

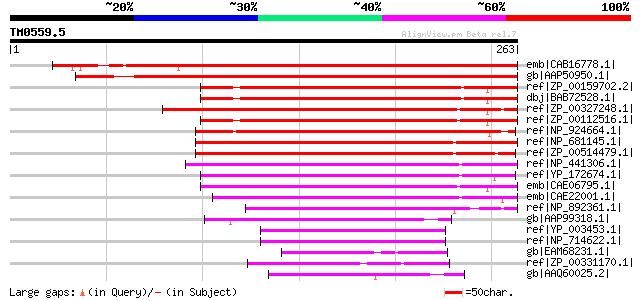
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0559.5
(263 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB16778.1| thiol-disulfide interchange like protein [Arabid... 327 2e-88
gb|AAP50950.1| putative thioredoxin [Oryza sativa (japonica cult... 310 3e-83
ref|ZP_00159702.2| COG0526: Thiol-disulfide isomerase and thiore... 153 4e-36
dbj|BAB72528.1| thioredoxin-like protein [Nostoc sp. PCC 7120] g... 150 5e-35
ref|ZP_00327248.1| COG0526: Thiol-disulfide isomerase and thiore... 148 2e-34
ref|ZP_00112516.1| COG0526: Thiol-disulfide isomerase and thiore... 146 5e-34
ref|NP_924664.1| thiol:disulfide interchange protein [Gloeobacte... 140 4e-32
ref|NP_681145.1| thiol:disulfide interchange protein [Thermosyne... 135 2e-30
ref|ZP_00514479.1| Thioredoxin-related [Crocosphaera watsonii WH... 132 8e-30
ref|NP_441306.1| thiol:disulfide interchange protein; TrxA [Syne... 131 2e-29
ref|YP_172674.1| thioredoxin [Synechococcus elongatus PCC 6301] ... 121 2e-26
emb|CAE06795.1| thioredoxin-like protein TxlA [Synechococcus sp.... 114 4e-24
emb|CAE22001.1| thioredoxin-like protein TxlA [Prochlorococcus m... 108 1e-22
ref|NP_892361.1| thioredoxin-like protein TxlA [Prochlorococcus ... 105 1e-21
gb|AAP99318.1| Thioredoxin family protein [Prochlorococcus marin... 99 2e-19
ref|YP_003453.1| thiol-disulfide interchange like protein [Lepto... 66 1e-09
ref|NP_714622.1| TPR-repeat-containing protein [Leptospira inter... 66 1e-09
gb|EAM68231.1| Thioredoxin [Jannaschia sp. CCS1] 64 4e-09
ref|ZP_00331170.1| COG0526: Thiol-disulfide isomerase and thiore... 63 7e-09
gb|AAQ60025.2| Thiol:disulfide interchange protein DsbD [Chromob... 63 1e-08
>emb|CAB16778.1| thiol-disulfide interchange like protein [Arabidopsis thaliana]
gi|7270669|emb|CAB80386.1| thiol-disulfide interchange
like protein [Arabidopsis thaliana]
gi|12049653|emb|CAC19858.1| thioredoxin-like protein
[Arabidopsis thaliana] gi|20453407|gb|AAM19942.1|
AT4g37200/C7A10_160 [Arabidopsis thaliana]
gi|18700133|gb|AAL77678.1| AT4g37200/C7A10_160
[Arabidopsis thaliana] gi|15235475|ref|NP_195437.1|
thioredoxin family protein [Arabidopsis thaliana]
gi|25407797|pir||E85439 thiol-disulfide interchange like
protein [imported] - Arabidopsis thaliana
Length = 261
Score = 327 bits (838), Expect = 2e-88
Identities = 166/252 (65%), Positives = 202/252 (79%), Gaps = 17/252 (6%)
Query: 23 FTVNPRHLH--FNTR----KFHTLACQTNPNLDEKDASATSQEIKIVVEPSTVNGESETC 76
F ++PR+L F T+ +F + C+ NP ++S T QE K+V++ + S+
Sbjct: 16 FNLSPRNLQSFFVTQTGAPRFRAVRCKPNP-----ESSETKQE-KLVIDNGETSSASKEV 69
Query: 77 KPTSSTADAS-----GLPQLPTKDINRKVAIASTLAALGLFLATRLDFGVSLKDLTANAL 131
+ +SS AD+S G P+ P KDINR+VA + +AAL LF++TRLDFG+SLKDLTA+AL
Sbjct: 70 ESSSSVADSSSSSSSGFPESPNKDINRRVAAVTVIAALSLFVSTRLDFGISLKDLTASAL 129
Query: 132 PYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDE 191
PYE+ALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKD+VNFVMLNVDNT WEQELDE
Sbjct: 130 PYEEALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDKVNFVMLNVDNTKWEQELDE 189
Query: 192 FGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEAR 251
FGVEGIPHFAFLD+EGNEEGNVVGRLPRQ L+ENV+ALA G+ S+P+AR VG+YSS+E+R
Sbjct: 190 FGVEGIPHFAFLDREGNEEGNVVGRLPRQYLVENVNALAAGKQSIPYARAVGQYSSSESR 249
Query: 252 KVHQVVDPRSHG 263
KVHQV DP SHG
Sbjct: 250 KVHQVTDPLSHG 261
>gb|AAP50950.1| putative thioredoxin [Oryza sativa (japonica cultivar-group)]
gi|50919029|ref|XP_469911.1| putative thioredoxin [Oryza
sativa (japonica cultivar-group)]
gi|40539065|gb|AAR87322.1| putative thioredoxin-like
protein [Oryza sativa (japonica cultivar-group)]
Length = 262
Score = 310 bits (793), Expect = 3e-83
Identities = 155/230 (67%), Positives = 179/230 (77%), Gaps = 11/230 (4%)
Query: 35 RKFHTLACQTNPNLDEKDASATSQEIKIVVEPSTVNGESETCKPTSSTADASGLPQLPTK 94
R+ +L+C P+ E EP+ + +E SS + +G P P K
Sbjct: 43 RRLGSLSCIAPPDSAEPQTD----------EPAAKDDSTEDKAEASSASQDAGNPTFPNK 92
Query: 95 DINRKVAIASTLAALGLFLATRLDFG-VSLKDLTANALPYEQALSNGKPTVVEFYADWCE 153
D++R++A+AST+ A+GLF RLDFG VSLKDL ANA PYE+ALSNGKPTVVEFYADWCE
Sbjct: 93 DLSRRIALASTIGAVGLFAYQRLDFGGVSLKDLAANATPYEEALSNGKPTVVEFYADWCE 152
Query: 154 VCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNV 213
VCRELAPDVYK+EQQYKDRVNFVMLNVDNT WEQELDEFGVEGIPHFAFLDKEGNEEGNV
Sbjct: 153 VCRELAPDVYKVEQQYKDRVNFVMLNVDNTKWEQELDEFGVEGIPHFAFLDKEGNEEGNV 212
Query: 214 VGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
VGRLP+Q L+NV ALA GE +VPHARVVG++SSAE+RKVHQV DPRSHG
Sbjct: 213 VGRLPKQYFLDNVVALASGEPTVPHARVVGQFSSAESRKVHQVADPRSHG 262
>ref|ZP_00159702.2| COG0526: Thiol-disulfide isomerase and thioredoxins [Anabaena
variabilis ATCC 29413]
Length = 192
Score = 153 bits (387), Expect = 4e-36
Identities = 83/167 (49%), Positives = 109/167 (64%), Gaps = 7/167 (4%)
Query: 100 VAIASTLA-ALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
VAIA T+A LGL T VSL L A + P E A+SNGKP++VEFYA+WC VC+++
Sbjct: 25 VAIALTVALVLGLRTETA---AVSLDKLYAASTPLEVAISNGKPSMVEFYANWCTVCQKM 81
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
APD+ + EQQY D+VNFVMLNVDNT W E+ ++ V+GIPHF FL+KEG +G P
Sbjct: 82 APDIAEFEQQYADKVNFVMLNVDNTKWLPEMLKYRVDGIPHFVFLNKEGESLAQAIGDQP 141
Query: 219 RQLLLENVDALARGEASVPHARVVGKYS--SAEARKVHQVVDPRSHG 263
++ N++AL G A +PHA+ G+ S S + DPRSHG
Sbjct: 142 HTIMASNLEALVTGSA-LPHAQTSGQVSQVSTPVAPIGSQDDPRSHG 187
>dbj|BAB72528.1| thioredoxin-like protein [Nostoc sp. PCC 7120]
gi|17228066|ref|NP_484614.1| thioredoxin-like protein
[Nostoc sp. PCC 7120] gi|25310547|pir||AI1877
thioredoxin-like protein [imported] - Nostoc sp. (strain
PCC 7120)
Length = 192
Score = 150 bits (378), Expect = 5e-35
Identities = 81/167 (48%), Positives = 108/167 (64%), Gaps = 7/167 (4%)
Query: 100 VAIASTLA-ALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
VAIA T+A LGL T VSL L A + P E A+SNGKP++VEFYA+WC VC+++
Sbjct: 25 VAIALTVALVLGLRTETA---AVSLDKLYAASTPLEVAISNGKPSMVEFYANWCTVCQKM 81
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
APD+ + EQQY D++NFVMLNVDNT W E+ ++ V+GIPHF FL+K G +G P
Sbjct: 82 APDIAEFEQQYADKMNFVMLNVDNTKWLPEMLKYRVDGIPHFVFLNKTGESLAQAIGDQP 141
Query: 219 RQLLLENVDALARGEASVPHARVVGKYS--SAEARKVHQVVDPRSHG 263
++ N++AL G A +PHA+ G+ S S + DPRSHG
Sbjct: 142 HTIMASNLEALVTGSA-LPHAQTSGQVSQVSTPVAPIGSQDDPRSHG 187
>ref|ZP_00327248.1| COG0526: Thiol-disulfide isomerase and thioredoxins [Trichodesmium
erythraeum IMS101]
Length = 193
Score = 148 bits (373), Expect = 2e-34
Identities = 76/189 (40%), Positives = 116/189 (61%), Gaps = 7/189 (3%)
Query: 80 SSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLD-FGVSLKDLTANALPYEQALS 138
++ ++++ + L T+ N + + ++ +F+A R D VSL ++ A P + ALS
Sbjct: 3 ANQSNSTNISSLVTRLRNFMIVSVAIALSVSIFMALRTDKLSVSLNEMAQAATPLDVALS 62
Query: 139 NGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIP 198
N KPT+VEFYA+WC C+ +AP++ +I Q+Y NFVMLNVDN+ W EL E+ V+GIP
Sbjct: 63 NNKPTLVEFYANWCTTCQAMAPELEEIRQKYTQNTNFVMLNVDNSRWLPELVEYRVDGIP 122
Query: 199 HFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARVVGKYS----SAEARKVH 254
HF FLD+EG E G +G +P+ ++ N++AL G + +P+ VG+ S E K
Sbjct: 123 HFVFLDREGTEVGQTIGEVPKSIMAANIEALVAG-SYLPYNSTVGQGSHFNAPVEPTKAS 181
Query: 255 QVVDPRSHG 263
Q DPR HG
Sbjct: 182 Q-ADPRVHG 189
>ref|ZP_00112516.1| COG0526: Thiol-disulfide isomerase and thioredoxins [Nostoc
punctiforme PCC 73102]
Length = 196
Score = 146 bits (369), Expect = 5e-34
Identities = 78/167 (46%), Positives = 110/167 (65%), Gaps = 7/167 (4%)
Query: 100 VAIASTLA-ALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
VAIA ++A LGL T SL L+ ++ P E A+SNGKP++VEFYADWC VC+++
Sbjct: 29 VAIALSVALVLGLRTETT---SASLAKLSNSSTPLEVAVSNGKPSLVEFYADWCTVCQKM 85
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
APD+ ++E +Y D++NFVMLNVDNT W E+ ++ V+GIPHF FL ++G +G P
Sbjct: 86 APDITQLETEYADKMNFVMLNVDNTKWLPEMLKYRVDGIPHFVFLSQQGETIAQAIGDQP 145
Query: 219 RQLLLENVDALARGEASVPHARVVGKYS--SAEARKVHQVVDPRSHG 263
R ++ N++AL G +S+P+A+ GK S SA DPRSHG
Sbjct: 146 RTIMASNLEALVTG-SSLPYAQASGKVSKFSAPVAPTASQDDPRSHG 191
>ref|NP_924664.1| thiol:disulfide interchange protein [Gloeobacter violaceus PCC
7421] gi|35212284|dbj|BAC89659.1| thiol:disulfide
interchange protein [Gloeobacter violaceus PCC 7421]
Length = 179
Score = 140 bits (353), Expect = 4e-32
Identities = 74/168 (44%), Positives = 107/168 (63%), Gaps = 6/168 (3%)
Query: 97 NRKVAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCR 156
N VA+AS L A+ +F A R + +++ L A A+P ++AL+NGKPT+VEFYADWC C+
Sbjct: 15 NGIVALASVLIAVLVFFAAR-NQAPTMQQLAAEAVPLDEALANGKPTLVEFYADWCASCQ 73
Query: 157 ELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGR 216
+AP V +++++Y ++ NFVMLNVDN W EL ++ V GIPH+AFLD G+ +G
Sbjct: 74 TMAPTVARLKERYGEQANFVMLNVDNPRWLPELQQYKVSGIPHYAFLDAAARPLGSAIGL 133
Query: 217 LPRQLLLENVDALARGEASVPHARVVGKYSS--AEARKVHQVVDPRSH 262
P Q+L +N+ ALA G ++P A +S +KV Q PR H
Sbjct: 134 QPSQILEDNLTALAAGAETLPKAGAPAGQTSKFTPPKKVDQ---PRDH 178
>ref|NP_681145.1| thiol:disulfide interchange protein [Thermosynechococcus elongatus
BP-1] gi|22294076|dbj|BAC07907.1| thiol:disulfide
interchange protein [Thermosynechococcus elongatus BP-1]
Length = 182
Score = 135 bits (339), Expect = 2e-30
Identities = 65/169 (38%), Positives = 109/169 (64%), Gaps = 3/169 (1%)
Query: 97 NRKVAIASTLAALGLFLATRLDFG-VSLKDLTANALPYEQALSNGKPTVVEFYADWCEVC 155
N +A+ + L + L D ++L + +A P+E+A NGKPT++EFYA+WC C
Sbjct: 14 NFVIAVVAILISFALLAGLGSDNQPLTLAEQAKHATPWEEAQVNGKPTLLEFYANWCSTC 73
Query: 156 RELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVG 215
R +A D+ +++ QY+DRVNFVMLNVDN+ W E+++F V+GIPHF FL+++ + +G
Sbjct: 74 RSMAKDLGELKAQYRDRVNFVMLNVDNSKWLPEIEQFNVDGIPHFVFLNRQLEPQAVAIG 133
Query: 216 RLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVHQ-VVDPRSHG 263
P++++ +N++AL+ + +P+ ++ G+ S + Q DPRSHG
Sbjct: 134 LQPKEVMAKNLEALS-NDLPLPYGQLTGEMSQLPSPLARQRSDDPRSHG 181
>ref|ZP_00514479.1| Thioredoxin-related [Crocosphaera watsonii WH 8501]
gi|67857077|gb|EAM52317.1| Thioredoxin-related
[Crocosphaera watsonii WH 8501]
Length = 183
Score = 132 bits (333), Expect = 8e-30
Identities = 64/167 (38%), Positives = 103/167 (61%), Gaps = 3/167 (1%)
Query: 97 NRKVAIASTLAALGLFLATRLDFG-VSLKDLTANALPYEQALSNGKPTVVEFYADWCEVC 155
N +A+A+ ++ +F + G VSL+ ++ P E A++NGKPT+ EFYADWC C
Sbjct: 12 NTLIAVAAITLSIAIFFGFQTQTGSVSLESQAKDSTPIEVAIANGKPTLTEFYADWCTSC 71
Query: 156 RELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVG 215
+ +AP++ +++Q+Y D VNFVM NVDN W E+ + V+GIPHF F+D +G E +G
Sbjct: 72 QAMAPELAQLKQKYGDSVNFVMFNVDNNKWLPEILRYRVDGIPHFVFIDDQGKEIAQAIG 131
Query: 216 RLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSH 262
P ++ N++AL ++P+A G+ S ++ + + DPRSH
Sbjct: 132 EQPSSVMQANLEALI-DHQTLPYAASTGETSDLKSPN-NPISDPRSH 176
>ref|NP_441306.1| thiol:disulfide interchange protein; TrxA [Synechocystis sp. PCC
6803] gi|1653069|dbj|BAA17986.1| thiol:disulfide
interchange protein; TrxA [Synechocystis sp. PCC 6803]
gi|2501215|sp|P73920|TXLA_SYNY3 Thiol:disulfide
interchange protein txlA homolog
Length = 180
Score = 131 bits (329), Expect = 2e-29
Identities = 68/174 (39%), Positives = 100/174 (57%), Gaps = 3/174 (1%)
Query: 92 PTKDINRKVAIASTLAALGLFLATRLDF-GVSLKDLTANALPYEQALSNGKPTVVEFYAD 150
P K N +A+ + + ++L + G+SL+ A+P AL NG+PT+VEFYAD
Sbjct: 3 PAKIRNALLAVVAIALSAAVYLGFQTQTQGISLEAQAQRAIPLATALDNGRPTLVEFYAD 62
Query: 151 WCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEE 210
WC C+ +APD+ ++++ Y VNF MLNVDN W E+ + V+GIPHF +LD G
Sbjct: 63 WCTSCQAMAPDLAELKKNYGGSVNFAMLNVDNNKWLPEVLRYRVDGIPHFVYLDDTGTAI 122
Query: 211 GNVVGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVH-QVVDPRSHG 263
+G P ++L +N+ AL E +P+A V G+ S E R + PRSHG
Sbjct: 123 AESIGEQPLRVLEQNITALVAHE-PIPYANVTGQTSVVENRTIEADPTSPRSHG 175
>ref|YP_172674.1| thioredoxin [Synechococcus elongatus PCC 6301]
gi|56686932|dbj|BAD80154.1| thioredoxin [Synechococcus
elongatus PCC 6301] gi|2126520|pir||A57254
thioredoxin-like protein txlA - Synechococcus sp
gi|454074|gb|AAA89104.1| TxlA
gi|464973|sp|P35088|TXLA_SYNP7 Thiol:disulfide
interchange protein txlA gi|46130315|ref|ZP_00165137.2|
COG0526: Thiol-disulfide isomerase and thioredoxins
[Synechococcus elongatus PCC 7942]
Length = 191
Score = 121 bits (303), Expect = 2e-26
Identities = 62/166 (37%), Positives = 93/166 (55%), Gaps = 4/166 (2%)
Query: 100 VAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELA 159
+A A L L + + + SL L A PYE A++N +P ++EFYADWC C+ +A
Sbjct: 17 IAAALVLTILVVLGSRQPSAAASLASLAEQATPYEVAIANDRPMLLEFYADWCTSCQAMA 76
Query: 160 PDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPR 219
+ ++Q Y DR++FVMLN+DN W E+ ++ V+GIP F +L+ +G +G +G LPR
Sbjct: 77 GRIAALKQDYSDRLDFVMLNIDNDKWLPEVLDYNVDGIPQFVYLNGQGQPQGISIGELPR 136
Query: 220 QLLLENVDALARGEASVPHARVVGKYSSAEA---RKVHQVVDPRSH 262
+L N+DAL + +P+ G S A DPRSH
Sbjct: 137 SVLAANLDALVEAQ-PLPYTNARGNLSEFSADLQPSRSSQTDPRSH 181
>emb|CAE06795.1| thioredoxin-like protein TxlA [Synechococcus sp. WH 8102]
gi|33864816|ref|NP_896375.1| thioredoxin-like protein
TxlA [Synechococcus sp. WH 8102]
Length = 184
Score = 114 bits (284), Expect = 4e-24
Identities = 61/168 (36%), Positives = 97/168 (57%), Gaps = 5/168 (2%)
Query: 100 VAIASTLAALGL-FLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
+ +A+ + A+GL + L G L+ L ++ + ALSNG+PT++EFYADWC+VCRE+
Sbjct: 18 LVVAAIVLAVGLTMMRGGLQSGSPLEQLARRSMTPDVALSNGRPTMIEFYADWCQVCREM 77
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
AP + K+EQ + +++ VM+NVDN W+ +D + V GIP + EG+ G +G
Sbjct: 78 APAMLKLEQSMQQQLDVVMVNVDNPRWQDLVDRYDVNGIPQLNLFNAEGDAVGRSIGLRR 137
Query: 219 RQLLLENVDALARGEASVPHARVVGKYS---SAEARKVHQVVDPRSHG 263
Q L DAL +G + +P + +G+ S ++ PRSHG
Sbjct: 138 EQELSLLADALIQG-SPLPSLQGLGRVSERTDVAPQQASLTAGPRSHG 184
>emb|CAE22001.1| thioredoxin-like protein TxlA [Prochlorococcus marinus str. MIT
9313] gi|33864093|ref|NP_895653.1| thioredoxin-like
protein TxlA [Prochlorococcus marinus str. MIT 9313]
Length = 188
Score = 108 bits (271), Expect = 1e-22
Identities = 58/165 (35%), Positives = 93/165 (56%), Gaps = 8/165 (4%)
Query: 106 LAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKI 165
LA + L L+ L+ L +L + AL+NG+PTV+EFYADWC+ CRE+AP + K
Sbjct: 25 LAVVLFLLRGGLNAEAPLEQLARRSLDPDVALTNGRPTVIEFYADWCQACREMAPAMLKT 84
Query: 166 EQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLEN 225
E+ +D+++ V++NVDN W+ +D + V GIP F D +G +G +G ++ L +
Sbjct: 85 ERDREDQLDVVLVNVDNPRWQDLVDRYDVNGIPQLNFFDNKGKLQGLSLGARTQEQLQQL 144
Query: 226 VDALARGEASVPHARVVGKYSSAEARKVH-------QVVDPRSHG 263
DAL + + +P+ VG S+ A + + + P SHG
Sbjct: 145 TDALIQNQ-PLPNFAGVGAISNLNASQERTEKQNDLEAIRPMSHG 188
>ref|NP_892361.1| thioredoxin-like protein TxlA [Prochlorococcus marinus subsp.
pastoris str. CCMP1986] gi|33633742|emb|CAE18701.1|
thioredoxin-like protein TxlA [Prochlorococcus marinus
subsp. pastoris str. CCMP1986]
Length = 181
Score = 105 bits (262), Expect = 1e-21
Identities = 56/144 (38%), Positives = 84/144 (57%), Gaps = 8/144 (5%)
Query: 123 LKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDN 182
LK +++ E A +N KPT +EFYADWCEVC+E+AP V I+ +Y+ +NFV LNVDN
Sbjct: 43 LKKFGQSSIDPEIAFTNNKPTFLEFYADWCEVCKEMAPKVEDIKNKYEKDINFVFLNVDN 102
Query: 183 TNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDAL---ARGEASVPHA 239
WE+ + F V GIP D + N E +G+ + E++D L RG++ + ++
Sbjct: 103 PKWEKYIRNFNVNGIPQINIFDNDSNLELTFIGKQEEDSIKESLDNLYDKFRGDSQILNS 162
Query: 240 RVVGKYSSAEARKVHQVVDPRSHG 263
++S + K +Q PRSHG
Sbjct: 163 ----EFSIIKDNKNYQ-SSPRSHG 181
>gb|AAP99318.1| Thioredoxin family protein [Prochlorococcus marinus subsp. marinus
str. CCMP1375] gi|33239724|ref|NP_874666.1| Thioredoxin
family protein [Prochlorococcus marinus subsp. marinus
str. CCMP1375]
Length = 189
Score = 98.6 bits (244), Expect = 2e-19
Identities = 52/131 (39%), Positives = 79/131 (59%), Gaps = 10/131 (7%)
Query: 102 IASTLAALGLFL---ATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCREL 158
IAS + A LFL L+ SL L ++L E AL+NG PT+ EFYADWCE C+E+
Sbjct: 19 IASFVLACFLFLLRGGLGLNSEKSLNILALSSLQPEVALNNGHPTIFEFYADWCEACQEM 78
Query: 159 APDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLP 218
AP + I+++Y+D+++ V+LNVDN W ++++ V GIP D+ G +G +G
Sbjct: 79 APSIKSIKEKYQDQIDIVLLNVDNEKWLDLIEKYEVNGIPQLNLFDQFGVIKGQSIG--- 135
Query: 219 RQLLLENVDAL 229
L+N+D +
Sbjct: 136 ----LKNIDQI 142
>ref|YP_003453.1| thiol-disulfide interchange like protein [Leptospira interrogans
serovar Copenhageni str. Fiocruz L1-130]
gi|45602615|gb|AAS72090.1| thiol-disulfide interchange
like protein [Leptospira interrogans serovar Copenhageni
str. Fiocruz L1-130]
Length = 357
Score = 65.9 bits (159), Expect = 1e-09
Identities = 27/96 (28%), Positives = 55/96 (57%)
Query: 131 LPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELD 190
+ +E+A S+GKP ++ YADWC C+ L ++Y ++ + FV L++D +
Sbjct: 116 IAFEKARSSGKPIFIDVYADWCSYCKTLKKEIYPKKEVQLELSKFVALSLDGDKFPNLKR 175
Query: 191 EFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENV 226
++G++G P FLD+ G+ + G +++L+++
Sbjct: 176 KYGIKGYPSILFLDRNGSLIDKITGMPDSKMILKSL 211
>ref|NP_714622.1| TPR-repeat-containing protein [Leptospira interrogans serovar Lai
str. 56601] gi|24202175|gb|AAN51637.1|
TPR-repeat-containing protein [Leptospira interrogans
serovar lai str. 56601]
Length = 357
Score = 65.9 bits (159), Expect = 1e-09
Identities = 27/96 (28%), Positives = 55/96 (57%)
Query: 131 LPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELD 190
+ +E+A S+GKP ++ YADWC C+ L ++Y ++ + FV L++D +
Sbjct: 116 IAFEKARSSGKPIFIDVYADWCSYCKTLKKEIYPKKEVQLELSKFVALSLDGDKFPNLKR 175
Query: 191 EFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENV 226
++G++G P FLD+ G+ + G +++L+++
Sbjct: 176 KYGIKGYPSILFLDRNGSLIDKITGMPDSKMILKSL 211
>gb|EAM68231.1| Thioredoxin [Jannaschia sp. CCS1]
Length = 106
Score = 63.9 bits (154), Expect = 4e-09
Identities = 34/87 (39%), Positives = 52/87 (59%), Gaps = 5/87 (5%)
Query: 142 PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVD-NTNWEQELDEFGVEGIPHF 200
P VV+F+A+WC C+++ P + ++ ++Y+ RV V LNVD N N +L GV GIP
Sbjct: 21 PVVVDFWAEWCGPCKQIGPALEELSEEYEGRVKIVKLNVDENANAPMQL---GVRGIPAL 77
Query: 201 AFLDKEGNEEGNVVGRLPRQLLLENVD 227
F+ K+G N VG P+ L + +D
Sbjct: 78 -FMFKDGEVVSNKVGAAPKAALQKWID 103
>ref|ZP_00331170.1| COG0526: Thiol-disulfide isomerase and thioredoxins [Moorella
thermoacetica ATCC 39073]
Length = 108
Score = 63.2 bits (152), Expect = 7e-09
Identities = 38/107 (35%), Positives = 57/107 (52%), Gaps = 6/107 (5%)
Query: 124 KDLTANALPYE-QALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDN 182
K + N +E + LS P VV+F+A WC CR +AP + ++ Y V F +NVD
Sbjct: 3 KPINVNQNDFEAEVLSAPLPVVVDFWAVWCGPCRMMAPVLEQLAADYDGNVKFAKVNVDE 62
Query: 183 TNWEQEL-DEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDA 228
QEL +G+ IP K+G E G +VG +P++ L E ++A
Sbjct: 63 ---NQELAARYGIMSIPTLVIF-KDGAEAGRIVGYMPKEKLQEQLEA 105
>gb|AAQ60025.2| Thiol:disulfide interchange protein DsbD [Chromobacterium violaceum
ATCC 12472] gi|34497808|ref|NP_902023.1| Thiol:disulfide
interchange protein DsbD [Chromobacterium violaceum ATCC
12472]
Length = 596
Score = 62.8 bits (151), Expect = 1e-08
Identities = 38/106 (35%), Positives = 50/106 (46%), Gaps = 11/106 (10%)
Query: 135 QALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQE----LD 190
+A + GKP +++FYADWC C+E+ DV+ +Q FV+L D T E L
Sbjct: 496 EAKAAGKPLLLDFYADWCVACKEMEADVFPQQQAAAQMQRFVLLRADVTANTPEHQALLK 555
Query: 191 EFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASV 236
FG+ G P D + E VVG P DA AR A V
Sbjct: 556 RFGLFGPPGIILFDSQSRESNRVVGYTP-------ADAFARELAKV 594
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 446,638,100
Number of Sequences: 2540612
Number of extensions: 18660422
Number of successful extensions: 49606
Number of sequences better than 10.0: 1911
Number of HSP's better than 10.0 without gapping: 1179
Number of HSP's successfully gapped in prelim test: 732
Number of HSP's that attempted gapping in prelim test: 47722
Number of HSP's gapped (non-prelim): 2386
length of query: 263
length of database: 863,360,394
effective HSP length: 125
effective length of query: 138
effective length of database: 545,783,894
effective search space: 75318177372
effective search space used: 75318177372
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0559.5


