQAYEQEVESQIQDMLNQGIIRTSNSPYNSPIWVVPKKQDASGKQKFRIVIDYRKLN 272
Query: 628 KVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYL 687
++T+ +R+P+P +D+++ +L F+ IDL G+HQI + E + KTAF T++GHYEYL
Sbjct: 273 EITVGDRHPIPNMDEILGKLGRCNYFTTIDLAKGFHQIEMDPESVSKTAFSTKHGHYEYL 332
Query: 688 VMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDK 747
MPFG+ NAPA F MN I P L++ +V++DDI+++S + +EH + L V + L
Sbjct: 333 RMPFGLKNAPATFQRCMNDILRPLLNKHCLVYLDDIIVFSTSLDEHLQSLGLVFEKLAKA 392
Query: 748 VLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYY 807
L KCEF +E FLGHV++ +GI +P K+EA+ + P +I++F+GL GYY
Sbjct: 393 NLKLQLDKCEFLKQETTFLGHVLTPDGIKPNPEKIEAIQKYPIPTKPKEIKAFLGLTGYY 452
Query: 808 RRFIEGFAKIAGPLTKLTRKNQPFAWTE-DCEQSFQDMKKRLTTAPVLTLPQEKEPYEVY 866
R+FI FA IA P+TK +KN T + + +F+ +K ++ P+L +P + + +
Sbjct: 453 RKFIPNFADIAKPMTKCLKKNMKIDTTNPEYDSAFKKLKYLISEDPILKVPDFTKKFTLT 512
Query: 867 CDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFT 926
DAS LG VL Q ++Y SR L HE NY T + EL A+V+A K +RHYL G F
Sbjct: 513 TDASDVALGAVLSQDGHPLSYISRTLNEHEINYSTIEKELLAIVWATKTFRHYLLGRHFE 572
Query: 927 VFS 929
+ S
Sbjct: 573 ISS 575
>POL2_DROME (P20825) Retrovirus-related Pol polyprotein from
transposon 297 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1059
Score = 281 bits (718), Expect = 7e-75
Identities = 147/370 (39%), Positives = 225/370 (60%), Gaps = 8/370 (2%)
Query: 567 PISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRS-----RLCV 621
PI Y +A E+++Q++++ +G IR S SP+ +P +V KK S R+ +
Sbjct: 206 PIYSKQYPLAQTHEIEVENQVQEMLNQGLIRESNSPYNSPTWVVPKKPDASGANKYRVVI 265
Query: 622 DYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRY 681
DYR+LN++TI +RYP+P +D+++ +L F+ IDL G+HQI + +E I KTAF T+
Sbjct: 266 DYRKLNEITIPDRYPIPNMDEILGKLGKCQYFTTIDLAKGFHQIEMDEESISKTAFSTKS 325
Query: 682 GHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVL 741
GHYEYL MPFG+ NAPA F MN I P L++ +V++DDI+I+S + EH ++ V
Sbjct: 326 GHYEYLRMPFGLRNAPATFQRCMNNILRPLLNKHCLVYLDDIIIFSTSLTEHLNSIQLVF 385
Query: 742 QVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFI 801
L D L KCEF +E FLGH+++ +GI +P KV+A++++ P +IR+F+
Sbjct: 386 TKLADANLKLQLDKCEFLKKEANFLGHIVTPDGIKPNPIKVKAIVSYPIPTKDKEIRAFL 445
Query: 802 GLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCE--QSFQDMKKRLTTAPVLTLPQE 859
GL GYYR+FI +A IA P+T +K T+ E ++F+ +K + P+L LP
Sbjct: 446 GLTGYYRKFIPNYADIAKPMTSCLKKRTKID-TQKLEYIEAFEKLKALIIRDPILQLPDF 504
Query: 860 KEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHY 919
++ + + DAS LG VL Q+ +++ SR L HE NY + EL A+V+A K +RHY
Sbjct: 505 EKKFVLTTDASNLALGAVLSQNGHPISFISRTLNDHELNYSAIEKELLAIVWATKTFRHY 564
Query: 920 LYGSTFTVFS 929
L G F + S
Sbjct: 565 LLGRQFLIAS 574
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 259 bits (663), Expect = 2e-68
Identities = 140/394 (35%), Positives = 226/394 (56%), Gaps = 7/394 (1%)
Query: 534 VVQDFEDVFPE-DVPGIP-PVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLT 591
+ ++F+D+ E + +P P++ +EF +++ + I Y + P ++ + ++
Sbjct: 377 IYKEFKDITAETNTEKLPKPIKGLEFEVELTQENYRLPIRNYPLPPGKMQAMNDEINQGL 436
Query: 592 KKGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAA 651
K G IR S + PV+ V KK+G R+ VDY+ LNK N YPLP I+ L+ +++G+
Sbjct: 437 KSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGST 496
Query: 652 IFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPF 711
IF+K+DL+S YH IRV+ D K AFR G +EYLVMP+G++ APA F ++N I
Sbjct: 497 IFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISTAPAHFQYFINTILGEA 556
Query: 712 LDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS 771
+ VV ++DDILI+S++ EH +H++ VLQ L++ L N KCEF +VKF+G+ IS
Sbjct: 557 KESHVVCYMDDILIHSKSESEHVKHVKDVLQKLKNANLIINQAKCEFHQSQVKFIGYHIS 616
Query: 772 *EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPF 831
+G ++ VL W++PK ++R F+G Y R+FI +++ PL L +K+ +
Sbjct: 617 EKGFTPCQENIDKVLQWKQPKNRKELRQFLGSVNYLRKFIPKTSQLTHPLNNLLKKDVRW 676
Query: 832 AWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRK-----AVA 886
WT Q+ +++K+ L + PVL + + DAS +G VL Q V
Sbjct: 677 KWTPTQTQAIENIKQCLVSPPVLRHFDFSKKILLETDASDVAVGAVLSQKHDDDKYYPVG 736
Query: 887 YSSRQLKIHERNYPTHDLELAAVVFALKIWRHYL 920
Y S ++ + NY D E+ A++ +LK WRHYL
Sbjct: 737 YYSAKMSKAQLNYSVSDKEMLAIIKSLKHWRHYL 770
>YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III
Length = 2186
Score = 257 bits (656), Expect = 1e-67
Identities = 138/407 (33%), Positives = 230/407 (55%), Gaps = 11/407 (2%)
Query: 534 VVQDFEDVFPEDVPGIPPVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKK 593
V++ F+DVF + E I++ G PI P + A E++ ++ + +
Sbjct: 909 VIEQFQDVFAISDDELGRNSGTECVIELKEGAEPIRQKPRPIPLALKPEIRKMIQKMLNQ 968
Query: 594 GFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIF 653
IR S SPW +PV+LVKKKDG R+C+DYR++NKV N +PLP I+ + L G ++
Sbjct: 969 KVIRESKSPWSSPVVLVKKKDGSIRMCIDYRKVNKVVKNNAHPLPNIEATLQSLAGKKLY 1028
Query: 654 SKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLD 713
+ D+ +G+ QI + ++ + TAF +E+ V+PFG+ +PA+F M I L
Sbjct: 1029 TVFDMIAGFWQIPLDEKSKEITAFAIGSELFEWNVLPFGLVISPALFQGTMEEIIGDLLG 1088
Query: 714 RFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*E 773
V++DD+LI S++ E+H + +++ L +R + A+KC +EV++LGH ++ +
Sbjct: 1089 VCAFVYVDDLLIASKDMEQHLQDVKEALTRIRKSGMKLRASKCHIAKKEVEYLGHKVTLD 1148
Query: 774 GIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAW 833
G+ K + + + RP V +++SF+GL GYYR+FI FA+IA LT L + W
Sbjct: 1149 GVETQEVKTDKMKQFSRPTNVKELQSFLGLVGYYRKFILNFAQIASSLTSLISAKVAWIW 1208
Query: 834 TEDCEQSFQDMKKRLTTAPVLTLPQEK------EPYEVYCDASYQGLGCVLMQ-----HR 882
++ E +FQ++KK + PVL P + P+ +Y DAS +G+G VL Q +
Sbjct: 1209 EKEQEIAFQELKKLVCQTPVLAQPDVEAALKGDRPFMIYTDASRKGIGAVLAQEGPDGQQ 1268
Query: 883 KAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTVFS 929
+A++S+ L E Y DLE A++FAL+ ++ +YG+ TVF+
Sbjct: 1269 HPIAFASKALSPAETRYHITDLEALAMMFALRRFKTIIYGTAITVFT 1315
Score = 33.5 bits (75), Expect = 2.7
Identities = 25/96 (26%), Positives = 38/96 (39%), Gaps = 14/96 (14%)
Query: 311 CFRCNKKGHYANHCSESLAACWNCNKPGHTAAECRIPKVEAAANVAGARRPTAGGRVYSI 370
CFRCN+ GH A WNC K +E P V + G R + +
Sbjct: 591 CFRCNEMGHIA----------WNCPKKNENTSEKEAP-VAKVETIEGVRMKDC---LLMV 636
Query: 371 SGTEAEEDDGLIRSTCEIAGNSLIALFDSGATHSFI 406
++E + +I ++ L DSGA+ S +
Sbjct: 637 KSEKSESEVTRSLEKGQIGKANVEILLDSGASISLM 672
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 257 bits (656), Expect = 1e-67
Identities = 139/394 (35%), Positives = 226/394 (57%), Gaps = 7/394 (1%)
Query: 534 VVQDFEDVFPE-DVPGIP-PVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLT 591
+ ++F+D+ E + +P P++ +EF +++ + I Y + P ++ + ++
Sbjct: 377 IYKEFKDITAETNTEKLPKPIKGLEFEVELTQENYRLPIRNYPLPPGKMQAMNDEINQGL 436
Query: 592 KKGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAA 651
K G IR S + PV+ V KK+G R+ VDY+ LNK N YPLP I+ L+ +++G+
Sbjct: 437 KSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGST 496
Query: 652 IFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPF 711
IF+K+DL+S YH IRV+ D K AFR G +EYLVMP+G++ APA F ++N I
Sbjct: 497 IFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAPAHFQYFINTILGEV 556
Query: 712 LDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS 771
+ VV ++D+ILI+S++ EH +H++ VLQ L++ L N KCEF +VKF+G+ IS
Sbjct: 557 KESHVVCYMDNILIHSKSESEHVKHVKDVLQKLKNANLIINQAKCEFHQSQVKFIGYHIS 616
Query: 772 *EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPF 831
+G ++ VL W++PK ++R F+G Y R+FI +++ PL L +K+ +
Sbjct: 617 EKGFTPCQENIDKVLQWKQPKNRKELRQFLGSVNYLRKFIPKTSQLTHPLNNLLKKDVRW 676
Query: 832 AWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRK-----AVA 886
WT Q+ +++K+ L + PVL + + DAS +G VL Q V
Sbjct: 677 KWTPTQTQAIENIKQCLVSPPVLRHFDFSKKILLETDASDVAVGAVLSQKHDDDKYYPVG 736
Query: 887 YSSRQLKIHERNYPTHDLELAAVVFALKIWRHYL 920
Y S ++ + NY D E+ A++ +LK WRHYL
Sbjct: 737 YYSAKMSKAQLNYSVSDKEMLAIIKSLKHWRHYL 770
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 257 bits (656), Expect = 1e-67
Identities = 139/394 (35%), Positives = 226/394 (57%), Gaps = 7/394 (1%)
Query: 534 VVQDFEDVFPE-DVPGIP-PVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLT 591
+ ++F+D+ E + +P P++ +EF +++ + I Y + P ++ + ++
Sbjct: 377 IYKEFKDITAETNTEKLPKPIKGLEFEVELTQENYRLPIRNYPLPPGKMQAMNDEINQGL 436
Query: 592 KKGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAA 651
K G IR S + PV+ V KK+G R+ VDY+ LNK N YPLP I+ L+ +++G+
Sbjct: 437 KSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGST 496
Query: 652 IFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPF 711
IF+K+DL+S YH IRV+ D K AFR G +EYLVMP+G++ APA F ++N I
Sbjct: 497 IFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAPAHFQYFINTILGEV 556
Query: 712 LDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS 771
+ VV ++D+ILI+S++ EH +H++ VLQ L++ L N KCEF +VKF+G+ IS
Sbjct: 557 KESHVVCYMDNILIHSKSESEHVKHVKDVLQKLKNANLIINQAKCEFHQSQVKFIGYHIS 616
Query: 772 *EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPF 831
+G ++ VL W++PK ++R F+G Y R+FI +++ PL L +K+ +
Sbjct: 617 EKGFTPCQENIDKVLQWKQPKNRKELRQFLGSVNYLRKFIPKTSQLTHPLNNLLKKDVRW 676
Query: 832 AWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRK-----AVA 886
WT Q+ +++K+ L + PVL + + DAS +G VL Q V
Sbjct: 677 KWTPTQTQAIENIKQCLVSPPVLRHFDFSKKILLETDASDVAVGAVLSQKHDDDKYYPVG 736
Query: 887 YSSRQLKIHERNYPTHDLELAAVVFALKIWRHYL 920
Y S ++ + NY D E+ A++ +LK WRHYL
Sbjct: 737 YYSAKMSKAQLNYSVSDKEMLAIIKSLKHWRHYL 770
>POLY_DROME (P10401) Retrovirus-related Pol polyprotein from
transposon gypsy [Contains: Reverse transcriptase (EC
2.7.7.49); Endonuclease]
Length = 1035
Score = 256 bits (653), Expect = 3e-67
Identities = 173/557 (31%), Positives = 287/557 (51%), Gaps = 41/557 (7%)
Query: 388 IAGNSLIALFDSGATHSFIDIACAARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWM 447
+AG +L L D+ A ++I V +L + V++P S S + + ++ +
Sbjct: 19 LAGRTLKMLIDTDAAKNYIR---------PVKELKNVMPVASPFSVSSIHGSTEIKHKCL 69
Query: 448 YLDKKFVANLICLP-LKGLDVIIGMDWLSHHHVLLDCANKVVIFPDAGLAEFLNSYFSKL 506
K ++ L L D IIG+D L+ V L+ A + + G+AE L+ YFS
Sbjct: 70 MKVFKHISPFFLLDSLNAFDAIIGLDLLTQAGVKLNLAEDSLEYQ--GIAEKLH-YFSCP 126
Query: 507 SLRKGALSSLMSTTVV--EAKENGVHGIAVVQDFEDVFPEDVPGIPPVRDMEFTIDIVPG 564
S+ ++ ++ V E K+ + + P + +R T+D
Sbjct: 127 SVNFTDVNDIVVPDSVKKEFKDTIIRRKKAFSTTNEALPFNTAVTATIR----TVD---- 178
Query: 565 TGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKK------DGRSR 618
P+ Y + ++++ L K G IRPS SP+ +P +V KK + R
Sbjct: 179 NEPVYSRAYPTLMGVSDFVNNEVKQLLKDGIIRPSRSPYNSPTWVVDKKGTDAFGNPNKR 238
Query: 619 LCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFR 678
L +D+R+LN+ TI +RYP+P I ++ L A F+ +DL+SGYHQI + + D +KT+F
Sbjct: 239 LVIDFRKLNEKTIPDRYPMPSIPMILANLGKAKFFTTLDLKSGYHQIYLAEHDREKTSFS 298
Query: 679 TRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLR 738
G YE+ +PFG+ NA ++F ++ + + + V++DD++I+S N +H H+
Sbjct: 299 VNGGKYEFCRLPFGLRNASSIFQRALDDVLREQIGKICYVYVDDVIIFSENESDHVRHID 358
Query: 739 QVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIR 798
VL+ L D + + K F+ E V++LG ++S +G DP KV+A+ + P V +R
Sbjct: 359 TVLKCLIDANMRVSQEKTRFFKESVEYLGFIVSKDGTKSDPEKVKAIQEYPEPDCVYKVR 418
Query: 799 SFIGLAGYYRRFIEGFAKIAGPLTKLTR-----------KNQPFAWTEDCEQSFQDMKKR 847
SF+GLA YYR FI+ FA IA P+T + + K P + E +FQ ++
Sbjct: 419 SFLGLASYYRVFIKDFAAIARPITDILKGENGSVSKHMSKKIPVEFNETQRNAFQRLRNI 478
Query: 848 LTTAPV-LTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLEL 906
L + V L P K+P+++ DAS G+G VL Q + + SR LK E+NY T++ EL
Sbjct: 479 LASEDVILKYPDFKKPFDLTTDASASGIGAVLSQEGRPITMISRTLKQPEQNYATNEREL 538
Query: 907 AAVVFALKIWRHYLYGS 923
A+V+AL +++LYGS
Sbjct: 539 LAIVWALGKLQNFLYGS 555
>POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from
transposon opus [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1003
Score = 254 bits (648), Expect = 1e-66
Identities = 137/362 (37%), Positives = 213/362 (57%), Gaps = 20/362 (5%)
Query: 582 ELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKK-----DGRSRLCVDYRQLNKVTIKNRYP 636
E++ Q+++L + G IRPS SP+ +P+ +V KK + + R+ VD+++LN VTI + YP
Sbjct: 138 EVERQIDELLQDGIIRPSNSPYNSPIWIVPKKPKPNGEKQYRMVVDFKRLNTVTIPDTYP 197
Query: 637 LPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNA 696
+P I+ + L A F+ +DL SG+HQI +K+ DI KTAF T G YE+L +PFG+ NA
Sbjct: 198 IPDINATLASLGNAKYFTTLDLTSGFHQIHMKESDIPKTAFSTLNGKYEFLRLPFGLKNA 257
Query: 697 PAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKC 756
PA+F ++ I + + V+IDDI+++S + + H ++LR VL L L N K
Sbjct: 258 PAIFQRMIDDILREHIGKVCYVYIDDIIVFSEDYDTHWKNLRLVLASLSKANLQVNLEKS 317
Query: 757 EFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAK 816
F +V+FLG++++ +GI DP KV A+ P +V +++ F+G+ YYR+FI+ +AK
Sbjct: 318 HFLDTQVEFLGYIVTADGIKADPKKVRAISEMPPPTSVKELKRFLGMTSYYRKFIQDYAK 377
Query: 817 IAGPLTKLTR-----------KNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEV 865
+A PLT LTR P E QSF D+K L ++ +L P +P+ +
Sbjct: 378 VAKPLTNLTRGLYANIKSSQSSKVPITLDETALQSFNDLKSILCSSEILAFPCFTKPFHL 437
Query: 866 YCDASYQGLGCVLMQ----HRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLY 921
DAS +G VL Q + +AY SR L E NY T + E+ A++++L R YLY
Sbjct: 438 TTDASNWAIGAVLSQDDQGRDRPIAYISRSLNKTEENYATIEKEMLAIIWSLDNLRAYLY 497
Query: 922 GS 923
G+
Sbjct: 498 GA 499
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 238 bits (608), Expect = 4e-62
Identities = 131/371 (35%), Positives = 206/371 (55%), Gaps = 10/371 (2%)
Query: 567 PISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRS------RLC 620
P+ YR +++ E+++Q++ L K + PSVS + +P+LLV KK + RL
Sbjct: 314 PVYTKNYRSPHSQVEEIQAQVQKLIKDKIVEPSVSQYNSPLLLVPKKSSPNSDKKKWRLV 373
Query: 621 VDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTR 680
+DYRQ+NK + +++PLPRIDD++DQL A FS +DL SG+HQI + + T+F T
Sbjct: 374 IDYRQINKKLLADKFPLPRIDDILDQLGRAKYFSCLDLMSGFHQIELDEGSRDITSFSTS 433
Query: 681 YGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQV 740
G Y + +PFG+ AP F M F +++DD+++ + + ++L +V
Sbjct: 434 NGSYRFTRLPFGLKIAPNSFQRMMTIAFSGIEPSQAFLYMDDLIVIGCSEKHMLKNLTEV 493
Query: 741 LQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSF 800
R+ L + KC F++ EV FLGH + +GI D K + + + P R F
Sbjct: 494 FGKCREYNLKLHPEKCSFFMHEVTFLGHKCTDKGILPDDKKYDVIQNYPVPHDADSARRF 553
Query: 801 IGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEK 860
+ YYRRFI+ FA + +T+L +KN PF WT++C+++F +K +L +L P
Sbjct: 554 VAFCNYYRRFIKNFADYSRHITRLCKKNVPFEWTDECQKAFIHLKSQLINPTLLQYPDFS 613
Query: 861 EPYEVYCDASYQGLGCVLMQ----HRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIW 916
+ + + DAS Q G VL Q H+ VAY+SR E N T + ELAA+ +A+ +
Sbjct: 614 KEFCITTDASKQACGAVLTQNHNGHQLPVAYASRAFTKGESNKSTTEQELAAIHWAIIHF 673
Query: 917 RHYLYGSTFTV 927
R Y+YG FTV
Sbjct: 674 RPYIYGKHFTV 684
>POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 184 bits (466), Expect = 1e-45
Identities = 119/381 (31%), Positives = 194/381 (50%), Gaps = 18/381 (4%)
Query: 555 MEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLV---- 610
M+ +I + + I + P + +P + E Q+++L I+PS SP AP LV
Sbjct: 234 MKASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEA 293
Query: 611 KKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDE 670
+K+ G+ R+ V+Y+ +NK TI + Y LP D+L+ ++G IFS D +SG+ Q+ + E
Sbjct: 294 EKRRGKKRMVVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQE 353
Query: 671 DIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNR 730
TAF GHYE+ V+PFG+ AP++F +M+ F F +F V++DDIL++S N
Sbjct: 354 SRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAFRVF-RKFCCVYVDDILVFSNNE 412
Query: 731 EEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWER 790
E+H H+ +LQ + + K + + +++ FLG I EG + + +
Sbjct: 413 EDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFLGLEID-EGTHKPQGHILEHIN-KF 470
Query: 791 PKTVTD---IRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKR 847
P T+ D ++ F+G+ Y +I A+I PL ++N P+ WT++ Q +KK
Sbjct: 471 PDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKN 530
Query: 848 LTTAPVLTLPQEKEPYEVYCDASYQGLGCVL--------MQHRKAVAYSSRQLKIHERNY 899
L P L P +E + DAS G +L Y+S K ERNY
Sbjct: 531 LQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAERNY 590
Query: 900 PTHDLELAAVVFALKIWRHYL 920
++D E AV+ +K + YL
Sbjct: 591 HSNDKETLAVINTIKKFSIYL 611
>POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 184 bits (466), Expect = 1e-45
Identities = 119/381 (31%), Positives = 194/381 (50%), Gaps = 18/381 (4%)
Query: 555 MEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLV---- 610
M+ +I + + I + P + +P + E Q+++L I+PS SP AP LV
Sbjct: 234 MKASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEA 293
Query: 611 KKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDE 670
+K+ G+ R+ V+Y+ +NK TI + Y LP D+L+ ++G IFS D +SG+ Q+ + E
Sbjct: 294 EKRRGKKRMVVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQE 353
Query: 671 DIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNR 730
TAF GHYE+ V+PFG+ AP++F +M+ F F +F V++DDIL++S N
Sbjct: 354 SRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAFRVF-RKFCCVYVDDILVFSNNE 412
Query: 731 EEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWER 790
E+H H+ +LQ + + K + + +++ FLG I EG + + +
Sbjct: 413 EDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFLGLEID-EGTHKPQGHILEHIN-KF 470
Query: 791 PKTVTD---IRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKR 847
P T+ D ++ F+G+ Y +I A+I PL ++N P+ WT++ Q +KK
Sbjct: 471 PDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKN 530
Query: 848 LTTAPVLTLPQEKEPYEVYCDASYQGLGCVL--------MQHRKAVAYSSRQLKIHERNY 899
L P L P +E + DAS G +L Y+S K ERNY
Sbjct: 531 LQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAERNY 590
Query: 900 PTHDLELAAVVFALKIWRHYL 920
++D E AV+ +K + YL
Sbjct: 591 HSNDKETLAVINTIKKFSIYL 611
>POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 182 bits (462), Expect = 4e-45
Identities = 117/381 (30%), Positives = 194/381 (50%), Gaps = 18/381 (4%)
Query: 555 MEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLV---- 610
M+ +I + + I + P + +P + E Q+++L I+PS SP AP LV
Sbjct: 234 MKASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEA 293
Query: 611 KKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDE 670
+K+ G+ R+ V+Y+ +NK T+ + Y LP D+L+ ++G IFS D +SG+ Q+ + E
Sbjct: 294 EKRRGKKRMVVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQE 353
Query: 671 DIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNR 730
TAF GHYE+ V+PFG+ AP++F +M+ F F +F V++DDIL++S N
Sbjct: 354 SRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAFRVF-RKFCCVYVDDILVFSNNE 412
Query: 731 EEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWER 790
E+H H+ +LQ + + K + + +++ FLG I EG + + +
Sbjct: 413 EDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFLGLEID-EGTHKPQGHILEHIN-KF 470
Query: 791 PKTVTD---IRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKR 847
P T+ D ++ F+G+ Y +I A+I PL ++N P+ WT++ Q +KK
Sbjct: 471 PDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKLKENVPWRWTKEDTLYMQKVKKN 530
Query: 848 LTTAPVLTLPQEKEPYEVYCDASYQGLGCVL--------MQHRKAVAYSSRQLKIHERNY 899
L P L P +E + DAS G +L Y+S K E+NY
Sbjct: 531 LQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAEKNY 590
Query: 900 PTHDLELAAVVFALKIWRHYL 920
++D E AV+ +K + YL
Sbjct: 591 HSNDKETLAVINTIKKFSIYL 611
>POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 674
Score = 179 bits (455), Expect = 2e-44
Identities = 116/381 (30%), Positives = 193/381 (50%), Gaps = 18/381 (4%)
Query: 555 MEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLV---- 610
M+ +I + + I + P + +P + E Q+++L I+PS SP AP LV
Sbjct: 229 MKASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEA 288
Query: 611 KKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDE 670
+K+ G+ R+ V+Y+ +NK T+ + Y P D+L+ ++G IFS D +SG+ Q+ + E
Sbjct: 289 EKRRGKKRMVVNYKAMNKATVGDAYNPPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQE 348
Query: 671 DIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNR 730
TAF GHYE+ V+PFG+ AP++F +M+ F F +F V++DDIL++S N
Sbjct: 349 SRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAFRVF-RKFCCVYVDDILVFSNNE 407
Query: 731 EEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWER 790
E+H H+ +LQ + + K + + +++ FLG I EG + + +
Sbjct: 408 EDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFLGLEID-EGTHKPQGHILEHIN-KF 465
Query: 791 PKTVTD---IRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKR 847
P T+ D ++ F+G+ Y +I A+I PL ++N P+ WT++ Q +KK
Sbjct: 466 PDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKN 525
Query: 848 LTTAPVLTLPQEKEPYEVYCDASYQGLGCVL--------MQHRKAVAYSSRQLKIHERNY 899
L P L P +E + DAS G +L Y+S K E+NY
Sbjct: 526 LQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAEKNY 585
Query: 900 PTHDLELAAVVFALKIWRHYL 920
++D E AV+ +K + YL
Sbjct: 586 HSNDKETLAVINTIKKFSIYL 606
>POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 680
Score = 177 bits (450), Expect = 9e-44
Identities = 115/381 (30%), Positives = 190/381 (49%), Gaps = 18/381 (4%)
Query: 555 MEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKD 614
M+ +I + + I + P + +P + E Q+++L I+PS SP AP LV +
Sbjct: 235 MKASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEA 294
Query: 615 ----GRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDE 670
G R+ V+Y+ +NK T+ + Y LP D+L+ ++G IFS D +SG+ Q+ + E
Sbjct: 295 ENGRGNKRMVVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQE 354
Query: 671 DIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNR 730
TAF GHYE+ V+PFG+ AP++F +M+ F F +F V++DDI+++S N
Sbjct: 355 SRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAFRVF-RKFCCVYVDDIVVFSNNE 413
Query: 731 EEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWER 790
E+H H+ +LQ + + K + + +++ FLG I EG + + +
Sbjct: 414 EDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFLGLEID-EGTHKPQGHILEHIN-KF 471
Query: 791 PKTVTD---IRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKR 847
P T+ D ++ F+G+ Y +I A++ PL ++N P+ WT++ Q +KK
Sbjct: 472 PDTLEDKKQLQRFLGILTYASDYIPNLAQMRQPLQAKLKENVPWKWTKEDTLYMQKVKKN 531
Query: 848 LTTAPVLTLPQEKEPYEVYCDASYQGLGCVL--------MQHRKAVAYSSRQLKIHERNY 899
L P L P +E + DAS G +L Y S K ERNY
Sbjct: 532 LQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYRSGSFKAAERNY 591
Query: 900 PTHDLELAAVVFALKIWRHYL 920
++D E AV+ +K + YL
Sbjct: 592 HSNDKETLAVINTIKKFSIYL 612
>POL_FMVD (P09523) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 666
Score = 168 bits (425), Expect = 7e-41
Identities = 110/392 (28%), Positives = 199/392 (50%), Gaps = 14/392 (3%)
Query: 549 IPPVRD---MEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGA 605
I P++ M+ +I ++ I + P +P + Q+++L G I PS S +
Sbjct: 218 IDPIKSKQWMKASIKLIDPLKVIRVKPMSYSPQDREGFAKQIKELLDLGLIIPSKSQHMS 277
Query: 606 PVLLVK----KKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSG 661
P LV+ ++ G+ R+ V+Y+ +N+ TI + + LP + +L+ L+G +IFS D +SG
Sbjct: 278 PAFLVENEAERRRGKKRMVVNYKAINQATIGDSHNLPNMQELLTLLRGKSIFSSFDCKSG 337
Query: 662 YHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFID 721
+ Q+ + +E + TAF GH+++ V+PFG+ AP++F +M + D+F +V++D
Sbjct: 338 FWQVVLDEESQKLTAFTCPQGHFQWKVVPFGLKQAPSIFQRHMQTALNG-ADKFCMVYVD 396
Query: 722 DILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSK 781
DI+++S + +H H+ VL+++ + + K + E++ FLG I
Sbjct: 397 DIIVFSNSELDHYNHVYAVLKIVEKYGIILSKKKANLFKEKINFLGLEIDKGTHCPQNHI 456
Query: 782 VEAVLAW-ERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQS 840
+E + + +R + ++ F+G+ Y +I A+I PL +K+ + WT+
Sbjct: 457 LENIHKFPDRLEDKKHLQRFLGVLTYAETYIPKLAEIRKPLQVKLKKDVTWNWTQSDSDY 516
Query: 841 FQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQH-----RKAVAYSSRQLKIH 895
+ +KK L + P L LP+ ++ + DAS G VL YSS K
Sbjct: 517 VKKIKKNLGSFPKLYLPKPEDHLIIETDASDSFWGGVLKARALDGVELICRYSSGSFKQA 576
Query: 896 ERNYPTHDLELAAVVFALKIWRHYLYGSTFTV 927
E+NY ++D EL AV + + YL FTV
Sbjct: 577 EKNYHSNDKELLAVKQVITKFSAYLTPVRFTV 608
>POL_CERV (P05400) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 659
Score = 162 bits (409), Expect = 5e-39
Identities = 106/384 (27%), Positives = 192/384 (49%), Gaps = 12/384 (3%)
Query: 555 MEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVK--- 611
M TI+++ + + P +P++ E Q+++L + I+PS S +P LV+
Sbjct: 216 MTATIELIDPKTVVKVKPMSYSPSDREEFDRQIKELLELKVIKPSKSTHMSPAFLVENEA 275
Query: 612 -KKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDE 670
++ G+ R+ V+Y+ +NK T + + LP D+L+ ++G I+S D +SG Q+ + E
Sbjct: 276 ERRRGKKRMVVNYKAMNKATKGDAHNLPNKDELLTLVRGKKIYSSFDCKSGLWQVLLDKE 335
Query: 671 DIQKTAFRTRYGHYEYLVMPFGVTNAPAVF-MDYMNRIFHPFLDRFVVVFIDDILIYSR- 728
TAF GHY++ V+PFG+ AP++F Y N + + ++ V++DDIL++S
Sbjct: 336 SQLLTAFTCPQGHYQWNVVPFGLKQAPSIFPKTYANSHSNQY-SKYCCVYVDDILVFSNT 394
Query: 729 NREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAW 788
R+EH H+ +L+ + + K + + E++ FLG I +E + +
Sbjct: 395 GRKEHYIHVLNILRRCEKLGIILSKKKAQLFKEKINFLGLEIDQGTHCPQNHILEHIHKF 454
Query: 789 -ERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKR 847
+R + ++ F+G+ Y +I A I PL +++ + W + Q +KK
Sbjct: 455 PDRIEDKKQLQRFLGILTYASDYIPKLASIRKPLQSKLKEDSTWTWNDTDSQYMAKIKKN 514
Query: 848 LTTAPVLTLPQEKEPYEVYCDASYQGLGCVLM----QHRKAVAYSSRQLKIHERNYPTHD 903
L + P L P+ + + DAS + G +L H Y+S K ERNY +++
Sbjct: 515 LKSFPKLYHPEPNDKLVIETDASEEFWGGILKAIHNSHEYICRYASGSFKAAERNYHSNE 574
Query: 904 LELAAVVFALKIWRHYLYGSTFTV 927
EL AV+ +K + YL S F +
Sbjct: 575 KELLAVIRVIKKFSIYLTPSRFLI 598
>POL_COYMV (P19199) Putative polyprotein [Contains: Coat protein;
Protease (EC 3.4.23.-); Reverse transcriptase (EC
2.7.7.49); Ribonuclease H (EC 3.1.26.4)]
Length = 1886
Score = 153 bits (387), Expect = 2e-36
Identities = 91/316 (28%), Positives = 162/316 (50%), Gaps = 14/316 (4%)
Query: 575 MAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLV-----------KKKDGRSRLCVDY 623
+ P + + Q+ L + IRPS S + +V K+K G+ R+ +Y
Sbjct: 1410 VTPGDEEAMTRQINLLLQMKVIRPSESKHRSTAFIVRSGTEIDPITGKEKKGKERMVFNY 1469
Query: 624 RQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGH 683
+ LN+ T ++Y LP I+ ++ ++ + I+SK DL+SG+ Q+ +++E + TAF
Sbjct: 1470 KLLNENTESDQYSLPGINTIISKVGRSKIYSKFDLKSGFWQVAMEEESVPWTAFLAGNKL 1529
Query: 684 YEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQV 743
YE+LVMPFG+ NAPA+F M+ +F ++F+ V+IDDIL++S E+H +HL +LQ+
Sbjct: 1530 YEWLVMPFGLKNAPAIFQRKMDNVFKG-TEKFIAVYIDDILVFSETAEQHSQHLYTMLQL 1588
Query: 744 LRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVT--DIRSFI 801
++ L + TK + E+ FLG + I + P + + + K T +RS++
Sbjct: 1589 CKENGLILSPTKMKIGTPEIDFLGASLGCTKIKLQPHIISKICDFSDEKLATPEGMRSWL 1648
Query: 802 GLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKE 861
G+ Y R +I+ K+ PL + + + + +K+++ P L LP +
Sbjct: 1649 GILSYARNYIQDIGKLVQPLRQKMAPTGDKRMNPETWKMVRQIKEKVKNLPDLQLPPKDS 1708
Query: 862 PYEVYCDASYQGLGCV 877
+ D G G V
Sbjct: 1709 FIIIETDGCMTGWGAV 1724
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 152 bits (383), Expect = 5e-36
Identities = 115/434 (26%), Positives = 205/434 (46%), Gaps = 24/434 (5%)
Query: 520 TVVEAKENGVHGIAVVQDFEDVFPEDVP-------GIPPVR-DMEFTIDIVPGTGPISIA 571
TV E+ + I V DV+ +D P G+ + ID+ P P+SI
Sbjct: 121 TVSLQDEHRLFDIPVTTSLPDVWLQDFPQAWAETGGLGRAKCQAPIIIDLKPTAVPVSIK 180
Query: 572 PYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRS-RLCVDYRQLNKVT 630
Y M+ ++ + + G +RP SPW P+L VKK + R D R++NK T
Sbjct: 181 QYPMSLEAHMGIRQHIIKFLELGVLRPCRSPWNTPLLPVKKPGTQDYRPVQDLREINKRT 240
Query: 631 IKNRYPLPRIDDLMDQLK-GAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTR------YGH 683
+ +P +L+ LK + ++ +DL+ + + + + + AF + G
Sbjct: 241 VDIHPTVPNPYNLLSTLKPDYSWYTVLDLKDAFFCLPLAPQSQELFAFEWKDPERGISGQ 300
Query: 684 YEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVV----FIDDILIYSRNREEHEEHLRQ 739
+ +P G N+P +F + ++R F + V ++DD+L+ + ++ + R
Sbjct: 301 LTWTRLPQGFKNSPTLFDEALHRDLTDFRTQHPEVTLLQYVDDLLLAAPTKKACTQGTRH 360
Query: 740 VLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRS 799
+LQ L +K A+A K + +V +LG+++S + P ++E V P+ ++R
Sbjct: 361 LLQELGEKGYRASAKKAQICQTKVTYLGYILSEGKRWLTPGRIETVARIPPPRNPREVRE 420
Query: 800 FIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQE 859
F+G AG+ R +I GFA++A PL LT+++ PF W + + +F+ +KK L +AP L LP
Sbjct: 421 FLGTAGFCRLWIPGFAELAAPLYALTKESTPFTWQTEHQLAFEALKKALLSAPALGLPDT 480
Query: 860 KEPYEVYCDASYQGLGCVLMQH----RKAVAYSSRQLKIHERNYPTHDLELAAVVFALKI 915
+P+ ++ D VL Q ++ VAY S++L +P +AA +K
Sbjct: 481 SKPFTLFLDERQGIAKGVLTQKLGPWKRPVAYLSKKLDPVAAGWPPCLRIMAATAMLVKD 540
Query: 916 WRHYLYGSTFTVFS 929
G TV +
Sbjct: 541 SAKLTLGQPLTVIT 554
>POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)]
Length = 1165
Score = 151 bits (382), Expect = 7e-36
Identities = 112/397 (28%), Positives = 191/397 (47%), Gaps = 21/397 (5%)
Query: 549 IPPVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVL 608
+PPV +++ G P+++ Y M+ ++ ++ G + P SPW P+L
Sbjct: 162 VPPV-----VVELRSGASPVAVRQYPMSKEAREGIRPHIQKFLDLGVLVPCRSPWNTPLL 216
Query: 609 LVKKKDGRS-RLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAI-FSKIDLRSGYHQIR 666
VKK R D R++NK +P +L+ L + +S +DL+ + +R
Sbjct: 217 PVKKPGTNDYRPVQDLREINKRVQDIHPTVPNPYNLLSSLPPSYTWYSVLDLKDAFFCLR 276
Query: 667 VKDEDIQKTAFRTR------YGHYEYLVMPFGVTNAPAVFMDYMNRIFHPF--LDRFVVV 718
+ AF + G + +P G N+P +F + ++R PF L+ VV+
Sbjct: 277 LHPNSQPLFAFEWKDPEKGNTGQLTWTRLPQGFKNSPTLFDEALHRDLAPFRALNPQVVL 336
Query: 719 --FIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIA 776
++DD+L+ + E+ ++ +++LQ L +A K + EV +LG+++
Sbjct: 337 LQYVDDLLVAAPTYEDCKKGTQKLLQELSKLGYRVSAKKAQLCQREVTYLGYLLKEGKRW 396
Query: 777 VDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTED 836
+ P++ V+ P T +R F+G AG+ R +I GFA +A PL LT+++ PF WTE+
Sbjct: 397 LTPARKATVMKIPVPTTPRQVREFLGTAGFCRLWIPGFASLAAPLYPLTKESIPFIWTEE 456
Query: 837 CEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQ----HRKAVAYSSRQL 892
+Q+F +KK L +AP L LP +P+ +Y D VL Q R+ VAY S++L
Sbjct: 457 HQQAFDHIKKALLSAPALALPDLTKPFTLYIDERAGVARGVLTQTLGPWRRPVAYLSKKL 516
Query: 893 KIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTVFS 929
+PT +AAV LK G TV +
Sbjct: 517 DPVASGWPTCLKAVAAVALLLKDADKLTLGQNVTVIA 553
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 149 bits (376), Expect = 3e-35
Identities = 111/412 (26%), Positives = 196/412 (46%), Gaps = 18/412 (4%)
Query: 535 VQDFEDVFPEDVPGIPPVR-DMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKK 593
+QDF + E G+ + + ID+ P P+SI Y M+ ++ + +
Sbjct: 1 LQDFPQAWAE-TGGLGRAKCQVPIIIDLKPTAMPVSIRQYPMSKEAHMGIQPHITRFLEL 59
Query: 594 GFIRPSVSPWGAPVLLVKKKDGRS-RLCVDYRQLNKVTIKNRYPLPRIDDLMDQLK-GAA 651
G +RP SPW P+L VKK R R D R++NK T+ +P +L+ L
Sbjct: 60 GVLRPCRSPWNTPLLPVKKPGTRDYRPVQDLREVNKRTMDIHPTVPNPYNLLSTLSPDRT 119
Query: 652 IFSKIDLRSGYHQIRVKDEDIQKTAFRTR------YGHYEYLVMPFGVTNAPAVFMDYMN 705
++ +DL+ + + + + + AF R G + +P G N+P +F + ++
Sbjct: 120 WYTVLDLKDAFFCLPLAPQSQELFAFEWRDPERGISGQLTWTRLPQGFKNSPTLFDEALH 179
Query: 706 RIFHPFLDRFVVV----FIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLE 761
R F + V ++DD+L+ + +E + +L+ L DK A+A K +
Sbjct: 180 RDLTDFRTQHPEVTLLQYVDDLLLAAPTKEACIRGTKHLLRELGDKGYRASAKKAQICQT 239
Query: 762 EVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPL 821
+V +LG+++S + P ++E V P+ ++R F+G AG+ R +I GFA++A PL
Sbjct: 240 KVTYLGYILSEGKRWLTPGRIETVAHIPPPQNPREVREFLGTAGFCRLWIPGFAELAAPL 299
Query: 822 TKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQH 881
LT+++ PF W E + +F+ +K+ L +AP L LP +P+ ++ D VL Q
Sbjct: 300 YALTKESAPFTWQEKHQSAFEALKEALLSAPALGLPDTSKPFTLFIDEKQGIAKGVLTQK 359
Query: 882 ----RKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTVFS 929
++ VAY S++L +P +AA +K G TV +
Sbjct: 360 LGPWKRPVAYLSKKLDPVAAGWPPCLRIMAATAMLVKDSAKLTLGQPLTVIT 411
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,114,555
Number of Sequences: 164201
Number of extensions: 4746996
Number of successful extensions: 12922
Number of sequences better than 10.0: 244
Number of HSP's better than 10.0 without gapping: 121
Number of HSP's successfully gapped in prelim test: 123
Number of HSP's that attempted gapping in prelim test: 12230
Number of HSP's gapped (non-prelim): 517
length of query: 929
length of database: 59,974,054
effective HSP length: 120
effective length of query: 809
effective length of database: 40,269,934
effective search space: 32578376606
effective search space used: 32578376606
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0004b.4


