

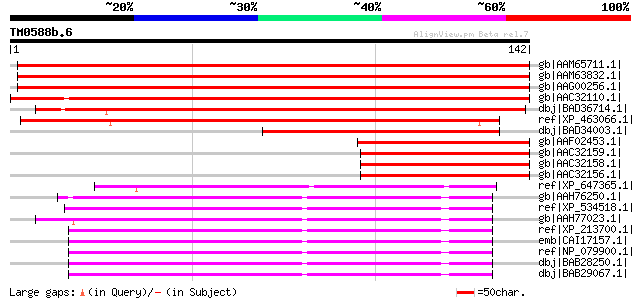
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588b.6
(142 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM65711.1| unknown [Arabidopsis thaliana] gi|21281209|gb|AAM... 214 3e-55
gb|AAM63832.1| unknown [Arabidopsis thaliana] gi|26449510|dbj|BA... 214 5e-55
gb|AAG00256.1| F1N21.7 [Arabidopsis thaliana] 212 1e-54
gb|AAC32110.1| hypothetical protein [Picea mariana] gi|25493097|... 197 6e-50
dbj|BAD36714.1| proteasome maturation factor-like [Oryza sativa ... 157 7e-38
ref|XP_463066.1| putative proteasome maturation factor [Oryza sa... 123 8e-28
dbj|BAD34003.1| proteasome maturation factor-like protein [Oryza... 89 2e-17
gb|AAF02453.1| unknown [Picea abies] gi|6049098|gb|AAF02452.1| u... 88 5e-17
gb|AAC32159.1| hypothetical protein [Picea mariana] gi|2996130|g... 86 2e-16
gb|AAC32158.1| hypothetical protein [Picea mariana] 84 6e-16
gb|AAC32156.1| hypothetical protein [Picea mariana] 83 2e-15
ref|XP_647365.1| hypothetical protein DDB0189600 [Dictyostelium ... 70 1e-11
gb|AAH76250.1| Zgc:92775 [Danio rerio] gi|51010931|ref|NP_001003... 67 7e-11
ref|XP_534518.1| PREDICTED: similar to chromosome 13 open readin... 67 9e-11
gb|AAH77023.1| MGC89819 protein [Xenopus tropicalis] gi|52346100... 65 3e-10
ref|XP_213700.1| PREDICTED: similar to chromosome 13 open readin... 64 6e-10
emb|CAI17157.1| chromosome 13 open reading frame 12 [Homo sapien... 62 4e-09
ref|NP_079900.1| hypothetical protein LOC66537 [Mus musculus] gi... 61 5e-09
dbj|BAB28250.1| unnamed protein product [Mus musculus] 61 5e-09
dbj|BAB29067.1| unnamed protein product [Mus musculus] 61 7e-09
>gb|AAM65711.1| unknown [Arabidopsis thaliana] gi|21281209|gb|AAM45059.1| unknown
protein [Arabidopsis thaliana]
gi|19347883|gb|AAL85998.1| unknown protein [Arabidopsis
thaliana] gi|10176833|dbj|BAB10155.1| unnamed protein
product [Arabidopsis thaliana]
gi|15240969|ref|NP_198681.1| proteasome maturation
factor UMP1 family protein [Arabidopsis thaliana]
Length = 141
Score = 214 bits (546), Expect = 3e-55
Identities = 102/140 (72%), Positives = 120/140 (84%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKSDI+ SHPL++A ES + E MKR+ + + YGA
Sbjct: 2 ESQKKIAHEIGGMKNDALRFGLHGVKSDILRSHPLETAYESGKQSQEEMKRRVITHTYGA 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMDLDRQILSRFQRP G IPSSMLGLE +TG+LD+FGFEDYL+DPRESET++P+D
Sbjct: 62 ALPLKMDLDRQILSRFQRPPGPIPSSMLGLEVYTGALDNFGFEDYLNDPRESETLKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLGLSKGP PSFM
Sbjct: 122 HHGMEVRLGLSKGPASPSFM 141
>gb|AAM63832.1| unknown [Arabidopsis thaliana] gi|26449510|dbj|BAC41881.1| unknown
protein [Arabidopsis thaliana]
gi|28372850|gb|AAO39907.1| At1g67250 [Arabidopsis
thaliana] gi|18408726|ref|NP_564892.1| proteasome
maturation factor UMP1 family protein [Arabidopsis
thaliana] gi|14994239|gb|AAK73254.1| Unknown protein
[Arabidopsis thaliana]
Length = 141
Score = 214 bits (544), Expect = 5e-55
Identities = 98/140 (70%), Positives = 119/140 (85%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKS+I+GSHPL+S+ ES K E +KR + + YG
Sbjct: 2 ESEKKIAHEIGGVKNDALRFGLHGVKSNIIGSHPLESSYESEKKSKEALKRTVIAHAYGT 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMD+DRQILSRFQRP G IPSSMLGLE +TG++DDFGFEDYL+DPR+SET +P+D
Sbjct: 62 ALPLKMDMDRQILSRFQRPPGPIPSSMLGLEVYTGAVDDFGFEDYLNDPRDSETFKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLG+SKGP+ PSFM
Sbjct: 122 HHGMEVRLGISKGPIYPSFM 141
>gb|AAG00256.1| F1N21.7 [Arabidopsis thaliana]
Length = 303
Score = 212 bits (540), Expect = 1e-54
Identities = 97/140 (69%), Positives = 119/140 (84%)
Query: 3 EAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
E+ K IAH+IGG++NDALRFGL GVKS+I+GSHPL+S+ ES K E +KR + + YG
Sbjct: 2 ESEKKIAHEIGGVKNDALRFGLHGVKSNIIGSHPLESSYESEKKSKEALKRTVIAHAYGT 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
A PLKMD+DRQILSRFQRP G IPSSMLGLE +TG++DDFGFEDYL+DPR+SET +P+D
Sbjct: 62 ALPLKMDMDRQILSRFQRPPGPIPSSMLGLEVYTGAVDDFGFEDYLNDPRDSETFKPVDF 121
Query: 123 HHGMEVRLGLSKGPVCPSFM 142
HHGMEVRLG+SKGP+ PSF+
Sbjct: 122 HHGMEVRLGISKGPIYPSFI 141
>gb|AAC32110.1| hypothetical protein [Picea mariana] gi|25493097|pir||T51959
hypothetical protein [imported] - Picea mariana
Length = 141
Score = 197 bits (500), Expect = 6e-50
Identities = 97/142 (68%), Positives = 113/142 (79%), Gaps = 1/142 (0%)
Query: 1 MEEAAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLY 60
M+ +S+ H+IGGI D LRFGL GVKSDI+ SHP+Q QE+A K MKR + Y
Sbjct: 1 MDPVQQSLPHEIGGI-TDTLRFGLAGVKSDILISHPVQVIQENAQKQQREMKRNILAKTY 59
Query: 61 GAAFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPL 120
G+AFPLKMD+++QILSRFQRP G +PSSMLGLE+ TGSLD+F FEDYL+DPRESET P
Sbjct: 60 GSAFPLKMDIEQQILSRFQRPPGELPSSMLGLESLTGSLDEFSFEDYLNDPRESETFVPA 119
Query: 121 DMHHGMEVRLGLSKGPVCPSFM 142
DMHHGMEVRLGLSKGPVC SFM
Sbjct: 120 DMHHGMEVRLGLSKGPVCRSFM 141
>dbj|BAD36714.1| proteasome maturation factor-like [Oryza sativa (japonica
cultivar-group)]
Length = 141
Score = 157 bits (396), Expect = 7e-38
Identities = 81/135 (60%), Positives = 99/135 (73%), Gaps = 2/135 (1%)
Query: 8 IAHQIGGIQNDALRFGLQ-GVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPL 66
+ +IGG +D LRFG+ VK D+ HPLQ++ +K KR +YG+AF +
Sbjct: 7 VKKEIGG-NHDVLRFGVNDSVKGDLAPPHPLQASVHKDAKFWADKKRFGAEAIYGSAFNI 65
Query: 67 KMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGM 126
+ DLD QILS+FQRP GA+PSSMLG EA TGSLDDFGFEDYL+ P++SE+ R DMHHGM
Sbjct: 66 RKDLDAQILSKFQRPPGALPSSMLGYEALTGSLDDFGFEDYLNLPQDSESFRAPDMHHGM 125
Query: 127 EVRLGLSKGPVCPSF 141
EVRLGLSKGPVCPSF
Sbjct: 126 EVRLGLSKGPVCPSF 140
>ref|XP_463066.1| putative proteasome maturation factor [Oryza sativa (japonica
cultivar-group)] gi|41469323|gb|AAS07179.1| putative
proteasome maturation factor [Oryza sativa (japonica
cultivar-group)]
Length = 281
Score = 123 bits (309), Expect = 8e-28
Identities = 66/135 (48%), Positives = 89/135 (65%), Gaps = 4/135 (2%)
Query: 4 AAKSIAHQIGGIQNDALRFGLQG-VKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGA 62
A+ SI + G +D L FG+ VK D+ HP+Q+ +K KR +YG+
Sbjct: 2 ASGSIVKKEVGENHDVLLFGVNNSVKGDLAPQHPIQATVHKEAKFWADKKRFGAEAIYGS 61
Query: 63 AFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDM 122
AF ++ DLD QILS+FQRP GA+PSSMLG EA TGSLDDFGFEDYL+ P++S++ DM
Sbjct: 62 AFNIRKDLDAQILSKFQRPPGALPSSMLGYEALTGSLDDFGFEDYLNLPQDSDSFHAPDM 121
Query: 123 HHGME---VRLGLSK 134
HHGME +++G+ K
Sbjct: 122 HHGMEKNLLKIGICK 136
>dbj|BAD34003.1| proteasome maturation factor-like protein [Oryza sativa (japonica
cultivar-group)] gi|51091633|dbj|BAD36402.1| proteasome
maturation factor-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 258
Score = 89.4 bits (220), Expect = 2e-17
Identities = 42/65 (64%), Positives = 51/65 (77%)
Query: 70 LDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVR 129
+D + RFQRP GA+PSSMLG EA TGSLDDFGFEDYL+ P++S++ DMHHGMEV
Sbjct: 29 VDDLSVCRFQRPPGALPSSMLGYEALTGSLDDFGFEDYLNLPQDSDSFHAPDMHHGMEVL 88
Query: 130 LGLSK 134
LG+ K
Sbjct: 89 LGICK 93
>gb|AAF02453.1| unknown [Picea abies] gi|6049098|gb|AAF02452.1| unknown [Picea
abies]
Length = 48
Score = 87.8 bits (216), Expect = 5e-17
Identities = 40/47 (85%), Positives = 42/47 (89%)
Query: 96 TGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLSKGPVCPSFM 142
TGSLD+F FEDYL+DPRESET P DMHHGMEVRLGLSKGPVC SFM
Sbjct: 2 TGSLDEFSFEDYLNDPRESETFVPADMHHGMEVRLGLSKGPVCRSFM 48
>gb|AAC32159.1| hypothetical protein [Picea mariana] gi|2996130|gb|AAC32157.1|
hypothetical protein [Picea mariana]
Length = 46
Score = 85.9 bits (211), Expect = 2e-16
Identities = 39/46 (84%), Positives = 41/46 (88%)
Query: 97 GSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLSKGPVCPSFM 142
GSLD+F FEDYL+DPRESET P DMHHGMEVRLGLSKGPVC SFM
Sbjct: 1 GSLDEFSFEDYLNDPRESETFVPADMHHGMEVRLGLSKGPVCRSFM 46
>gb|AAC32158.1| hypothetical protein [Picea mariana]
Length = 46
Score = 84.3 bits (207), Expect = 6e-16
Identities = 38/46 (82%), Positives = 41/46 (88%)
Query: 97 GSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLSKGPVCPSFM 142
GSLD+F FEDYL+DPRESET P DMHHGMEVRLGLSKGPVC SF+
Sbjct: 1 GSLDEFSFEDYLNDPRESETFVPADMHHGMEVRLGLSKGPVCRSFI 46
>gb|AAC32156.1| hypothetical protein [Picea mariana]
Length = 46
Score = 82.8 bits (203), Expect = 2e-15
Identities = 38/46 (82%), Positives = 40/46 (86%)
Query: 97 GSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLSKGPVCPSFM 142
GSLD+F FEDYL+DPRESET P DMHHGMEVRLGLSKG VC SFM
Sbjct: 1 GSLDEFSFEDYLNDPRESETFVPADMHHGMEVRLGLSKGSVCRSFM 46
>ref|XP_647365.1| hypothetical protein DDB0189600 [Dictyostelium discoideum]
gi|60475451|gb|EAL73386.1| hypothetical protein
DDB0189600 [Dictyostelium discoideum]
Length = 128
Score = 69.7 bits (169), Expect = 1e-11
Identities = 42/111 (37%), Positives = 63/111 (55%), Gaps = 3/111 (2%)
Query: 24 LQGVKSDIVG-SHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILSRFQRPS 82
L+ S++ G SHP++S Q K +K M N++G + ++++QI S+F+R
Sbjct: 14 LRNESSNVKGLSHPVESIQLDQGKTEIKLKNFAMKNVFGTHMVMNNEIEKQIYSQFKRLP 73
Query: 83 GAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGLS 133
+ SSM+GLE G +DF F DYL DP SE P +H ME RLG++
Sbjct: 74 -TLQSSMVGLETILGLDEDFDFGDYLCDPATSEAPLP-QLHTSMEHRLGMT 122
>gb|AAH76250.1| Zgc:92775 [Danio rerio] gi|51010931|ref|NP_001003424.1|
hypothetical protein LOC445030 [Danio rerio]
Length = 142
Score = 67.4 bits (163), Expect = 7e-11
Identities = 42/119 (35%), Positives = 67/119 (56%), Gaps = 4/119 (3%)
Query: 14 GIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQ 73
G+Q D+LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ +
Sbjct: 27 GVQ-DSLRRGFSSVKNELLPSHPLELSEKNFQLNQDKMNFNTLRNIQGLHAPLKLQMEYR 85
Query: 74 ILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR +PSS L L+ GS D GFED L+DP + E + D H E +LGL
Sbjct: 86 AAKQIQR-LPFLPSSNLALDTLRGSDDTIGFEDILNDPVQCEMMG--DPHIMTEYKLGL 141
>ref|XP_534518.1| PREDICTED: similar to chromosome 13 open reading frame 12 [Canis
familiaris]
Length = 141
Score = 67.0 bits (162), Expect = 9e-11
Identities = 40/117 (34%), Positives = 69/117 (58%), Gaps = 3/117 (2%)
Query: 16 QNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQIL 75
Q+D LR GL VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 27 QHDLLRKGLPCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAV 86
Query: 76 SRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 87 QQVQR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSELMG--EPHLMVEYKLGL 140
>gb|AAH77023.1| MGC89819 protein [Xenopus tropicalis]
gi|52346100|ref|NP_001005093.1| MGC89819 protein
[Xenopus tropicalis]
Length = 142
Score = 65.5 bits (158), Expect = 3e-10
Identities = 41/126 (32%), Positives = 72/126 (56%), Gaps = 4/126 (3%)
Query: 8 IAHQIGGIQ-NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPL 66
++ Q GG +D LR G V+S+++ SHPL+ ++++ E + + N+ G PL
Sbjct: 19 LSTQYGGYGVHDNLRTGFTSVRSELMPSHPLELSEKNFQINQEKVNLSTVRNIQGLHAPL 78
Query: 67 KMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGM 126
K+ ++ + + + QR +PSS + L+ G+ + GFED L+DP +SE + + H M
Sbjct: 79 KLQMEFKAVRQVQR-LPFLPSSNIALDTLRGTDESIGFEDILNDPSQSEGMG--EPHLMM 135
Query: 127 EVRLGL 132
E +LGL
Sbjct: 136 EYKLGL 141
>ref|XP_213700.1| PREDICTED: similar to chromosome 13 open reading frame 12 [Rattus
norvegicus]
Length = 141
Score = 64.3 bits (155), Expect = 6e-10
Identities = 38/116 (32%), Positives = 67/116 (57%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVQR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSELMG--EPHMMVEYKLGL 140
>emb|CAI17157.1| chromosome 13 open reading frame 12 [Homo sapiens]
gi|55729624|emb|CAH91541.1| hypothetical protein [Pongo
pygmaeus] gi|15680038|gb|AAH14334.1| Chromosome 13 open
reading frame 12 [Homo sapiens]
gi|13097270|gb|AAH03390.1| Chromosome 13 open reading
frame 12 [Homo sapiens] gi|14279182|gb|AAK58521.1|
voltage-gated K channel beta subunit 4.1 [Homo sapiens]
gi|5106990|gb|AAD39914.1| HSPC036 protein [Homo sapiens]
gi|4679014|gb|AAD26995.1| HSPC014 [Homo sapiens]
gi|7705429|ref|NP_057016.1| hypothetical protein
LOC51371 [Homo sapiens]
Length = 141
Score = 61.6 bits (148), Expect = 4e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ QR + SS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVQR-LPFLSSSNLSLDVLRGNDETIGFEDILNDPSQSEVMG--EPHLMVEYKLGL 140
>ref|NP_079900.1| hypothetical protein LOC66537 [Mus musculus]
gi|20072527|gb|AAH27016.1| RIKEN cDNA 2510048O06 [Mus
musculus] gi|12852049|dbj|BAB29252.1| unnamed protein
product [Mus musculus] gi|12847031|dbj|BAB27410.1|
unnamed protein product [Mus musculus]
gi|12834112|dbj|BAB22791.1| unnamed protein product [Mus
musculus]
Length = 141
Score = 61.2 bits (147), Expect = 5e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ R +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVHR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSELMG--EPHVMVEHKLGL 140
>dbj|BAB28250.1| unnamed protein product [Mus musculus]
Length = 141
Score = 61.2 bits (147), Expect = 5e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSEKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ R +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVHR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSEQMG--EPHVMVEHKLGL 140
>dbj|BAB29067.1| unnamed protein product [Mus musculus]
Length = 141
Score = 60.8 bits (146), Expect = 7e-09
Identities = 37/116 (31%), Positives = 66/116 (56%), Gaps = 3/116 (2%)
Query: 17 NDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILS 76
+D LR G VK++++ SHPL+ ++++ + M + N+ G PLK+ ++ + +
Sbjct: 28 HDLLRKGFSCVKNELLPSHPLELSKKNFQLNQDKMNFSTLRNIQGLFAPLKLQMEFKAVQ 87
Query: 77 RFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMHHGMEVRLGL 132
+ R +PSS L L+ G+ + GFED L+DP +SE + + H +E +LGL
Sbjct: 88 QVHR-LPFLPSSNLSLDILRGNDETIGFEDILNDPSQSELMG--EPHVMVEHKLGL 140
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 221,496,848
Number of Sequences: 2540612
Number of extensions: 8197152
Number of successful extensions: 16160
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 16102
Number of HSP's gapped (non-prelim): 48
length of query: 142
length of database: 863,360,394
effective HSP length: 118
effective length of query: 24
effective length of database: 563,568,178
effective search space: 13525636272
effective search space used: 13525636272
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0588b.6


