

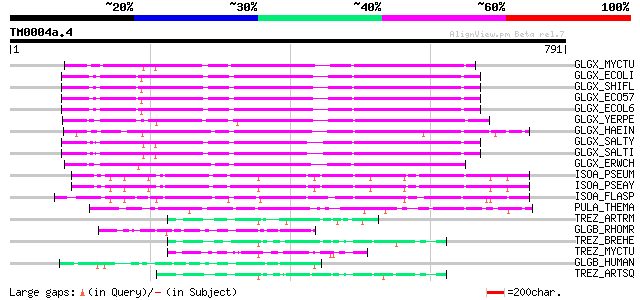
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004a.4
(791 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
GLGX_MYCTU (Q10767) Glycogen operon protein glgX homolog (EC 3.2... 442 e-123
GLGX_ECOLI (P15067) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 383 e-106
GLGX_SHIFL (Q83J89) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 382 e-105
GLGX_ECO57 (Q8X6X8) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 382 e-105
GLGX_ECOL6 (Q8FCR8) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 377 e-104
GLGX_YERPE (Q8ZA76) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 376 e-103
GLGX_HAEIN (P45178) Glycogen operon protein glgX homolog (EC 3.2... 373 e-102
GLGX_SALTY (Q8ZLG6) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 372 e-102
GLGX_SALTI (Q8Z234) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 371 e-102
GLGX_ERWCH (Q8KR69) Glycogen debranching enzyme (EC 3.2.1.-) (Gl... 365 e-100
ISOA_PSEUM (P26501) Isoamylase precursor (EC 3.2.1.68) 270 1e-71
ISOA_PSEAY (P10342) Isoamylase precursor (EC 3.2.1.68) 270 1e-71
ISOA_FLASP (O32611) Isoamylase precursor (EC 3.2.1.68) 270 1e-71
PULA_THEMA (O33840) Pullulanase precursor (EC 3.2.1.41) (Alpha-d... 167 1e-40
TREZ_ARTRM (Q9AJN6) Malto-oligosyltrehalose trehalohydrolase (EC... 81 1e-14
GLGB_RHOMR (Q93HU3) 1,4-alpha-glucan branching enzyme (EC 2.4.1.... 79 4e-14
TREZ_BREHE (O52520) Malto-oligosyltrehalose trehalohydrolase (EC... 77 1e-13
TREZ_MYCTU (Q10769) Malto-oligosyltrehalose trehalohydrolase (EC... 74 2e-12
GLGB_HUMAN (Q04446) 1,4-alpha-glucan branching enzyme (EC 2.4.1.... 74 2e-12
TREZ_ARTSQ (Q44316) Malto-oligosyltrehalose trehalohydrolase (EC... 72 7e-12
>GLGX_MYCTU (Q10767) Glycogen operon protein glgX homolog (EC
3.2.1.-)
Length = 721
Score = 442 bits (1136), Expect = e-123
Identities = 254/606 (41%), Positives = 337/606 (54%), Gaps = 60/606 (9%)
Query: 79 GYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTGSVWH 138
G P GAT G NF+++S A LCL ++ V I LD + G VWH
Sbjct: 22 GNAYPLGATYDGAGTNFSLFSEIAEKVELCLI-----DEDGVESRIPLDEV---DGYVWH 73
Query: 139 VFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEFGSL-------GP 191
+L YG++ G F P GH D S +LLDPY K+ FG
Sbjct: 74 AYLPNITPGQRYGFRVHGPFDPAAGHRCDPSKLLLDPYGKSFHGDFTFGQALYSYDVNAV 133
Query: 192 DGNCWPQMAGMVPSN------DDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTK 244
D + P M + + FDW D + P + +IYE HV+G T+ H S +
Sbjct: 134 DPDSTPPMVDSLGHTMTSVVINPFFDWAYDRSPRTPYHETVIYEAHVKGMTQTHPSIPPE 193
Query: 245 FPGTYLGVVEK--LDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRV-NFWGYSTVN 301
GTY G+ +DHL EL V +EL+P H+F ++S D + N+WGY+T
Sbjct: 194 LRGTYAGLAHPVIIDHLNELNVTAVELMPVHQF------LHDSRLLDLGLRNYWGYNTFG 247
Query: 302 YFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFR 361
+F+P +Y+S + G + E K +++ H+ GIEVI+DVV+NHT EGN GP I+FR
Sbjct: 248 FFAPHHQYAST--RQAG-SAVAEFKTMVRSLHEAGIEVILDVVYNHTAEGNHLGPTINFR 304
Query: 362 GVDNSIYYMIAPKG-EFY-NYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLA 419
G+DN+ YY + FY +++G GN+LN HP Q I+D LRYWV EMHVDGFRFDLA
Sbjct: 305 GIDNTAYYRLMDHDLRFYKDFTGTGNSLNARHPHTLQLIMDSLRYWVIEMHVDGFRFDLA 364
Query: 420 SIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDAG- 478
S + R + ++ F DL+ DP++ VKLIAE WD G
Sbjct: 365 STLARELHDVDRLSAF--------------------FDLVQQDPVVSQVKLIAEPWDVGE 404
Query: 479 GLYQVGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNS 538
G YQVG FP G+W+EWNGKYRDTVR + +G G FA L GS ++Y+ GR+P S
Sbjct: 405 GGYQVGNFP--GLWTEWNGKYRDTVRDYWRGEPATLGEFASRLTGSSDLYEATGRRPSAS 462
Query: 539 INFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQ 598
INFV HDGFTL DLV+YN+KHN NGE+N DGE++N SWNCG EG + LR RQ
Sbjct: 463 INFVTAHDGFTLNDLVSYNDKHNEANGENNRDGESYNRSWNCGVEGPTDDPDILALRARQ 522
Query: 599 MRNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCC 658
MRN + +LMVSQG PMI GDE G T+ GNNN YC D+ L++ W + ++ ++D F
Sbjct: 523 MRNMWATLMVSQGTPMIAHGDEIGRTQYGNNNVYCQDSELSWMDWSLVDK-NADLLAFAR 581
Query: 659 LMTKFR 664
T R
Sbjct: 582 KATTLR 587
>GLGX_ECOLI (P15067) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 657
Score = 383 bits (984), Expect = e-106
Identities = 231/608 (37%), Positives = 320/608 (51%), Gaps = 58/608 (9%)
Query: 75 QVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTG 134
Q++ G PAP GA GVNF ++S +A LC+F + Q Y L +G
Sbjct: 3 QLAIGKPAPLGAHYDGQGVNFTLFSAHAERVELCVFDAN----GQEHRY----DLPGHSG 54
Query: 135 SVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEF-------- 186
+WH +L + YGY+ G + P EGH ++ + +L+DP A+ + GEF
Sbjct: 55 DIWHGYLPDARPGLRYGYRVHGPWQPAEGHRFNPAKLLIDPCARQI--DGEFKDNPLLHA 112
Query: 187 GSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTKF 245
G PD +A D +DWE D P + P IIYE HV+G T H +
Sbjct: 113 GHNEPDYRDNAAIAPKCVVVVDHYDWEDDAPPRTPWGSTIIYEAHVKGLTYLHPEIPVEI 172
Query: 246 PGTY--LGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYF 303
GTY LG +++LK+LG+ +ELLP +F S +Q N+WGY+ V F
Sbjct: 173 RGTYKALGHPVMINYLKQLGITALELLPVAQFA-----SEPRLQRMGLSNYWGYNPVAMF 227
Query: 304 SPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGV 363
+ Y+ + ++E + IK HK GIEVI+D+V NH+ E + +GP+ S RG+
Sbjct: 228 ALHPAYACSP-----ETALDEFRDAIKALHKAGIEVILDIVLNHSAELDLDGPLFSLRGI 282
Query: 364 DNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMT 423
DN YY I G+++N++GCGNTLN +HP V + CLRYWV HVDGFRFDLA++M
Sbjct: 283 DNRSYYWIREDGDYHNWTGCGNTLNLSHPAVVDYASACLRYWVETCHVDGFRFDLAAVMG 342
Query: 424 RSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWD-AGGLYQ 482
R+ PL I N P+L VKLIAE WD A G YQ
Sbjct: 343 RTPEFRQDA---------------------PLFTAIQNCPVLSQVKLIAEPWDIAPGGYQ 381
Query: 483 VGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFV 542
VG FP +++EWN +RD R+F D GAFA S +V++ GR P +IN V
Sbjct: 382 VGNFPP--LFAEWNDHFRDAARRFWLHYDLPLGAFAGRFAASSDVFKRNGRLPSAAINLV 439
Query: 543 CTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNF 602
HDGFTL D V +N+KHN NGE+N DG N+N S N G+EG S + + R+ +
Sbjct: 440 TAHDGFTLRDCVCFNHKHNEANGEENRDGTNNNYSNNHGKEGLGGSLDLVERRRDSIHAL 499
Query: 603 FLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTK 662
+L++SQG PM+ GDE+GH++ GNNN YC DN L + W ++SS F +
Sbjct: 500 LTTLLLSQGTPMLLAGDEHGHSQHGNNNAYCQDNQLTWLDW---SQASSGLTAFTAALIH 556
Query: 663 FRYECESL 670
R +L
Sbjct: 557 LRKRIPAL 564
>GLGX_SHIFL (Q83J89) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 657
Score = 382 bits (982), Expect = e-105
Identities = 230/608 (37%), Positives = 320/608 (51%), Gaps = 58/608 (9%)
Query: 75 QVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTG 134
Q++ G PAP GA GVNF ++S +A LC+F + Q Y L +G
Sbjct: 3 QLAIGKPAPLGAHYDGQGVNFTLFSAHAERVELCVFDAN----GQEHRY----DLPGNSG 54
Query: 135 SVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEF-------- 186
+WH +L + YGY+ G + P EGH ++ + +L+DP A+ + GEF
Sbjct: 55 DIWHGYLPDARPGLRYGYRVHGPWQPAEGHRFNPAKLLIDPCARQI--DGEFKDNPLLHA 112
Query: 187 GSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTKF 245
G PD +A D +DWE D P + P IIYE HV+G T H +
Sbjct: 113 GHNEPDYRDNAAIAPKCVVVVDHYDWEDDAPPRTPWGSTIIYEAHVKGLTYLHPEIPVEI 172
Query: 246 PGTY--LGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYF 303
GTY LG +++LK+LG+ +ELLP +F S +Q N+WGY+ V F
Sbjct: 173 RGTYKALGHPVMINYLKQLGITALELLPVAQFA-----SEPRLQRMGLSNYWGYNPVAMF 227
Query: 304 SPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGV 363
+ Y+ + ++E + IK HK GIEVI+D+V NH+ E + +GP+ S RG+
Sbjct: 228 ALHPAYACSP-----ETALDEFRDAIKALHKAGIEVILDIVLNHSAELDLDGPLFSLRGI 282
Query: 364 DNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMT 423
DN YY I G+++N++GCGNTLN +HP V + CLRYWV HVDGFRFDLA++M
Sbjct: 283 DNRSYYWIREDGDYHNWTGCGNTLNLSHPAVVDYASACLRYWVETCHVDGFRFDLAAVMG 342
Query: 424 RSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWD-AGGLYQ 482
R+ PL I N P+L VKLIAE WD A YQ
Sbjct: 343 RTPEFRQDA---------------------PLFTAIQNCPVLSQVKLIAEPWDIAPSGYQ 381
Query: 483 VGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFV 542
VG FP +++EWN +RD R+F D GAFA S +V++ GR P +IN V
Sbjct: 382 VGNFPP--LFAEWNDHFRDAARRFWLHYDLPLGAFAGRFAASSDVFKRNGRLPSAAINLV 439
Query: 543 CTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNF 602
HDGFTL D V +N+KHN NGE+N DG N+N S N G+EG + + + R+ +
Sbjct: 440 TAHDGFTLRDCVCFNHKHNEANGEENRDGTNNNYSNNHGKEGLGGTLDLVERRRDSIHAL 499
Query: 603 FLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTK 662
+L++SQG PM+ GDE+GH++ GNNN YC DN LN+ W ++SS F +
Sbjct: 500 LTTLLLSQGTPMLLAGDEHGHSQHGNNNAYCQDNQLNWLDW---SQASSGLTAFTAALIH 556
Query: 663 FRYECESL 670
R +L
Sbjct: 557 LRKRIPAL 564
>GLGX_ECO57 (Q8X6X8) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 657
Score = 382 bits (982), Expect = e-105
Identities = 230/608 (37%), Positives = 320/608 (51%), Gaps = 58/608 (9%)
Query: 75 QVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTG 134
Q++ G PAP GA GVNF ++S +A LC+F + Q Y L +G
Sbjct: 3 QLAIGKPAPLGAHYDGQGVNFTLFSAHAERVELCVFDAN----GQEHRY----DLPGHSG 54
Query: 135 SVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEF-------- 186
+WH +L + YGY+ G + P EGH ++ + +L+DP A+ + GEF
Sbjct: 55 DIWHGYLPDARPGLRYGYRVHGPWQPAEGHRFNPAKLLIDPCARQI--DGEFKDNPLLHA 112
Query: 187 GSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTKF 245
G PD +A D +DWE D P + P IIYE HV+G T H +
Sbjct: 113 GHNEPDYRDNAAIAPKCVVVVDHYDWEDDAPPRTPWGSTIIYEAHVKGLTYLHPEIPVEI 172
Query: 246 PGTY--LGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYF 303
GTY LG +++LK+LG+ +ELLP +F S +Q N+WGY+ V F
Sbjct: 173 RGTYKALGHPVMINYLKQLGITALELLPVAQFA-----SEPRLQRMGLSNYWGYNPVAMF 227
Query: 304 SPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGV 363
+ Y+ + ++E + IK HK GIEVI+D+V NH+ E + +GP+ S RG+
Sbjct: 228 ALHPAYACSP-----ETALHEFRDAIKALHKAGIEVILDIVLNHSAELDLDGPLFSLRGI 282
Query: 364 DNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMT 423
DN YY I G+++N++GCGNTLN +HP V + CLRYWV HVDGFRFDLA++M
Sbjct: 283 DNRSYYWIREDGDYHNWTGCGNTLNLSHPAVVDYASACLRYWVETCHVDGFRFDLAAVMG 342
Query: 424 RSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWD-AGGLYQ 482
R+ PL I N P+L VKLIAE WD A G YQ
Sbjct: 343 RTPEFRQDA---------------------PLFTAIQNCPVLSQVKLIAEPWDIAPGGYQ 381
Query: 483 VGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFV 542
VG FP +++EWN +RD R+F D GAFA S +V++ GR P +IN V
Sbjct: 382 VGNFPP--LFAEWNDHFRDAARRFWLHYDLPLGAFAGRFAASSDVFKRNGRLPSAAINLV 439
Query: 543 CTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNF 602
HDGFTL D V +N+KHN NGE+N DG N+N S N G+EG + + + R+ +
Sbjct: 440 TAHDGFTLRDCVCFNHKHNEANGEENRDGTNNNYSNNHGKEGVGGTLDLVERRRDSIHAL 499
Query: 603 FLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTK 662
+L++SQG PM+ GDE+GH++ GNNN YC DN L + W ++SS F +
Sbjct: 500 LTTLLLSQGTPMLLAGDEHGHSQHGNNNAYCQDNQLTWLDW---SQASSGLTAFTAALIH 556
Query: 663 FRYECESL 670
R +L
Sbjct: 557 LRKRIPAL 564
>GLGX_ECOL6 (Q8FCR8) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 657
Score = 377 bits (967), Expect = e-104
Identities = 227/608 (37%), Positives = 317/608 (51%), Gaps = 58/608 (9%)
Query: 75 QVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTG 134
Q++ G P P GA GVNF ++S +A LC+F + Q Y L +G
Sbjct: 3 QLAIGKPTPLGAHYDGQGVNFTLFSAHAERVELCVFDAN----GQEHRY----DLPGHSG 54
Query: 135 SVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEF-------- 186
+WH +L + YGY+ G + P EGH ++ + +L+DP A+ + GEF
Sbjct: 55 DIWHGYLPDARPGLRYGYRVHGPWQPAEGHRFNPAKLLIDPCARQI--DGEFKDNPLLHA 112
Query: 187 GSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTKF 245
G PD +A D +DWE D P + P IIYE HV+G T H +
Sbjct: 113 GHNEPDYRDNASIAPKCVVVVDHYDWEDDAPPRTPWGSTIIYEAHVKGLTYLHPEIPVEI 172
Query: 246 PGTY--LGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYF 303
GTY LG +++LK+LG+ +ELLP +F S +Q N+WGY+ V F
Sbjct: 173 RGTYKALGHPVMINYLKQLGITALELLPVAQFA-----SEPRLQRMGLSNYWGYNPVAMF 227
Query: 304 SPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGV 363
+ Y+ + ++E + IK HK GIEVI+D+V NH+ E + +GP+ S RG+
Sbjct: 228 ALHPAYACSP-----ETALDEFRDAIKALHKAGIEVILDIVLNHSAELDLDGPLFSLRGI 282
Query: 364 DNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMT 423
DN YY I G+++N++GCGNTLN +HP V + CLRYWV HVDGFRFDLA++M
Sbjct: 283 DNRSYYWIREDGDYHNWTGCGNTLNLSHPAVVDYASACLRYWVETCHVDGFRFDLAAVMG 342
Query: 424 RSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWD-AGGLYQ 482
R+ PL I N P+L VKLIAE WD A G YQ
Sbjct: 343 RTPEFRQDA---------------------PLFTAIQNCPVLSQVKLIAEPWDIAPGGYQ 381
Query: 483 VGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFV 542
VG FP +++EWN +RD R+F D G FA S +V++ R P +IN V
Sbjct: 382 VGNFPP--LFAEWNDHFRDAARRFWLHYDLPLGVFAGRFAASSDVFKRNDRLPSAAINLV 439
Query: 543 CTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNF 602
HDGFTL D V +N+KHN NGE+N DG N+N S N G+EG + + + R+ +
Sbjct: 440 TAHDGFTLRDCVCFNHKHNEANGEENRDGTNNNYSNNHGKEGLGGTLDLVERRRDSIHAL 499
Query: 603 FLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTK 662
+L++SQG PM+ GDE+GH++ GNNN YC DN L + W ++SS F +
Sbjct: 500 LTTLLLSQGTPMLLAGDEHGHSQHGNNNAYCQDNQLTWLDW---SQASSGLTAFTAALIH 556
Query: 663 FRYECESL 670
R +L
Sbjct: 557 LRKRIPAL 564
>GLGX_YERPE (Q8ZA76) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 662
Score = 376 bits (966), Expect = e-103
Identities = 225/632 (35%), Positives = 326/632 (50%), Gaps = 78/632 (12%)
Query: 76 VSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTGS 135
++ G P P GA G+NF ++S +A TLCLF Q+ Q+ + +TG
Sbjct: 4 LTHGSPTPSGAHFDGKGINFTLFSAHAEQVTLCLFD-EQGQERQIA-------MPARTGD 55
Query: 136 VWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEFGSLGPDGNC 195
+WH +L G YGY+ G F P GH ++ +L+DP +A+ G +G D
Sbjct: 56 IWHGYLPGGKPGQRYGYRVSGPFEPSRGHRFNPHKLLIDPRTRALE-----GKVGDD--- 107
Query: 196 WPQMAGMVPSND---------------DEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HE 239
P+ G V D +E+DW+GD P P + +IYE HVRG T+ H
Sbjct: 108 -PRFTGGVSQPDVRDSAAALPKCLVIHEEYDWQGDKPPAIPWGNTVIYEAHVRGLTQLHP 166
Query: 240 SSKTKFPGTYLGVVEK--LDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGY 297
+ GTY + ++HLK LG+ +ELLP + +Q N+WGY
Sbjct: 167 DIPPELRGTYAALAHPALIEHLKTLGITTLELLPVQF-----HIDEPRLQKMGLSNYWGY 221
Query: 298 STVNYFSPMIRYSSAGIQNCGRDGIN---EVKFLIKEAHKRGIEVIMDVVFNHTVEGNEN 354
+ + F+ Y+S GR+GI+ E++ +K H GIEVI+DVVFNH+ E +
Sbjct: 222 NVLAPFAVDPDYAS------GREGISPLRELRDAVKALHNAGIEVILDVVFNHSAELDVF 275
Query: 355 GPIISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGF 414
GP + RG+DN+ YY + P GE+ N +GCGN L +HP V Q+++DCL YW HVDGF
Sbjct: 276 GPTLCQRGIDNASYYWLTPDGEYDNITGCGNALRLSHPYVTQWVIDCLNYWRDSCHVDGF 335
Query: 415 RFDLASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEA 474
RFDL +++ R+ + PL ++ D L KLIAE
Sbjct: 336 RFDLGTVLGRTPAFDQHA---------------------PLFAALAADERLSACKLIAEP 374
Query: 475 WDAG-GLYQVGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGR 533
WD G G YQ+G FP +SEWN +YRD +R F + G FA+ S +++ GR
Sbjct: 375 WDIGLGGYQLGNFPTG--FSEWNDQYRDAMRGFWLRGEVPRGTFAQHFAASSRLFEQRGR 432
Query: 534 KPWNSINFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKK 593
P SIN + HDGFTL DL+ +N KHN NGE+N DG ++N+S N G EG A +++ +
Sbjct: 433 LPSASINQITAHDGFTLLDLLCFNQKHNQMNGEENRDGSDNNHSNNFGCEGLVADAAIWQ 492
Query: 594 LRKRQMRNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDF 653
RK R +L++SQG PM+ GDE GH++ GNNN YC +N L + W + +
Sbjct: 493 RRKACQRALLTTLLLSQGTPMLLAGDEQGHSQQGNNNAYCQNNILTWLDWGSADRA---L 549
Query: 654 FRFCCLMTKFRYECESLGLDDFPTS--DRLQW 683
F + + R + +L D + S +QW
Sbjct: 550 MTFTADLIRLRQQIPALTQDQWWQSGDSNVQW 581
>GLGX_HAEIN (P45178) Glycogen operon protein glgX homolog (EC
3.2.1.-)
Length = 659
Score = 373 bits (957), Expect = e-102
Identities = 235/690 (34%), Positives = 349/690 (50%), Gaps = 84/690 (12%)
Query: 77 SRGYPAPFG-ATVRDGGV---NFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNK 132
+ G P P G + + V NFA++S A+ LCLF + NQ T M +
Sbjct: 6 NNGNPIPMGYSQAVENNVQITNFALFSAAAIGVELCLFD----EQNQETRLP-----MVR 56
Query: 133 TGSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEFGS---- 188
T +VWH+ + G Y ++ G+F+ + ++LDPYAKAV + + S
Sbjct: 57 TENVWHLAVTGVKTGTEYAFRIHGEFA-------NPQKLILDPYAKAVNGKPDLSSEESR 109
Query: 189 ---LGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTK 244
L D +A +EFDWE D P + I+YE+HV+GF++ +E
Sbjct: 110 SWFLLSDNRDNAHLAPRAVVISEEFDWENDTSPNTPWAETIVYELHVKGFSQLNEKIPAA 169
Query: 245 FPGTYLGVVE--KLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNY 302
GTY G+ L +LKELGV +ELLP + + +Q N+WGY+ +
Sbjct: 170 LRGTYTGLAHPVNLAYLKELGVTAVELLPVNFHINEPHLQARGLQ-----NYWGYNPLAM 224
Query: 303 FSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRG 362
F+ +Y++ + + E K ++K HK GIEVI+DVVFNH+ E + P S RG
Sbjct: 225 FAVEPKYAATN------NPLAEFKTMVKAFHKAGIEVILDVVFNHSAESEQTYPTFSQRG 278
Query: 363 VDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIM 422
+D+ YY +G + N++GCGN LN + V R+++VDCLRYWV + H+DGFRFDLA+++
Sbjct: 279 IDDQTYYWRNDQGRYINWTGCGNMLNLSSDVGRKWVVDCLRYWVEQCHIDGFRFDLATVL 338
Query: 423 TRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDAGGL-Y 481
R + +N S L I N+P L +KLIAE WD G Y
Sbjct: 339 GRDTPDFNS--------------------SAQLFTDIKNEPSLQNIKLIAEPWDIGHYGY 378
Query: 482 QVGTFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINF 541
QVG FP + ++EWN ++RD + +F G GAFAE GS ++++ R P ++NF
Sbjct: 379 QVGNFPSY--FAEWNDRFRDDLCRFWLWKSGEIGAFAERFAGSSDLFKKNDRLPHTTLNF 436
Query: 542 VCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFAS------SSVKKLR 595
+ HDGFTL DLV+YN KHN NGE+N DG N N S+N G EG S S+V+ R
Sbjct: 437 ITAHDGFTLKDLVSYNQKHNETNGEENRDGRNENYSYNHGVEGSTESLSEPQKSAVENNR 496
Query: 596 KRQMRNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFR 655
+SL+++ G PM+ GDE+G+T+ GNNN YC DN + + +W + + F
Sbjct: 497 TFAQSGLLMSLLLANGTPMLLAGDEFGNTQYGNNNAYCQDNEITWLKW---ANFNEELFE 553
Query: 656 FCCLMTKFRYECESLGLDDFPTSDRLQWHGHFPGKP----DWSETSRFVAFTMVDSVKRE 711
R + SL D + + + +QW + G+P DW ++D+
Sbjct: 554 LTKQTIALRKQIGSLNKDQWWSDENVQWL-NIVGEPMTVEDWQNQQTKALQVVLDN---R 609
Query: 712 IYIAFNTSHLPVTITLPERPGYRWEPLVDT 741
+ N LP R +W+P + T
Sbjct: 610 WLLLINAKAEGQMFHLPNR---KWKPQIGT 636
>GLGX_SALTY (Q8ZLG6) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 658
Score = 372 bits (956), Expect = e-102
Identities = 223/606 (36%), Positives = 320/606 (52%), Gaps = 54/606 (8%)
Query: 75 QVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTG 134
Q++ G P GAT GVNF ++S +A LC+F D + N+ Y L + G
Sbjct: 3 QLAIGEATPHGATYDGHGVNFTLFSAHAERVELCVF---DSRGNE-RRY----DLPGRRG 54
Query: 135 SVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEFGSL---GP 191
VWH +L G + YGY+ G + P +GH ++ + +LLDPYA+ V + L G
Sbjct: 55 DVWHGYLAGARPGLRYGYRVHGPWQPAQGHRFNPAKLLLDPYARRVEGELKDHPLLHGGH 114
Query: 192 DGNCWPQMAGMVPSN---DDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTKFPG 247
D + A + P + D +DWE D + P +IYE HV+G T H + G
Sbjct: 115 DEPDYRDNAAVAPKSVVISDHYDWEDDAAPRTPWGKTVIYEAHVKGLTYLHPELPQEIRG 174
Query: 248 TY--LGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYFSP 305
TY LG + + K+LG+ +ELLP +F S +Q N+WGY+ + F+
Sbjct: 175 TYKALGHPVMVAYFKQLGITALELLPVAQFA-----SEPRLQRMGLTNYWGYNPMAMFAL 229
Query: 306 MIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGVDN 365
++S+ ++E + +K H+ GIEVI+D+V NH+ E + +GP S RG+DN
Sbjct: 230 HPAWASSP-----ETALDEFRDAVKALHRAGIEVILDIVLNHSAELDLDGPTFSLRGIDN 284
Query: 366 SIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRS 425
YY I G+++N++GCGNTLN +HP V ++ +CLRYWV HVDGFRFDLAS+M R
Sbjct: 285 RSYYWIRDDGDYHNWTGCGNTLNLSHPGVVEYACECLRYWVETCHVDGFRFDLASVMGR- 343
Query: 426 SSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDAG-GLYQVG 484
T T PL I P+L VKLIAE WD G G YQVG
Sbjct: 344 --------------------TPTFRQDAPLFAAIKACPVLSTVKLIAEPWDIGEGGYQVG 383
Query: 485 TFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFVCT 544
FP ++EWN +RD R+F + G FA S +V++ GR P S+N +
Sbjct: 384 NFPP--PFAEWNDHFRDAARRFWLPRNLTTGEFACRFAASSDVFKRNGRAPGASVNLLTA 441
Query: 545 HDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFL 604
HDGFTL D V +N KHN NGE+N DG N N S N G+EG + + R+ +
Sbjct: 442 HDGFTLRDCVCFNQKHNEANGEENRDGTNSNYSDNHGKEGLGGPLDLMERRRDSIHALLA 501
Query: 605 SLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTKFR 664
+L++SQG PM+ GDE+GH++ GNNN YC DN L + W ++++ F + + R
Sbjct: 502 TLLLSQGTPMLLAGDEHGHSQHGNNNAYCQDNALTWLDW---QQANRGLTTFTAALIRLR 558
Query: 665 YECESL 670
+ +L
Sbjct: 559 QQIPAL 564
>GLGX_SALTI (Q8Z234) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 654
Score = 371 bits (953), Expect = e-102
Identities = 222/606 (36%), Positives = 320/606 (52%), Gaps = 54/606 (8%)
Query: 75 QVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTG 134
Q++ G P GAT GVNF ++S +A LC+F D + N+ Y L + G
Sbjct: 3 QLAIGEATPHGATYDGHGVNFTLFSAHAERVELCVF---DSRGNE-RRY----DLPGRRG 54
Query: 135 SVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEFGSL---GP 191
VWH +L G + YGY+ G + P +GH ++ + +LLDPYA+ V + L G
Sbjct: 55 DVWHGYLAGARPGLRYGYRVHGPWQPAQGHRFNPAKLLLDPYARQVEGELKDHPLLHGGH 114
Query: 192 DGNCWPQMAGMVPSN---DDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTKFPG 247
D + A + P + D +DWE D + P +IYE HV+G T H + G
Sbjct: 115 DEPDYRDNAAVAPKSVVISDHYDWEDDAAPRTPWGKTVIYEAHVKGLTYLHPELPQEIRG 174
Query: 248 TY--LGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYFSP 305
TY LG + + K+LG+ +ELLP +F S +Q N+WGY+ + F+
Sbjct: 175 TYKALGHPVMVAYFKQLGITALELLPVAQFA-----SEPRLQRMGLTNYWGYNPMAMFAL 229
Query: 306 MIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGVDN 365
++S+ ++E + +K H+ GIEVI+D+V NH+ E + +GP S RG+DN
Sbjct: 230 HPAWASSP-----ETALDEFRDAVKALHRAGIEVILDIVLNHSAELDLDGPTFSLRGIDN 284
Query: 366 SIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRS 425
YY I G+++N++GCGNTLN +HP V ++ +CLRYW+ HVDGFRFDLAS+M R
Sbjct: 285 RSYYWIRDDGDYHNWTGCGNTLNLSHPGVVEYACECLRYWMETCHVDGFRFDLASVMGR- 343
Query: 426 SSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDAG-GLYQVG 484
T T PL I P+L VKLIAE WD G G YQVG
Sbjct: 344 --------------------TPTFRQDAPLFAAIKACPVLSTVKLIAEPWDIGEGGYQVG 383
Query: 485 TFPHWGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFVCT 544
FP ++EWN +RD R+F + G FA S +V++ GR P S+N +
Sbjct: 384 NFPP--PFAEWNDHFRDAARRFWLPRNLTTGEFACRFAASSDVFKRNGRAPGASVNLLTA 441
Query: 545 HDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFL 604
HDGFTL D V +N KHN NGE+N DG N N S N G+EG + + R+ +
Sbjct: 442 HDGFTLRDCVCFNQKHNEANGEENRDGTNSNYSDNHGKEGLGGPLDLMERRRDSIHALLA 501
Query: 605 SLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTKFR 664
+L++SQG PM+ GDE+GH++ GNNN YC DN L + W ++++ F + + R
Sbjct: 502 TLLLSQGTPMLLAGDEHGHSQHGNNNAYCQDNALTWLDW---QQANRGLTTFTAALIRLR 558
Query: 665 YECESL 670
+ +L
Sbjct: 559 QQIPAL 564
>GLGX_ERWCH (Q8KR69) Glycogen debranching enzyme (EC 3.2.1.-)
(Glycogen operon protein glgX)
Length = 657
Score = 365 bits (937), Expect = e-100
Identities = 203/581 (34%), Positives = 309/581 (52%), Gaps = 53/581 (9%)
Query: 79 GYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTGSVWH 138
G P P G+ GVNFA++S A LC+F ++ ++ PL +TG +WH
Sbjct: 7 GRPRPLGSHFDGEGVNFALFSSGASRVELCIF--DGLREQRL-------PLTARTGDIWH 57
Query: 139 VFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVIS------RGEFGSLGPD 192
+L + YGY+ DG F P G ++++ +LLDP A+ + R G PD
Sbjct: 58 GYLPDAQPGLCYGYRVDGAFDPSRGQRFNANKLLLDPCARQMDGWVVDDQRLHGGYHQPD 117
Query: 193 GNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRGFTK-HESSKTKFPGTYLG 251
+ ++ D+ +DW+GD + P ++YE HVRG T+ H GTY
Sbjct: 118 PSDSAEVMPPSVVVDEHYDWQGDRLPRTPWSQTVLYEAHVRGLTRRHPGIPAAIRGTYAA 177
Query: 252 VVEK--LDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYFSPMIRY 309
+ LD+L +LGV +EL+P + + ++ N+WGY+T+ F+
Sbjct: 178 LAHPVMLDYLTQLGVTALELMPVQQHADEPRLQSMGLR-----NYWGYNTLLPFAVDNSL 232
Query: 310 SSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIISFRGVDNSIYY 369
+++ D +NE + ++ H+ GIEVI+DVVFNH+ E + +GP ++ RG+DN+ YY
Sbjct: 233 AASD------DPLNEFRDTVRALHQAGIEVILDVVFNHSAELDVDGPTLTLRGIDNASYY 286
Query: 370 MIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSLW 429
+ G+++N++GCGN L HP V ++++CL +W HVDGFRFDLA+I+ R
Sbjct: 287 WLTENGDYHNWAGCGNVLRLEHPAVLHWVIECLTFWHEVCHVDGFRFDLATILGRLPDFS 346
Query: 430 NGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDAG-GLYQVGTFPH 488
+ S P + N L KLIAE WD G YQ+G FP
Sbjct: 347 S---------------------SAPFFTALRNHRSLRDCKLIAEPWDISPGGYQLGQFP- 384
Query: 489 WGIWSEWNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFVCTHDGF 548
++EWN ++RD +R+F D G A S V++ G R+PW S+N + +HDGF
Sbjct: 385 -APFAEWNDRFRDDMRRFWLHGDLPIGVLARRFAASSEVFERGSRQPWASVNMLTSHDGF 443
Query: 549 TLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMV 608
TL DLV +N+KHN NGE N DG N N S+N G EG A + + R+ + +L++
Sbjct: 444 TLRDLVCFNHKHNDANGEQNRDGTNSNFSFNHGTEGLEADETTQARRRVSQQALLTTLLL 503
Query: 609 SQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEES 649
SQG PM+ GDE+G+++ GNNN YC DN L + WD +++
Sbjct: 504 SQGTPMLLAGDEFGNSQQGNNNAYCQDNALAWLHWDQADDA 544
>ISOA_PSEUM (P26501) Isoamylase precursor (EC 3.2.1.68)
Length = 776
Score = 270 bits (690), Expect = 1e-71
Identities = 226/726 (31%), Positives = 324/726 (44%), Gaps = 101/726 (13%)
Query: 89 RDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTGSVWHVFLK------ 142
+ + F +YS A L L++ T TL P + VW V +
Sbjct: 40 QQANITFRVYSSQATRIVLYLYSAGYGVQESAT--YTLSPAGS---GVWAVTVPVSSIKA 94
Query: 143 -GDFGDMLYGYKFDGKFSPI------------------EGHYYDSSLILLDPYAKAVISR 183
G G + YGY+ G P G ++ + +LLDPYA+ V S+
Sbjct: 95 AGITGAVYYGYRAWGPNWPYASNWGKGSQAGFVSDVDANGDRFNPNKLLLDPYAQEV-SQ 153
Query: 184 GEFGSLGPDGNCW---------------PQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIY 228
+GN + P+ +VPS G P + QKD +IY
Sbjct: 154 DPLNPSNQNGNVFASGASYRTTDSGIYAPKGVVLVPSTQST----GTKPTR-AQKDDVIY 208
Query: 229 EMHVRGFTKHESS-KTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEF-NELEYFSYNSV 286
E+HVRGFT+ ++S ++ GTY G K +L LGV +E LP E N+ NS
Sbjct: 209 EVHVRGFTEQDTSIPAQYRGTYYGAGLKASYLASLGVTAVEFLPVQETQNDANDVVPNS- 267
Query: 287 QGDYRVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFN 346
D N+WGY T NYFSP RY+ E + +++ H GI+V MDVV+N
Sbjct: 268 --DANQNYWGYMTENYFSPDRRYA---YNKAAGGPTAEFQAMVQAFHNAGIKVYMDVVYN 322
Query: 347 HTVEGNE-------NGPIISFRGVDNSIYYMIAPKGE-FYNYSGCGNTLNCNHPVVRQFI 398
HT EG I S+RG+DN+ YY + + FY+ +G G N + V + I
Sbjct: 323 HTAEGGTWTSSDPTTATIYSWRGLDNTTYYELTSGNQYFYDNTGIGANFNTYNTVAQNLI 382
Query: 399 VDCLRYWVTEMHVDGFRFDLASIMTRSSSLWNGV------NVFGTSIEGDLLATGTPLVS 452
VD L YW M VDGFRFDLAS++ +S NG N D A + +
Sbjct: 383 VDSLAYWANTMGVDGFRFDLASVL--GNSCLNGAYTASAPNCPNGGYNFD--AADSNVAI 438
Query: 453 PPLIDLISNDPIL--HGVKLIAEAWDAGG-LYQVGTFPHWGIWSEWNGKYRDTVRQFVKG 509
++ + P G+ L AE W GG YQ+G FP WSEWNG +RD++RQ
Sbjct: 439 NRILREFTVRPAAGGSGLDLFAEPWAIGGNSYQLGGFPQG--WSEWNGLFRDSLRQAQNE 496
Query: 510 TDG---FAGAFAECLCGSPNVYQGGGRKPWNSINFVCTHDGFTLADLVTYNNKHN---LP 563
+ A GS N++Q GR PWNSINF+ HDG TL D+ + N +N P
Sbjct: 497 LGSMTIYVTQDANDFSGSSNLFQSSGRSPWNSINFIDVHDGMTLKDVYSCNGANNSQAWP 556
Query: 564 NGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMVSQGVPMIYMGDEYGH 623
G ++ G + N SW+ +G A + ++R R M+S G P++ GDEY
Sbjct: 557 YG-PSDGGTSTNYSWD---QGMSAGTGAAVDQRRAARTGMAFEMLSAGTPLMQGGDEYLR 612
Query: 624 TKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTKFRYECESLGLDDFPTSDRLQW 683
T NNN Y D+ N+ + + S+F+ F + FR +L + + +L W
Sbjct: 613 TLQCNNNAYNLDSSANWLTYSWTTD-QSNFYTFAQRLIAFRKAHPALRPSSWYSGSQLTW 671
Query: 684 H---GHFPGKPDWSETSRFVAFTMVDSV----KREIYIAFNTSHLPVTITLPERP-GYRW 735
+ G W+ TS + ++ IY+A+N VT TLP P G +W
Sbjct: 672 YQPSGAVADSNYWNNTSNYAIAYAINGPSLGDSNSIYVAYNGWSSSVTFTLPAPPSGTQW 731
Query: 736 EPLVDT 741
+ DT
Sbjct: 732 YRVTDT 737
>ISOA_PSEAY (P10342) Isoamylase precursor (EC 3.2.1.68)
Length = 776
Score = 270 bits (690), Expect = 1e-71
Identities = 226/726 (31%), Positives = 324/726 (44%), Gaps = 101/726 (13%)
Query: 89 RDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTGSVWHVFLK------ 142
+ + F +YS A L L++ T TL P + VW V +
Sbjct: 40 QQANITFRVYSSQATRIVLYLYSAGYGVQESAT--YTLSPAGS---GVWAVTVPVSSIKA 94
Query: 143 -GDFGDMLYGYKFDGKFSPI------------------EGHYYDSSLILLDPYAKAVISR 183
G G + YGY+ G P G ++ + +LLDPYA+ V S+
Sbjct: 95 AGITGAVYYGYRAWGPNWPYASNWGKGSQAGFVSDVDANGDRFNPNKLLLDPYAQEV-SQ 153
Query: 184 GEFGSLGPDGNCW---------------PQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIY 228
+GN + P+ +VPS G P + QKD +IY
Sbjct: 154 DPLNPSNQNGNVFASGASYRTTDSGIYAPKGVVLVPSTQST----GTKPTR-AQKDDVIY 208
Query: 229 EMHVRGFTKHESS-KTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEF-NELEYFSYNSV 286
E+HVRGFT+ ++S ++ GTY G K +L LGV +E LP E N+ NS
Sbjct: 209 EVHVRGFTEQDTSIPAQYRGTYYGAGLKASYLASLGVTAVEFLPVQETQNDANDVVPNS- 267
Query: 287 QGDYRVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFN 346
D N+WGY T NYFSP RY+ E + +++ H GI+V MDVV+N
Sbjct: 268 --DANQNYWGYMTENYFSPDRRYA---YNKAAGGPTAEFQAMVQAFHNAGIKVYMDVVYN 322
Query: 347 HTVEGNE-------NGPIISFRGVDNSIYYMIAPKGE-FYNYSGCGNTLNCNHPVVRQFI 398
HT EG I S+RG+DN+ YY + + FY+ +G G N + V + I
Sbjct: 323 HTAEGGTWTSSDPTTATIYSWRGLDNATYYELTSGNQYFYDNTGIGANFNTYNTVAQNLI 382
Query: 399 VDCLRYWVTEMHVDGFRFDLASIMTRSSSLWNGV------NVFGTSIEGDLLATGTPLVS 452
VD L YW M VDGFRFDLAS++ +S NG N D A + +
Sbjct: 383 VDSLAYWANTMGVDGFRFDLASVL--GNSCLNGAYTASAPNCPNGGYNFD--AADSNVAI 438
Query: 453 PPLIDLISNDPIL--HGVKLIAEAWDAGG-LYQVGTFPHWGIWSEWNGKYRDTVRQFVKG 509
++ + P G+ L AE W GG YQ+G FP WSEWNG +RD++RQ
Sbjct: 439 NRILREFTVRPAAGGSGLDLFAEPWAIGGNSYQLGGFPQG--WSEWNGLFRDSLRQAQNE 496
Query: 510 TDG---FAGAFAECLCGSPNVYQGGGRKPWNSINFVCTHDGFTLADLVTYNNKHN---LP 563
+ A GS N++Q GR PWNSINF+ HDG TL D+ + N +N P
Sbjct: 497 LGSMTIYVTQDANDFSGSSNLFQSSGRSPWNSINFIDVHDGMTLKDVYSCNGANNSQAWP 556
Query: 564 NGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMVSQGVPMIYMGDEYGH 623
G ++ G + N SW+ +G A + ++R R M+S G P++ GDEY
Sbjct: 557 YG-PSDGGTSTNYSWD---QGMSAGTGAAVDQRRAARTGMAFEMLSAGTPLMQGGDEYLR 612
Query: 624 TKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTKFRYECESLGLDDFPTSDRLQW 683
T NNN Y D+ N+ + + S+F+ F + FR +L + + +L W
Sbjct: 613 TLQCNNNAYNLDSSANWLTYSWTTD-QSNFYTFAQRLIAFRKAHPALRPSSWYSGSQLTW 671
Query: 684 H---GHFPGKPDWSETSRFVAFTMVDSV----KREIYIAFNTSHLPVTITLPERP-GYRW 735
+ G W+ TS + ++ IY+A+N VT TLP P G +W
Sbjct: 672 YQPSGAVADSNYWNNTSNYAIAYAINGPSLGDSNSIYVAYNGWSSSVTFTLPAPPSGTQW 731
Query: 736 EPLVDT 741
+ DT
Sbjct: 732 YRVTDT 737
>ISOA_FLASP (O32611) Isoamylase precursor (EC 3.2.1.68)
Length = 777
Score = 270 bits (690), Expect = 1e-71
Identities = 242/760 (31%), Positives = 338/760 (43%), Gaps = 138/760 (18%)
Query: 65 VDKPQLGGRFQVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYI 124
+D QLG R+ ++ A F +YS A + L+ +
Sbjct: 34 IDAQQLGARYDAAQANLA------------FRVYSSRATRVEVFLYKNPTGSQEVARLAL 81
Query: 125 TLDPLMNKTGSVWHVFLK--------GDFGDMLYGYKFDGKFSPIE-------------- 162
+ DP VW + L G G + YGY+ G P +
Sbjct: 82 SKDPATQ----VWSLSLPTSTIKNTYGITGAVYYGYRAWGPNWPYDAAWTKGSATGFVSD 137
Query: 163 ----GHYYDSSLILLDPYAKAVISRGEFGSLGPDGNCWPQMAGMVPSNDD---------- 208
G+ ++ + +LLDPYA+ IS+ + DG + G N D
Sbjct: 138 VDNAGNRFNPNKLLLDPYARE-ISQDPNTATCADGTIYA--TGAAHRNKDSGLCASKGIA 194
Query: 209 ---EFDWEGDLPLKYPQKDLIIYEMHVRGFTKHESSKTKFP-GTYLGVVEKLDHLKELGV 264
+ G P + KD +IYE+HVRG T+++ S GTY G K L LGV
Sbjct: 195 LAADATSVGSKPTR-ALKDEVIYEVHVRGLTRNDDSVPAAERGTYKGAARKAAALAALGV 253
Query: 265 NCIELLPCHEF-NELEYFSYNSVQGDYRVNFWGYSTVNYFSPMIRYS---SAGIQNCGRD 320
+E LP E N+ NS GD N+WGY T+NYF+P RY+ SAG
Sbjct: 254 TAVEFLPVQETQNDQNDVDPNSTAGD---NYWGYMTLNYFAPDRRYAYDKSAG------G 304
Query: 321 GINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGP-----------IISFRGVDNSIYY 369
E K ++K H GI+V +DVV+NHT EG GP ++SFRG+DN YY
Sbjct: 305 PTREWKAMVKAFHDAGIKVYIDVVYNHTGEG---GPWSGTDGLSVYNLLSFRGLDNPAYY 361
Query: 370 MIAPKGEF-YNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSL 428
++ ++ ++ +G G N HP+ + IVD L YW + VDGFRFDLAS++ +S
Sbjct: 362 SLSSDYKYPWDNTGVGGNYNTRHPIAQNLIVDSLAYWRDALGVDGFRFDLASVL--GNSC 419
Query: 429 WNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGV--KLIAEAWDAGG-LYQVGT 485
+G F + G+ L + P P G LIAE W GG YQVG
Sbjct: 420 QHGCFNFDKNDSGNALNRIVAELPPR--------PAAGGAGADLIAEPWAIGGNSYQVGG 471
Query: 486 FPHWGIWSEWNGKYRDTVR--QFVKGTDGFA-GAFAECLCGSPNVYQGGGRKPWNSINFV 542
FP W+EWNG YRD +R Q G + G A GS ++Y GRKPW+SINFV
Sbjct: 472 FPAG--WAEWNGLYRDALRKKQNKLGVETVTPGTLATRFAGSNDLYGDDGRKPWHSINFV 529
Query: 543 CTHDGFTLADLVTYNNKHN---LPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQM 599
HDGFTL DL YN+K N P G ++ GE+HN SWN G + +++
Sbjct: 530 VAHDGFTLNDLYAYNDKQNNQPWPYG-PSDGGEDHNLSWNQG--------GIVAEQRKAA 580
Query: 600 RNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCL 659
R LM+S GVPMI GDE T+ GNNNTY D+ N+ W + +D +
Sbjct: 581 RTGLALLMLSAGVPMITGGDEALRTQFGNNNTYNLDSAANWLYWS-RSALEADHETYTKR 639
Query: 660 MTKFRYECESLGLDDFPTS--------DRLQW---------HGHFPGKPDWSETSRFVAF 702
+ FR +L +F ++ ++L+W +F G + + R
Sbjct: 640 LIAFRKAHPALRPANFYSASDTNGNVMEQLRWFKPDGAQADSAYFNGADNHALAWRIDGS 699
Query: 703 TMVDSVKREIYIAFNTSHLPVTITLP-ERPGYRWEPLVDT 741
DS IY+A+N V LP G +W + DT
Sbjct: 700 EFGDSAS-AIYVAYNGWSGAVDFKLPWPGTGKQWYRVTDT 738
>PULA_THEMA (O33840) Pullulanase precursor (EC 3.2.1.41)
(Alpha-dextrin endo-1,6-alpha-glucosidase) (Pullulan
6-glucanohydrolase)
Length = 843
Score = 167 bits (423), Expect = 1e-40
Identities = 179/664 (26%), Positives = 280/664 (41%), Gaps = 130/664 (19%)
Query: 115 FQDNQVTEYITLDPLMNKTGSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLD 174
F++ + TE + + K VW ++GD + Y Y+ + Y +D
Sbjct: 251 FKNGEDTEPYQVVNMEYKGNGVWEAVVEGDLDGVFYLYQLEN---------YGKIRTTVD 301
Query: 175 PYAKAVISRGEFGSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYP-QKDLIIYEMHVR 233
PY+KAV + + ++ + +N + WE D K +D IIYE+H+
Sbjct: 302 PYSKAVYANSKKSAV----------VNLARTNPE--GWENDRGPKIEGYEDAIIYEIHIA 349
Query: 234 GFTKHESSKTKFPGTYLGVVEK-----------LDHLKELGVNCIELLPCHEFNELEYFS 282
T E+S K G YLG+ E+ L HL ELGV + +LP +F ++
Sbjct: 350 DITGLENSGVKNKGLYLGLTEENTKGPGGVTTGLSHLVELGVTHVHILPFFDF-----YT 404
Query: 283 YNSVQGDYRVNF-WGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIM 341
+ + D+ + WGY + P RYS+ R I EVK ++K HK GI VIM
Sbjct: 405 GDELDKDFEKYYNWGYDPYLFMVPEGRYSTDPKNPHTR--IREVKEMVKALHKHGIGVIM 462
Query: 342 DVVFNHTVEGNENGPIISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDC 401
D+VF HT G + +F +Y I G + N SGCGN + P++R+FIVD
Sbjct: 463 DMVFPHTYG---IGELSAFDQTVPYYFYRIDKTGAYLNESGCGNVIASERPMMRKFIVDT 519
Query: 402 LRYWVTEMHVDGFRFDLASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISN 461
+ YWV E H+DGFRFD ++ + + L +E L + P +I
Sbjct: 520 VTYWVKEYHIDGFRFDQMGLIDKKTML---------EVERAL-----HKIDPTII----- 560
Query: 462 DPILHGVKLIAEAWDA-GGLYQVGTFPHWGIW-SEWNGKYRDTVR---------QFVKGT 510
L E W G + G G + +N ++RD +R FV G
Sbjct: 561 --------LYGEPWGGWGAPIRFGKSDVAGTHVAAFNDEFRDAIRGSVFNPSVKGFVMGG 612
Query: 511 DGFAGAFAECLCGSPNVYQGGGRK-----PWNSINFVCTHDGFTLADLVTYNNKHNLPNG 565
G + GS N Y G K P +IN+ HD TL D K+ L
Sbjct: 613 YGKETKIKRGVVGSIN-YDGKLIKSFALDPEETINYAACHDNHTLWD------KNYLAAK 665
Query: 566 EDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMVSQGVPMIYMGDEYGHTK 625
D ++ E+ +K +K L+ SQGVP ++ G ++ TK
Sbjct: 666 AD--------------KKKEWTEEELKNAQKLA----GAILLTSQGVPFLHGGQDFCRTK 707
Query: 626 GGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTKFRYECESLGLDDFPTSDRLQWHG 685
N+N+Y +N F ++ K + D F + + K R E + L + ++ ++ H
Sbjct: 708 NFNDNSYNAPISINGFDYERKLQ-FIDVFNYHKGLIKLRKEHPAFRLKN---AEEIKKHL 763
Query: 686 HF-PGKPDWSETSRFVAFTMVDSVK----REIYIAFNTSHLPVTITLPERPGYRWEPLVD 740
F PG R VAF + D ++I + +N + T LPE +W +V+
Sbjct: 764 EFLPG------GRRIVAFMLKDHAGGDPWKDIVVIYNGNLEKTTYKLPEG---KWNVVVN 814
Query: 741 TGKS 744
+ K+
Sbjct: 815 SQKA 818
>TREZ_ARTRM (Q9AJN6) Malto-oligosyltrehalose trehalohydrolase (EC
3.2.1.141) (MTHase)
(4-alpha-D-{(1->4)-alpha-D-glucano}trehalose
trehalohydrolase) (Maltooligosyl trehalose
trehalohydrolase)
Length = 575
Score = 80.9 bits (198), Expect = 1e-14
Identities = 93/323 (28%), Positives = 131/323 (39%), Gaps = 82/323 (25%)
Query: 226 IIYEMHVRGFTKHESSKTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNS 285
+IYE+HV FT GT + +LDHL LGV+ +ELLP + FN
Sbjct: 101 VIYELHVGTFTPE--------GTLDSAIRRLDHLVRLGVDAVELLPVNAFN--------- 143
Query: 286 VQGDYRVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVF 345
+ WGY V +++ Y G + + H RG+ V+ DVV+
Sbjct: 144 -----GTHGWGYDGVLWYAVHEPYG----------GPEAYQRFVDACHARGLAVVQDVVY 188
Query: 346 NHT-VEGN---ENGPIISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPV---VRQFI 398
NH GN + GP + G N+ G+ LN + P+ VR++I
Sbjct: 189 NHLGPSGNHLPDFGPYLG-SGAANT----------------WGDALNLDGPLSDEVRRYI 231
Query: 399 VDCLRYWVTEMHVDGFRFDLASIM--TRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLI 456
+D YW+ +MH DG R D + R+ L + + G+L G PL
Sbjct: 232 IDNAVYWLRDMHADGLRLDAVHALRDARALHLLEELAARVDELAGEL---GRPLTLIAES 288
Query: 457 DLISNDPIL------HGVKLIAEAWDAGGLYQVGTFPHWGIWSEWNGKYRD------TVR 504
DL NDP L HG L A+ WD + V H + E G Y D V+
Sbjct: 289 DL--NDPKLIRSRAAHGYGLDAQ-WDDDVHHAV----HANVTGETVGYYADFGGLGALVK 341
Query: 505 QFVKG--TDGFAGAFAECLCGSP 525
F +G DG +F E G P
Sbjct: 342 VFQRGWFHDGTWSSFRERHHGRP 364
>GLGB_RHOMR (Q93HU3) 1,4-alpha-glucan branching enzyme (EC 2.4.1.18)
(Glycogen branching enzyme) (BE)
(1,4-alpha-D-glucan:1,4-alpha-D-glucan
6-glucosyl-transferase)
Length = 621
Score = 79.3 bits (194), Expect = 4e-14
Identities = 85/320 (26%), Positives = 141/320 (43%), Gaps = 62/320 (19%)
Query: 127 DPLMNKTGSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLDPYAKAVISRGEF 186
+PL G +W ++ G Y Y+ I +Y + DPYA F
Sbjct: 59 NPLERYGGGLWAGYVPGARPGHTYKYR-------IRHGFYQADKT--DPYA--------F 101
Query: 187 GSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQ------KDLIIYEMHVRGFTKHES 240
P G+ +A ++ D + W D ++ + + + IYE+H+ G +H+
Sbjct: 102 AMEPPTGSPIEGLASIITRLD--YTWHDDEWMRRRKGPASLYEPVSIYEVHL-GSWRHKR 158
Query: 241 SKTKFPGTYLGVVEKL-DHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNFWGYST 299
F +Y + E L D+++E+G +ELLP +E+ Y S WGY
Sbjct: 159 PGESF--SYREIAEPLADYVQEMGFTHVELLPV-----MEHPYYGS---------WGYQV 202
Query: 300 VNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNENGPIIS 359
V Y++P RY S ++ +LI H+RGI VI+D V +H + ++
Sbjct: 203 VGYYAPTFRYGSP----------QDLMYLIDYLHQRGIGVILDWVPSHFAADPQG--LVF 250
Query: 360 FRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFD-L 418
F G +++ PK ++ G + N P VR F++ +W+ + HVDG R D +
Sbjct: 251 FDGT--TLFEYDDPKMRYHPDWGT-YVFDYNKPGVRNFLISNALFWLEKYHVDGLRVDAV 307
Query: 419 ASIMTR--SSSLWNGVNVFG 436
AS++ R S W N+FG
Sbjct: 308 ASMLYRDYSRKEWT-PNIFG 326
>TREZ_BREHE (O52520) Malto-oligosyltrehalose trehalohydrolase (EC
3.2.1.141) (MTHase)
(4-alpha-D-{(1->4)-alpha-D-glucano}trehalose
trehalohydrolase) (Maltooligosyl trehalose
trehalohydrolase)
Length = 589
Score = 77.4 bits (189), Expect = 1e-13
Identities = 105/411 (25%), Positives = 162/411 (38%), Gaps = 99/411 (24%)
Query: 226 IIYEMHVRGFTKHESSKTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNS 285
+IY++HV T GT EKL +L +LG++ IELLP + FN
Sbjct: 109 LIYQLHVGTSTPD--------GTLDAAGEKLSYLVDLGIDFIELLPVNGFN--------- 151
Query: 286 VQGDYRVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVF 345
+ WGY V +++ + G G + + AH G+ VI DVV+
Sbjct: 152 -----GTHNWGYDGVQWYT----------VHEGYGGPAAYQRFVDAAHAAGLGVIQDVVY 196
Query: 346 NHT-VEGN---ENGPIISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDC 401
NH + GN + GP + +G N++ G+ N G G+ V R++I+D
Sbjct: 197 NHLGLRGNYFPKLGPNLK-QGDANTL-------GDSVNLDGAGS------DVFREYILDN 242
Query: 402 LRYWVTEMHVDGFRFDLASIM--TRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLI 459
WV + HVDG FD + R+ + + G +I G+ TG P D
Sbjct: 243 AALWVGDYHVDGVGFDAVHAVRDERAVHILEDLGALGDAISGE---TGLPKTLIAESDF- 298
Query: 460 SNDPILHGVKLIAEAWDAGGLYQVGTFPHWGIWSEWNGKYRDTVRQFVKG-TDGFAGAFA 518
N+P L + D G +G+ +W+ + V V G T G+ F
Sbjct: 299 -NNPRLIYPR------DVNG---------YGLAGQWSDDFHTAVHVSVSGETTGYYSDF- 341
Query: 519 ECLCGSPNVYQGGGRKPWNSINFVCTHDGFTL-------ADLVTYNNKHNLPNGEDNNDG 571
E L V + G + +F H G + A LV N H+ D
Sbjct: 342 ESLAVLAKVLKDGFLHDGSYSSFRGRHHGRPINPSLANPAALVVCNQNHDQIGNRATGDR 401
Query: 572 ENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMVSQGVPMIYMGDEYG 622
+ + S+ G+ A ++V L S PM++MG+EYG
Sbjct: 402 LSQSLSY-----GQLAVAAVLTL-------------TSPFTPMLFMGEEYG 434
>TREZ_MYCTU (Q10769) Malto-oligosyltrehalose trehalohydrolase (EC
3.2.1.141) (MTHase)
(4-alpha-D-{(1->4)-alpha-D-glucano}trehalose
trehalohydrolase) (Maltooligosyl trehalose
trehalohydrolase)
Length = 580
Score = 73.9 bits (180), Expect = 2e-12
Identities = 81/295 (27%), Positives = 124/295 (41%), Gaps = 83/295 (28%)
Query: 226 IIYEMHVRGFTKHESSKTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNS 285
+IYE+H+ FT GT+ +EKLD+L +LG++ +EL+P + S+
Sbjct: 99 VIYELHIGTFTT--------AGTFDAAIEKLDYLVDLGIDFVELMPVN--------SFAG 142
Query: 286 VQGDYRVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVF 345
+G WGY V ++S Y G DG+ V+F I H R + V++D VF
Sbjct: 143 TRG------WGYDGVLWYSVHEPYG-------GPDGL--VRF-IDACHARRLGVLIDAVF 186
Query: 346 NHT-VEGN---ENGPIISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDC 401
NH GN GP +S P G+ N +G + VR +I+DC
Sbjct: 187 NHLGPSGNYLPRFGPYLSSAS---------NPWGDGINIAG------ADSDEVRHYIIDC 231
Query: 402 LRYWVTEMHVDGFRFDLASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPL---IDL 458
W+ + H DG R D + +++ V+V + LA T +S L + L
Sbjct: 232 ALRWMRDFHADGLRLDAVHALVDTTA----VHVL------EELANATRWLSGQLGRPLSL 281
Query: 459 IS----NDPILHGVKLIAEAWDAGGLYQVGTFPHWGIWSEWNGKYRDTVRQFVKG 509
I+ NDP +LI G +GI ++WN + V G
Sbjct: 282 IAETDRNDP-----RLITRPSHGG----------YGITAQWNDDIHHAIHTAVSG 321
>GLGB_HUMAN (Q04446) 1,4-alpha-glucan branching enzyme (EC 2.4.1.18)
(Glycogen branching enzyme) (Brancher enzyme)
Length = 702
Score = 73.9 bits (180), Expect = 2e-12
Identities = 96/396 (24%), Positives = 153/396 (38%), Gaps = 91/396 (22%)
Query: 71 GGRFQVSRGYPAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEY------- 123
GG + SRGY + DGG +YS +F DF Y
Sbjct: 64 GGIDKFSRGYESFGVHRCADGG----LYSKEWAPGAEGVFLTGDFNGWNPFSYPYKKLDY 119
Query: 124 ----ITLDPLMNKT-----GSVWHVFLKGDFGDMLYGYKFDGKFSPIEGHYYDSSLILLD 174
+ + P NK+ GS V + G++LY +
Sbjct: 120 GKWELYIPPKQNKSVLVPHGSKLKVVITSKSGEILYR---------------------IS 158
Query: 175 PYAKAVISRGEFGSLGPDGNCWPQMAGMVPSNDDEFDWEGDLPLKYPQKDLIIYEMHVRG 234
P+AK V+ G+ ++ D W D E +E + L IYE HV G
Sbjct: 159 PWAKYVVREGD--NVNYDWIHW----------DPEHSYEFKHSRPKKPRSLRIYESHV-G 205
Query: 235 FTKHESSKTKFPGTYLGVVEKLDHLKELGVNCIELLPCHEFNELEYFSYNSVQGDYRVNF 294
+ HE + V L +K LG NCI+L+ +E+ Y S
Sbjct: 206 ISSHEGKVASYKHFTCNV---LPRIKGLGYNCIQLMAI-----MEHAYYAS--------- 248
Query: 295 WGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTVEGNEN 354
+GY ++F+ RY + E++ L+ AH GI V++DVV +H + + +
Sbjct: 249 FGYQITSFFAASSRYGTP----------EELQELVDTAHSMGIIVLLDVVHSHASKNSAD 298
Query: 355 GPIISFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGF 414
G + F G D S Y+ P+G + + V +F++ +R+W+ E DGF
Sbjct: 299 G-LNMFDGTD-SCYFHSGPRGTHDLWDS--RLFAYSSWEVLRFLLSNIRWWLEEYRFDGF 354
Query: 415 RFD-LASIMTRSSSLWNGVN-----VFGTSIEGDLL 444
RFD + S++ + G + FG ++ D L
Sbjct: 355 RFDGVTSMLYHHHGVGQGFSGDYSEYFGLQVDEDAL 390
>TREZ_ARTSQ (Q44316) Malto-oligosyltrehalose trehalohydrolase (EC
3.2.1.141) (MTHase)
(4-alpha-D-{(1->4)-alpha-D-glucano}trehalose
trehalohydrolase) (Maltooligosyl trehalose
trehalohydrolase)
Length = 598
Score = 71.6 bits (174), Expect = 7e-12
Identities = 107/429 (24%), Positives = 158/429 (35%), Gaps = 103/429 (24%)
Query: 210 FDWEGDLPLKYPQKDLIIYEMHVRGFTKHESSKTKFPGTYLGVVEKLDHLKELGVNCIEL 269
+ W+ D + +IYE+H+ FT GT KLD+L LGV+ IEL
Sbjct: 102 YSWQDDAWQGRELQGAVIYELHLGTFTPE--------GTLEAAAGKLDYLAGLGVDFIEL 153
Query: 270 LPCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLI 329
LP + FN + WGY V +F+ Y G + +
Sbjct: 154 LPVNAFNG--------------THNWGYDGVQWFAVHEAYG----------GPEAYQRFV 189
Query: 330 KEAHKRGIEVIMDVVFNHT-VEGN---ENGPIISFRGVDNSIYYMIAPKGEFYNYSGCGN 385
AH G+ VI DVV+NH GN GP + +G N+ G+ N G G+
Sbjct: 190 DAAHAAGLGVIQDVVYNHLGPSGNYLPRFGPYLK-QGEGNTW-------GDSVNLDGPGS 241
Query: 386 TLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSLWNGVNVFGTSIEGDLLA 445
+H VR++I+D L W+ + VDG R D + ++ + + FG +
Sbjct: 242 ----DH--VRRYILDNLAMWLRDYRVDGLRLDAVHALKDERAV-HILEDFGALADQISAE 294
Query: 446 TGTPLVSPPLIDLISNDPILHGVKLIAEAWDAGGLYQVGTFPHWGIWSEWNGKYRDTVRQ 505
G PL DL N+P L + D G +G+ +W+ + V
Sbjct: 295 VGRPLTLIAESDL--NNPRLLYPR------DVNG---------YGLEGQWSDDFHHAVHV 337
Query: 506 FVKG-TDGFAGAFAECLCGSPNVYQGG-----------GRKPWNSINFVCTHDGFTLADL 553
V G T G+ F + L V + G R INF H A L
Sbjct: 338 NVTGETTGYYSDF-DSLAALAKVLRDGFFHDGSYSSFRERHHGRPINFSAVHP----AAL 392
Query: 554 VTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMVSQGVP 613
V + H+ D + + G A ++V L P
Sbjct: 393 VVCSQNHDQIGNRATGDRLSQTLPY-----GSLALAAVLTL-------------TGPFTP 434
Query: 614 MIYMGDEYG 622
M+ MG+EYG
Sbjct: 435 MLLMGEEYG 443
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.139 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 106,384,772
Number of Sequences: 164201
Number of extensions: 5073617
Number of successful extensions: 12119
Number of sequences better than 10.0: 181
Number of HSP's better than 10.0 without gapping: 72
Number of HSP's successfully gapped in prelim test: 109
Number of HSP's that attempted gapping in prelim test: 11549
Number of HSP's gapped (non-prelim): 351
length of query: 791
length of database: 59,974,054
effective HSP length: 118
effective length of query: 673
effective length of database: 40,598,336
effective search space: 27322680128
effective search space used: 27322680128
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0004a.4


