

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0550.4
(888 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
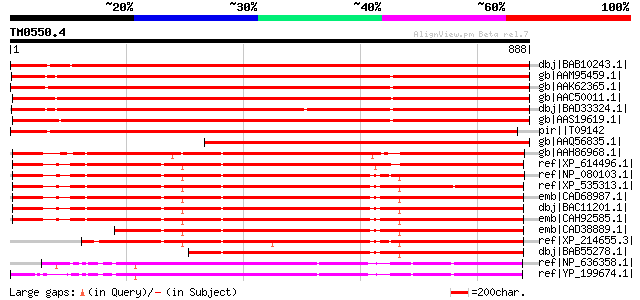
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB10243.1| mitochondrial Lon protease homolog 1 precursor [... 1425 0.0
gb|AAM95459.1| Lon protease [Oryza sativa (indica cultivar-group)] 1414 0.0
gb|AAK62365.1| Lon protease [Dichanthelium lanuginosum] 1407 0.0
gb|AAC50011.1| LON1 protease [Zea mays] gi|7428224|pir||T04321 e... 1406 0.0
dbj|BAD33324.1| putative Lon protease [Oryza sativa (japonica cu... 1394 0.0
gb|AAS19619.1| LON1 protease [Triticum aestivum] 1390 0.0
pir||T09142 endopeptidase La homolog (EC 3.4.21.-) - spinach gi|... 1380 0.0
gb|AAQ56835.1| At5g47040 [Arabidopsis thaliana] gi|27311537|gb|A... 928 0.0
gb|AAH86968.1| Zgc:92557 [Danio rerio] gi|56693217|ref|NP_001008... 704 0.0
ref|XP_614496.1| PREDICTED: similar to peroxisomal lon protease ... 692 0.0
ref|NP_080103.1| peroxisomal lon protease [Mus musculus] gi|2914... 688 0.0
ref|XP_535313.1| PREDICTED: similar to peroxisomal lon protease ... 687 0.0
emb|CAD68987.1| peroxisomal lon protease [Homo sapiens] gi|62740... 681 0.0
dbj|BAC11201.1| unnamed protein product [Homo sapiens] 680 0.0
emb|CAH92585.1| hypothetical protein [Pongo pygmaeus] 677 0.0
emb|CAD38889.1| hypothetical protein [Homo sapiens] 655 0.0
ref|XP_214655.3| PREDICTED: similar to RIKEN cDNA 1300002A08 [Ra... 644 0.0
dbj|BAB55278.1| unnamed protein product [Homo sapiens] 610 e-173
ref|NP_636358.1| ATP-dependent serine proteinase La [Xanthomonas... 585 e-165
ref|YP_199674.1| ATP-dependent serine proteinase La [Xanthomonas... 581 e-164
>dbj|BAB10243.1| mitochondrial Lon protease homolog 1 precursor [Arabidopsis
thaliana] gi|18422747|ref|NP_568675.1| Lon protease
homolog 1, mitochondrial (LON) [Arabidopsis thaliana]
gi|2935279|gb|AAC05085.1| Lon protease [Arabidopsis
thaliana] gi|3914002|sp|O64948|LONH1_ARATH Lon protease
homolog 1, mitochondrial precursor
Length = 888
Score = 1425 bits (3689), Expect = 0.0
Identities = 729/891 (81%), Positives = 807/891 (89%), Gaps = 6/891 (0%)
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRD 60
MAE+VELPSRLAILPFRNKVLLPGAIIRIRCTS +SV LVEQELWQ+E+KGLIGILPVRD
Sbjct: 1 MAETVELPSRLAILPFRNKVLLPGAIIRIRCTSHSSVTLVEQELWQKEEKGLIGILPVRD 60
Query: 61 AAETEVKPVGPTISQDG--DTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALH 118
AE +G I+ D+ ++S K ++D+ K DAK Q D+ WH RGVAARALH
Sbjct: 61 DAEGS--SIGTMINPGAGSDSGERSLKFLVGTTDAQKSDAKDQQDL-QWHTRGVAARALH 117
Query: 119 LSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFI 178
LSRGVEKPSGRVTY+VVLEGL RF+VQEL RG Y ARI+SLEM K E+EQV+ DPDF+
Sbjct: 118 LSRGVEKPSGRVTYVVVLEGLSRFNVQELGKRGPYSVARITSLEMTKAELEQVKQDPDFV 177
Query: 179 MLSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDS 238
LSRQFK TAMEL+SVLEQKQKTGGRTKVLLETVP+HKLADIFVASFE+SFEEQLSMLDS
Sbjct: 178 ALSRQFKTTAMELVSVLEQKQKTGGRTKVLLETVPIHKLADIFVASFEMSFEEQLSMLDS 237
Query: 239 VDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDD 298
VD K+RLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKE+LLRQQMRAIKEELGDNDD
Sbjct: 238 VDLKVRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEYLLRQQMRAIKEELGDNDD 297
Query: 299 DEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKA 358
DEDD+AALERKMQ+AGMP +IWKHA RELRRLKKMQPQQPGY+SSR YL+LLADLPW KA
Sbjct: 298 DEDDVAALERKMQAAGMPSNIWKHAQRELRRLKKMQPQQPGYNSSRVYLELLADLPWDKA 357
Query: 359 SEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 418
SEEHE+DLK+A+ERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL
Sbjct: 358 SEEHELDLKAAKERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 417
Query: 419 ASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEI 478
ASSIAAAL RKFVR+SLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRV VCNPVMLLDEI
Sbjct: 418 ASSIAAALGRKFVRLSLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVGVCNPVMLLDEI 477
Query: 479 DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDR 538
DKTGSDVRGDPASALLEVLDPEQNK+FNDHYLNVP+DLSKV+FVATANR Q IPPPLLDR
Sbjct: 478 DKTGSDVRGDPASALLEVLDPEQNKSFNDHYLNVPYDLSKVVFVATANRVQPIPPPLLDR 537
Query: 539 MEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNL 598
ME+IELPGYT EEKLKIAM+HLIPRVLDQHGLSSE+L+IPE MV+ +IQRYTREAGVR+L
Sbjct: 538 MELIELPGYTQEEKLKIAMRHLIPRVLDQHGLSSEFLKIPEAMVKNIIQRYTREAGVRSL 597
Query: 599 ERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDI 658
ERNLA+LARAAAV VAE EQ +PL+K+VQ LT+PLL R+ +G EVEMEVIPM VN +I
Sbjct: 598 ERNLAALARAAAVMVAEHEQSLPLSKDVQKLTSPLLNGRMAEGGEVEMEVIPMGVNDHEI 657
Query: 659 SNTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATE 718
TF+ S LVV+E +LEK+LGPP+FD EAA+RVA+ G+ VGLVWTTFGGEVQFVEAT
Sbjct: 658 GGTFQSPSALVVDETMLEKILGPPRFDDSEAADRVASAGVSVGLVWTTFGGEVQFVEATS 717
Query: 719 MVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAI 778
MVGKG++HLTGQLGDVIKESAQ+ALTWVRARA+D +LA A +N+L+GRDIHIHFPAGA+
Sbjct: 718 MVGKGEMHLTGQLGDVIKESAQLALTWVRARASDFKLALAGDMNVLDGRDIHIHFPAGAV 777
Query: 779 PKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKR 838
PKDGPSAGVTLVTALVSLFS+KRVR+DTAMTGEMTLRGLVLPVGGIKDKILAAHR GIKR
Sbjct: 778 PKDGPSAGVTLVTALVSLFSQKRVRADTAMTGEMTLRGLVLPVGGIKDKILAAHRYGIKR 837
Query: 839 VIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQ-SKL 888
VI+P+RN KDLVEVP++VL +LE++LAKRMEDVLE A EGGCPWR+ SKL
Sbjct: 838 VILPQRNSKDLVEVPAAVLSSLEVILAKRMEDVLENAFEGGCPWRNNYSKL 888
>gb|AAM95459.1| Lon protease [Oryza sativa (indica cultivar-group)]
Length = 884
Score = 1414 bits (3661), Expect = 0.0
Identities = 722/886 (81%), Positives = 795/886 (89%), Gaps = 7/886 (0%)
Query: 4 SVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRDAAE 63
+VELP RLAILPFRNKVLLPGAI+RIRCT+P+SVKLVEQELWQRE+KGLIG+LPV D+ E
Sbjct: 5 AVELPGRLAILPFRNKVLLPGAIVRIRCTNPSSVKLVEQELWQREEKGLIGVLPVHDS-E 63
Query: 64 TEVKPVGPTISQDGDTLDQSSKVHGASS-DSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+ P + D + SK G S+ +S K D K + +HWH+RGVAARALHLSRG
Sbjct: 64 AAGSLLSPGVGSDSG--EGGSKAPGGSAGESTKQDTKNGKETIHWHSRGVAARALHLSRG 121
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
VEKPSGRVTYIVVLEGLCRFSVQELS RG+YH AR+S L+M KTE+E E DPD I LSR
Sbjct: 122 VEKPSGRVTYIVVLEGLCRFSVQELSARGSYHVARVSRLDMTKTELEHAEQDPDLIALSR 181
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QFKATAMELISVLEQKQKT GRTKVLLETVPV++LADIFVASFEISFEEQLSMLDSVD K
Sbjct: 182 QFKATAMELISVLEQKQKTVGRTKVLLETVPVYRLADIFVASFEISFEEQLSMLDSVDLK 241
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDD 302
+RLSKATELVDRHLQSI VAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDD
Sbjct: 242 VRLSKATELVDRHLQSILVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDD 301
Query: 303 LAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEH 362
+AALERKMQ+AGMP +IWKHA RELRRL+KMQPQQPGYSSSR YL+LLA+LPWQK SEE
Sbjct: 302 VAALERKMQNAGMPANIWKHAQRELRRLRKMQPQQPGYSSSRTYLELLAELPWQKVSEER 361
Query: 363 EMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSI 422
E+DL++A+E LD DHYGL KVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSI
Sbjct: 362 ELDLRAAKESLDRDHYGLTKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSI 421
Query: 423 AAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTG 482
A ALDRKF+RISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRV+V NPVMLLDEIDKTG
Sbjct: 422 AKALDRKFIRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVSVSNPVMLLDEIDKTG 481
Query: 483 SDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVI 542
SDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANR Q IPPPLLDRMEVI
Sbjct: 482 SDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRMQPIPPPLLDRMEVI 541
Query: 543 ELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNL 602
ELPGYTPEEKLKIAMKHLIPRVL+QHGLSS YLQIPE MV+L+I+RYTREAGVRNLERNL
Sbjct: 542 ELPGYTPEEKLKIAMKHLIPRVLEQHGLSSTYLQIPEAMVRLIIERYTREAGVRNLERNL 601
Query: 603 ASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDISNTF 662
A+LARAAAV+VAEQ+ V+ L KE+Q +TT LL++RL DG EVEMEVIPM +DISNT+
Sbjct: 602 AALARAAAVKVAEQDSVLRLGKEIQPITTTLLDSRLADGGEVEMEVIPM---GQDISNTY 658
Query: 663 RITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATEMVGK 722
SP++V+EA+LEKVLGPP+FD EAA+RVA+PG+ VGLVWT+FGGEVQFVEAT MVGK
Sbjct: 659 ENPSPMIVDEAMLEKVLGPPRFDDSEAADRVASPGVSVGLVWTSFGGEVQFVEATAMVGK 718
Query: 723 GDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAIPKDG 782
GDLHLTGQLGDVIKESAQ+ALTWVRARAADL L+ INLLE RDIHIHFPAGA+PKDG
Sbjct: 719 GDLHLTGQLGDVIKESAQLALTWVRARAADLNLSPTSDINLLESRDIHIHFPAGAVPKDG 778
Query: 783 PSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRVIIP 842
PSAGVTLVT+LVSLFS ++VR+DTAMTGEMTLRGLVLPVGG+KDK+LAAHR GIKRVI+P
Sbjct: 779 PSAGVTLVTSLVSLFSHRKVRADTAMTGEMTLRGLVLPVGGVKDKVLAAHRYGIKRVILP 838
Query: 843 ERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQSKL 888
ERN+KDL EVP+ +L LEILL KR+E+VL+ A EGGCP R SKL
Sbjct: 839 ERNMKDLAEVPAPILSGLEILLVKRIEEVLDHAFEGGCPLRPHSKL 884
>gb|AAK62365.1| Lon protease [Dichanthelium lanuginosum]
Length = 884
Score = 1407 bits (3641), Expect = 0.0
Identities = 721/890 (81%), Positives = 794/890 (89%), Gaps = 8/890 (0%)
Query: 1 MAES-VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVR 59
MA+S VELP RLAILPFRNKVLLPGAI+RIRCT+P+SVKLVEQELWQ+E+KGLIG+LPVR
Sbjct: 1 MADSPVELPGRLAILPFRNKVLLPGAIVRIRCTNPSSVKLVEQELWQKEEKGLIGVLPVR 60
Query: 60 DAAETEVKPVGPTISQD-GDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALH 118
D+ E VG +S G + G S +S K D K + +HWH++GVAARALH
Sbjct: 61 DS---EAAAVGSLLSPGVGSDSGEGGSKAGGSGESSKQDTKNGKEPIHWHSKGVAARALH 117
Query: 119 LSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFI 178
LSRGVEKPSGRVTYIVVLEGLCRFSVQELS RG+YH AR+S L+M KTE+EQ E DPD I
Sbjct: 118 LSRGVEKPSGRVTYIVVLEGLCRFSVQELSARGSYHVARVSRLDMTKTELEQAEQDPDLI 177
Query: 179 MLSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDS 238
LSRQFKATAMELISVLEQKQKT GRTKVLL+TVPV++LADIFVASFEISFEEQLSMLDS
Sbjct: 178 ALSRQFKATAMELISVLEQKQKTVGRTKVLLDTVPVYRLADIFVASFEISFEEQLSMLDS 237
Query: 239 VDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDD 298
VD K+RLSKATELVDRHLQSI VAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDD
Sbjct: 238 VDLKVRLSKATELVDRHLQSILVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDD 297
Query: 299 DEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKA 358
DEDD+AALERKMQ+AGMP +IWKHA RELRRL+KMQPQQPGYSSSRAYL+LLADLPWQK
Sbjct: 298 DEDDVAALERKMQNAGMPANIWKHAQRELRRLRKMQPQQPGYSSSRAYLELLADLPWQKV 357
Query: 359 SEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 418
SEE E+DL++A+E LD DHYGL KVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL
Sbjct: 358 SEERELDLRAAKESLDQDHYGLPKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 417
Query: 419 ASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEI 478
ASSIA AL+RKF+RISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRV+V NPVMLLDEI
Sbjct: 418 ASSIAKALNRKFIRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVSVSNPVMLLDEI 477
Query: 479 DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDR 538
DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANR Q IPPPLLD
Sbjct: 478 DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRMQPIPPPLLDS 537
Query: 539 MEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNL 598
MEVIELPGYTPEEKLKIAMKHLIPRVL+QHGLSS YLQIPE +L+I+RYTREAGVRNL
Sbjct: 538 MEVIELPGYTPEEKLKIAMKHLIPRVLEQHGLSSAYLQIPEVRSKLIIERYTREAGVRNL 597
Query: 599 ERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDI 658
ERNLA+LARAAAV+VAEQ + L+KE+Q +TT LL++RL DG EVEMEVIPM DI
Sbjct: 598 ERNLAALARAAAVKVAEQVNTLRLSKEIQPITTTLLDSRLADGGEVEMEVIPM---GHDI 654
Query: 659 SNTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATE 718
SNT+ SP++V+E +LEKVLGPP+FD REAA+RVA+PG+ VGLVWT+FGGEVQFVEAT
Sbjct: 655 SNTYENPSPMIVDETMLEKVLGPPRFDDREAADRVASPGVSVGLVWTSFGGEVQFVEATA 714
Query: 719 MVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAI 778
MVGKGDLHLTGQLGDVIKESAQ+ALTWVRARAADL L+ INLLE RDIHIHFPAGA+
Sbjct: 715 MVGKGDLHLTGQLGDVIKESAQLALTWVRARAADLNLSPTSDINLLESRDIHIHFPAGAV 774
Query: 779 PKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKR 838
PKDGPSAGVTLVT+LVSLFS ++VR+DTAMTGEMTLRGLVLPVGG+KDK+LAAHR GIKR
Sbjct: 775 PKDGPSAGVTLVTSLVSLFSNRKVRADTAMTGEMTLRGLVLPVGGVKDKVLAAHRYGIKR 834
Query: 839 VIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQSKL 888
VI+PERNLKDL EVPS +L +EILL KR+E+VL+ A EGGC R +SKL
Sbjct: 835 VILPERNLKDLTEVPSPILSGMEILLVKRIEEVLDHAFEGGCLLRSRSKL 884
>gb|AAC50011.1| LON1 protease [Zea mays] gi|7428224|pir||T04321 endopeptidase La
homolog (EC 3.4.21.-) LON1 precursor, mitochondrial -
maize gi|3914005|sp|P93647|LONH1_MAIZE Lon protease
homolog 1, mitochondrial precursor
Length = 885
Score = 1406 bits (3639), Expect = 0.0
Identities = 717/885 (81%), Positives = 794/885 (89%), Gaps = 6/885 (0%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRDAAET 64
VELPSRLA+LPFRNKVLLPGAI+RIRCT+P+SVKLVEQELWQ+E+KGLIG+LPVRD+ T
Sbjct: 6 VELPSRLAVLPFRNKVLLPGAIVRIRCTNPSSVKLVEQELWQKEEKGLIGVLPVRDSEAT 65
Query: 65 EVKPV-GPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRGV 123
V + P + D + SKV G++ +S K D K + +HWH++GVAARALHLSRGV
Sbjct: 66 AVGSLLSPGVGSDSG--EGGSKVGGSAVESSKQDTKNGKEPIHWHSKGVAARALHLSRGV 123
Query: 124 EKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSRQ 183
EKPSGRVTYIVVLEGLCRFSVQELS RG YH AR+S L+M KTE+EQ E DPD I LSRQ
Sbjct: 124 EKPSGRVTYIVVLEGLCRFSVQELSARGPYHVARVSRLDMTKTELEQAEQDPDLIALSRQ 183
Query: 184 FKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAKL 243
FKATAMELISVLEQKQKT GRTKVLL+TVPV++LADIFVASFEISFEEQLSMLDSV K+
Sbjct: 184 FKATAMELISVLEQKQKTVGRTKVLLDTVPVYRLADIFVASFEISFEEQLSMLDSVHLKV 243
Query: 244 RLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDDL 303
RLSKATELVDRHLQSI VAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDD+
Sbjct: 244 RLSKATELVDRHLQSILVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDDV 303
Query: 304 AALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHE 363
AALERKMQ+AGMP +IWKHA RE+RRL+KMQPQQPGYSSSRAYL+LLADLPWQK SEE E
Sbjct: 304 AALERKMQNAGMPANIWKHAQREMRRLRKMQPQQPGYSSSRAYLELLADLPWQKVSEERE 363
Query: 364 MDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIA 423
+DL+ A+E LD DHYGL KVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIA
Sbjct: 364 LDLRVAKESLDQDHYGLTKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIA 423
Query: 424 AALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGS 483
AL+RKF+RISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRV+V NPVMLLDEIDKTGS
Sbjct: 424 KALNRKFIRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVSVSNPVMLLDEIDKTGS 483
Query: 484 DVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIE 543
DVRGDPASALLEVLDPEQNK FNDHYLNVPFDLSKVIFVATANR Q IPPPLLDRME+IE
Sbjct: 484 DVRGDPASALLEVLDPEQNKAFNDHYLNVPFDLSKVIFVATANRMQPIPPPLLDRMEIIE 543
Query: 544 LPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLA 603
LPGYTPEEKLKIAMKHLIPRVL+QHGLS+ LQIPE MV+LVI+RYTREAGVRNLERNLA
Sbjct: 544 LPGYTPEEKLKIAMKHLIPRVLEQHGLSTTNLQIPEAMVKLVIERYTREAGVRNLERNLA 603
Query: 604 SLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDISNTFR 663
+LARAAAV+VAEQ + + L KE+Q +TT LL++RL DG EVEMEVIPM+ DISNT+
Sbjct: 604 ALARAAAVKVAEQVKTLRLGKEIQPITTTLLDSRLADGGEVEMEVIPME---HDISNTYE 660
Query: 664 ITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATEMVGKG 723
SP++V+EA+LEKVLGPP+FD REAA+RVA+PG+ VGLVWT+ GGEVQFVEAT MVGKG
Sbjct: 661 NPSPMIVDEAMLEKVLGPPRFDDREAADRVASPGVSVGLVWTSVGGEVQFVEATAMVGKG 720
Query: 724 DLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAIPKDGP 783
DLHLTGQLGDVIKESAQ+ALTWVRARAADL L+ INLLE RDIHIHFPAGA+PKDGP
Sbjct: 721 DLHLTGQLGDVIKESAQLALTWVRARAADLNLSPTSDINLLESRDIHIHFPAGAVPKDGP 780
Query: 784 SAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRVIIPE 843
SAGVTLVTALVSLFS ++VR+DTAMTGEMTLRGLVLPVGG+KDK+LAAHR GIKRVI+PE
Sbjct: 781 SAGVTLVTALVSLFSNRKVRADTAMTGEMTLRGLVLPVGGVKDKVLAAHRYGIKRVILPE 840
Query: 844 RNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQSKL 888
RNLKDL EVP +L ++EILL KR+E+VL+ A EG CP R +SKL
Sbjct: 841 RNLKDLSEVPLPILSDMEILLVKRIEEVLDHAFEGRCPLRSRSKL 885
>dbj|BAD33324.1| putative Lon protease [Oryza sativa (japonica cultivar-group)]
gi|52075953|dbj|BAD46033.1| putative Lon protease [Oryza
sativa (japonica cultivar-group)]
Length = 880
Score = 1394 bits (3607), Expect = 0.0
Identities = 715/886 (80%), Positives = 789/886 (88%), Gaps = 11/886 (1%)
Query: 4 SVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRDAAE 63
+VELP RLAILPFRNKVLLPGAI+RIRCT+P+SVKLVEQELWQRE+KGLIG+LPV D+ E
Sbjct: 5 AVELPGRLAILPFRNKVLLPGAIVRIRCTNPSSVKLVEQELWQREEKGLIGVLPVHDS-E 63
Query: 64 TEVKPVGPTISQDGDTLDQSSKVHGASS-DSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+ P + D + SK G S+ +S K D K + +HWH+RGVAARALHLSRG
Sbjct: 64 AAGSLLSPGVGSDSG--EGGSKAPGGSAGESTKQDTKNGKETIHWHSRGVAARALHLSRG 121
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
VEKPSGRVTYIVVLEGLCRFSVQELS RG+YH AR+S L+M KTE+E E DPD I LSR
Sbjct: 122 VEKPSGRVTYIVVLEGLCRFSVQELSARGSYHVARVSRLDMTKTELEHAEQDPDLIALSR 181
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QFKATAMELISVLEQKQKT GRTKVLLETVPV++LADIFVASFEI FEEQLSMLDSVD K
Sbjct: 182 QFKATAMELISVLEQKQKTVGRTKVLLETVPVYRLADIFVASFEIGFEEQLSMLDSVDLK 241
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDD 302
+RLSKATELVDRHLQSI VAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDD
Sbjct: 242 VRLSKATELVDRHLQSILVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDD 301
Query: 303 LAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEH 362
+AALERKMQ+AGMP +IWKHA RELRRL+KMQPQQPGYSSSR YL+LLA+LPWQK SEE
Sbjct: 302 VAALERKMQNAGMPANIWKHAQRELRRLRKMQPQQPGYSSSRTYLELLAELPWQKVSEER 361
Query: 363 EMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSI 422
E+DL++A+E LD DHYGL KVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSI
Sbjct: 362 ELDLRAAKESLDRDHYGLTKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSI 421
Query: 423 AAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTG 482
A AL+RKF+RISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRV+V NPVMLLDEIDKTG
Sbjct: 422 AKALNRKFIRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVSVSNPVMLLDEIDKTG 481
Query: 483 SDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVI 542
SDVRGDPASALLEVLDPEQNKT YLNVPFDLSKVIFVATANR Q IPPPLLDRMEVI
Sbjct: 482 SDVRGDPASALLEVLDPEQNKT----YLNVPFDLSKVIFVATANRMQPIPPPLLDRMEVI 537
Query: 543 ELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNL 602
ELPGYTPEEKLKIAMKHLIPRVL+QHGLSS YLQIPE MV+L+I+RYTREAGVRNLERNL
Sbjct: 538 ELPGYTPEEKLKIAMKHLIPRVLEQHGLSSTYLQIPEAMVRLIIERYTREAGVRNLERNL 597
Query: 603 ASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDISNTF 662
A+LARAAAV+VAEQ+ + L KE+Q +TT LL++RL DG EVEMEVIPM +DISNT+
Sbjct: 598 AALARAAAVKVAEQDSALRLGKEIQPITTTLLDSRLADGGEVEMEVIPM---GQDISNTY 654
Query: 663 RITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATEMVGK 722
SP++V+EA+LEKVLGPP+FD EAA+RVA+PG+ VGLVWT+FGGEVQFVEAT MVGK
Sbjct: 655 ENPSPMIVDEAMLEKVLGPPRFDDSEAADRVASPGVSVGLVWTSFGGEVQFVEATAMVGK 714
Query: 723 GDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAIPKDG 782
GDLHLTGQLGDVIKESAQ+ALTWVRARAADL L+ INLLE RDIHIHFPAGA+PKDG
Sbjct: 715 GDLHLTGQLGDVIKESAQLALTWVRARAADLNLSPTSDINLLESRDIHIHFPAGAVPKDG 774
Query: 783 PSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRVIIP 842
PSAGVTLVT+LVSLFS ++VR+DTAMTGEMTLRGLVLPVGG+KDK+LAAHR GIKRVI+P
Sbjct: 775 PSAGVTLVTSLVSLFSHRKVRADTAMTGEMTLRGLVLPVGGVKDKVLAAHRYGIKRVILP 834
Query: 843 ERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQSKL 888
ERN+KDL EVP+ +L LEILL KR+E+VL+ A EGGCP R SKL
Sbjct: 835 ERNMKDLAEVPAPILSGLEILLVKRIEEVLDHAFEGGCPLRPHSKL 880
>gb|AAS19619.1| LON1 protease [Triticum aestivum]
Length = 886
Score = 1390 bits (3598), Expect = 0.0
Identities = 705/885 (79%), Positives = 789/885 (88%), Gaps = 5/885 (0%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRDAAET 64
VELP RLAILPFRNKVLLPGAI+RIRCT P+SVKLVEQELWQRED+GLIG+LPVRD+
Sbjct: 6 VELPGRLAILPFRNKVLLPGAIVRIRCTYPSSVKLVEQELWQREDRGLIGVLPVRDSEAA 65
Query: 65 EVKPV-GPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRGV 123
V + P + D + S G+ +S K DAK+ + +HWH+RGVAARALHLSRGV
Sbjct: 66 AVGSILSPGVGSDSGDGGRRSP-GGSGGESTKQDAKSGKEPIHWHSRGVAARALHLSRGV 124
Query: 124 EKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSRQ 183
EKPSGRVTYIVVLEGLCRFSV+EL+ RG+YH AR+S L+M KTE+EQ E DPD I LSRQ
Sbjct: 125 EKPSGRVTYIVVLEGLCRFSVEELNARGSYHVARVSRLDMTKTELEQAEQDPDLIALSRQ 184
Query: 184 FKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAKL 243
FKATAMELISVLEQKQKT GRTKVLLETVPV++LADIFVASFEISFEEQL+MLDSVD K+
Sbjct: 185 FKATAMELISVLEQKQKTVGRTKVLLETVPVYRLADIFVASFEISFEEQLAMLDSVDLKV 244
Query: 244 RLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDDL 303
RLSKATELVDRHLQSI VAEKITQKVEGQLSKSQKEFLLRQQMRAIK+ELGDNDDDEDD+
Sbjct: 245 RLSKATELVDRHLQSILVAEKITQKVEGQLSKSQKEFLLRQQMRAIKDELGDNDDDEDDI 304
Query: 304 AALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHE 363
AALERKMQ+AGMP +IWKHA RELRRL+KMQPQQPGYSSSRAYL+L+ADLPWQK SEE E
Sbjct: 305 AALERKMQNAGMPANIWKHAQRELRRLRKMQPQQPGYSSSRAYLELIADLPWQKVSEERE 364
Query: 364 MDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIA 423
+DL++A+E LD DHYGL KVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLA+SIA
Sbjct: 365 LDLRAAKESLDRDHYGLTKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLATSIA 424
Query: 424 AALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGS 483
AL+RKF+RISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRV+V NPVMLLDEIDKTGS
Sbjct: 425 KALNRKFIRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVSVNNPVMLLDEIDKTGS 484
Query: 484 DVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIE 543
DVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKV+FVATANR Q IPP LLDRMEVIE
Sbjct: 485 DVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVVFVATANRMQPIPPALLDRMEVIE 544
Query: 544 LPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLA 603
LPGYTPEEKLKIAMKHL+PRVL+QHGLSS YLQIPE +V+L+I+RYTREAGVRNLERNLA
Sbjct: 545 LPGYTPEEKLKIAMKHLLPRVLEQHGLSSAYLQIPEAVVKLIIERYTREAGVRNLERNLA 604
Query: 604 SLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDISNTFR 663
+LARAAAV+VAE + + L KE+Q +TT LL++RL DG EVEMEVIPM +DISNT+
Sbjct: 605 ALARAAAVKVAELDSTLRLGKEMQPITTTLLDSRLADGGEVEMEVIPM---GQDISNTYE 661
Query: 664 ITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATEMVGKG 723
SP++V+EA+LEKVLGPP+FD REAA+RV++PG+ VGLVWT+FGGEVQFVEAT M GKG
Sbjct: 662 NPSPMIVDEAMLEKVLGPPRFDDREAADRVSSPGVSVGLVWTSFGGEVQFVEATAMAGKG 721
Query: 724 DLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAIPKDGP 783
DLHLTGQLGDVIKESAQ+ALTWVRAR+ADL L+ IN+LE RDIHIHFPAGA+PKDGP
Sbjct: 722 DLHLTGQLGDVIKESAQLALTWVRARSADLNLSPTSDINILESRDIHIHFPAGAVPKDGP 781
Query: 784 SAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRVIIPE 843
SAGVTLVT+LVSLFS ++VR+DTAMTGEMTLRGLVLPVGG+KDK+LAAHR GIKRVI+PE
Sbjct: 782 SAGVTLVTSLVSLFSNRKVRADTAMTGEMTLRGLVLPVGGVKDKVLAAHRYGIKRVILPE 841
Query: 844 RNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQSKL 888
RNLKDL E+P+ +L +EILL KR+E+VL A E G P R S L
Sbjct: 842 RNLKDLAEIPAPILAGIEILLVKRIEEVLGHAFENGFPLRLHSSL 886
>pir||T09142 endopeptidase La homolog (EC 3.4.21.-) - spinach
gi|3913996|sp|O04979|LONH1_SPIOL Lon protease homolog 1,
mitochondrial precursor gi|2208927|dbj|BAA20482.1|
ATP-dependent protease Lon [Spinacia oleracea]
Length = 875
Score = 1380 bits (3572), Expect = 0.0
Identities = 697/871 (80%), Positives = 783/871 (89%), Gaps = 4/871 (0%)
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRD 60
MAE+VELPSRL IL FRNKVLLPGAIIRIRCTSP+SVKLVEQELWQRE+KGLIGI+PVRD
Sbjct: 1 MAEAVELPSRLGILAFRNKVLLPGAIIRIRCTSPSSVKLVEQELWQREEKGLIGIVPVRD 60
Query: 61 AAETEVKPVGPTISQDG--DTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALH 118
A+E+ V P + G D+ +++ K SDS K D K+Q + VHWH RGVAARALH
Sbjct: 61 ASESA--SVAPVLYPGGGTDSGERNVKSQPGLSDSRKADGKSQQEAVHWHTRGVAARALH 118
Query: 119 LSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFI 178
LSRGVEKPSGRVTY VVLEGLCRF V EL++RG Y+TARIS L++ K +MEQ + DPDF+
Sbjct: 119 LSRGVEKPSGRVTYTVVLEGLCRFRVMELNSRGNYYTARISPLDITKADMEQAQQDPDFV 178
Query: 179 MLSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDS 238
L+RQFK TA+ELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQL MLDS
Sbjct: 179 SLARQFKVTAVELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLCMLDS 238
Query: 239 VDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDD 298
+D K+RLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQ+EFLLRQQMRAIKEELGDNDD
Sbjct: 239 IDLKVRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQREFLLRQQMRAIKEELGDNDD 298
Query: 299 DEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKA 358
DEDD+A LERKMQSAGMP +IWKHA RELRRLKKMQPQQPGYSSSR YL+LLADLPWQ A
Sbjct: 299 DEDDVAVLERKMQSAGMPANIWKHAQRELRRLKKMQPQQPGYSSSRVYLELLADLPWQNA 358
Query: 359 SEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 418
+EE ++DL++A+ERLDSDHYGL KVKQRIIEYLAVRKLKPDARGP+LCFVGPPGVGKTSL
Sbjct: 359 TEEQKLDLRAAKERLDSDHYGLVKVKQRIIEYLAVRKLKPDARGPILCFVGPPGVGKTSL 418
Query: 419 ASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEI 478
A+SI+AAL RKF+RISLGGVKDEADIRGHRRTYIGSMPGRLIDG+KRV V NPVMLLDEI
Sbjct: 419 AASISAALGRKFIRISLGGVKDEADIRGHRRTYIGSMPGRLIDGIKRVGVSNPVMLLDEI 478
Query: 479 DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDR 538
DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVP+DLSKVIFVATAN+ Q IPPPLLDR
Sbjct: 479 DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPYDLSKVIFVATANKVQPIPPPLLDR 538
Query: 539 MEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNL 598
MEVIELPGYTPEEK +IAM++LIPRV+DQHGLSSE+LQI E MV+L+IQRYTREAGVRNL
Sbjct: 539 MEVIELPGYTPEEKARIAMQYLIPRVMDQHGLSSEFLQISEDMVKLIIQRYTREAGVRNL 598
Query: 599 ERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDI 658
ERNL++LARAAAV+VAEQ+ ++K+ T+P+ E+RL +GAEVEMEVIPM V++R+I
Sbjct: 599 ERNLSALARAAAVKVAEQDNATAVSKDFHQFTSPVEESRLAEGAEVEMEVIPMGVDNREI 658
Query: 659 SNTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATE 718
SN ++ SPL+V+E +LE VLGPP++D RE AERV+ PG+ VGLVWT FGGEVQFVEA+
Sbjct: 659 SNALQVMSPLIVDETMLENVLGPPRYDDRETAERVSNPGVSVGLVWTAFGGEVQFVEASV 718
Query: 719 MVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAI 778
M GKG+L LTGQLGDVIKESAQIALTWVRARA +L L + INL+EGRDIHIHFPAGA+
Sbjct: 719 MAGKGELRLTGQLGDVIKESAQIALTWVRARAMELNLVATGEINLMEGRDIHIHFPAGAV 778
Query: 779 PKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKR 838
PKDGPSAGVTLVTALVSL S+KR+R+DTAMTGEMTLRGLVLPVGG+KDK+LAAHR GIKR
Sbjct: 779 PKDGPSAGVTLVTALVSLLSQKRMRADTAMTGEMTLRGLVLPVGGVKDKVLAAHRYGIKR 838
Query: 839 VIIPERNLKDLVEVPSSVLVNLEILLAKRME 869
VI+PERNLKDLVEVPS+VL NLEI+ AKRME
Sbjct: 839 VILPERNLKDLVEVPSAVLSNLEIIYAKRME 869
>gb|AAQ56835.1| At5g47040 [Arabidopsis thaliana] gi|27311537|gb|AAO00734.1| Lon
protease homolog 1 precursor [Arabidopsis thaliana]
Length = 557
Score = 928 bits (2399), Expect = 0.0
Identities = 464/557 (83%), Positives = 515/557 (92%), Gaps = 1/557 (0%)
Query: 333 MQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLA 392
MQPQQPGY+SSR YL+LLADLPW KASEEHE+DLK+A+ERLDSDHYGLAKVKQRIIEYLA
Sbjct: 1 MQPQQPGYNSSRVYLELLADLPWDKASEEHELDLKAAKERLDSDHYGLAKVKQRIIEYLA 60
Query: 393 VRKLKPDARGPVLCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYI 452
VRKLKPDARGPVLCFVGPPGVGKTSLASSIAAAL RKFVR+SLGGVKDEADIRGHRRTYI
Sbjct: 61 VRKLKPDARGPVLCFVGPPGVGKTSLASSIAAALGRKFVRLSLGGVKDEADIRGHRRTYI 120
Query: 453 GSMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNV 512
GSMPGRLIDGLKRV VCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNK+FNDHYLNV
Sbjct: 121 GSMPGRLIDGLKRVGVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKSFNDHYLNV 180
Query: 513 PFDLSKVIFVATANRAQQIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSS 572
P+DLSKV+FVATANR Q IPPPLLDRME+IELPGYT EEKLKIAM+HLIPRVLDQHGLSS
Sbjct: 181 PYDLSKVVFVATANRVQPIPPPLLDRMELIELPGYTQEEKLKIAMRHLIPRVLDQHGLSS 240
Query: 573 EYLQIPEGMVQLVIQRYTREAGVRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTP 632
E+L+IPE MV+ +IQRYTREAGVR+LERNLA+LARAAAV VAE EQ +PL+K+VQ LT+P
Sbjct: 241 EFLKIPEAMVKNIIQRYTREAGVRSLERNLAALARAAAVMVAEHEQSLPLSKDVQKLTSP 300
Query: 633 LLENRLTDGAEVEMEVIPMDVNSRDISNTFRITSPLVVNEAILEKVLGPPKFDGREAAER 692
LL R+ +G EVEMEVIPM VN +I TF+ S LVV+E +LEK+LGPP+FD EAA+R
Sbjct: 301 LLNGRMAEGGEVEMEVIPMGVNDHEIGGTFQSPSALVVDETMLEKILGPPRFDDSEAADR 360
Query: 693 VAAPGICVGLVWTTFGGEVQFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAAD 752
VA+ G+ VGLVWTTFGGEVQFVEAT MVGKG++HLTGQLGDVIKESAQ+ALTWVRARA+D
Sbjct: 361 VASAGVSVGLVWTTFGGEVQFVEATSMVGKGEMHLTGQLGDVIKESAQLALTWVRARASD 420
Query: 753 LRLASAEGINLLEGRDIHIHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEM 812
+LA A +N+L+GRDIHIHFPAGA+PKDGPSAGVTLVTALVSLFS+KRVR+DTAMTGEM
Sbjct: 421 FKLALAGDMNVLDGRDIHIHFPAGAVPKDGPSAGVTLVTALVSLFSQKRVRADTAMTGEM 480
Query: 813 TLRGLVLPVGGIKDKILAAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVL 872
TLRGLVLPVGGIKDKILAAHR GIKRVI+P+RN KDLVEVP++VL +LE++LAKRMEDVL
Sbjct: 481 TLRGLVLPVGGIKDKILAAHRYGIKRVILPQRNSKDLVEVPAAVLSSLEVILAKRMEDVL 540
Query: 873 EQALEGGCPWRHQ-SKL 888
E A EGGCPWR+ SKL
Sbjct: 541 ENAFEGGCPWRNNYSKL 557
>gb|AAH86968.1| Zgc:92557 [Danio rerio] gi|56693217|ref|NP_001008573.1|
hypothetical protein LOC494030 [Danio rerio]
Length = 840
Score = 704 bits (1817), Expect = 0.0
Identities = 389/887 (43%), Positives = 563/887 (62%), Gaps = 75/887 (8%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRED--KGLIGILPVRDAA 62
+++PSRL +L + VLLPG+ +R+ + +++LV+ L + +IG++P
Sbjct: 7 IQIPSRLPLLCTHDGVLLPGSTMRVSVDTARNMQLVKSRLLKGTSLKSTIIGVIP----- 61
Query: 63 ETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
DS +L + H+ G A A+ + G
Sbjct: 62 ----------------------NTRDPEHDSDELPSL--------HSIGTAGLAVQVV-G 90
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
P Y +++ GLCRF V +L + A + L+ + E D + LS+
Sbjct: 91 SNWPKPH--YTLLITGLCRFRVSQLLRERPFPVAEVEQLDKLEQYTEGDPADGELGELSQ 148
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
+F A++L+ +L+ + + LL+++P L D+ A S +E+L +LD+VD +
Sbjct: 149 RFYQAAVQLVGMLDMSVPVVAKLRRLLDSLPKETLPDVLAAMIRTSNKEKLQVLDAVDLE 208
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKS----QKEFLLRQQMRAIKEELGDNDD 298
R KA L+ R ++ +++ +K T+K+ K +K + + ++ EE+ D+
Sbjct: 209 ERFKKALPLLTRQIEGLKLLQK-TRKLRPDDDKRVLSIRKGGVFPGRQFSLDEEV--EDE 265
Query: 299 DEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKA 358
D DD A LERK+++A MP++ + +ELRRLKKM P Y+ +R YL+++ +LPW K+
Sbjct: 266 DSDDTALLERKVKAAAMPEAALRVCLKELRRLKKMPQSMPEYALTRNYLEMMVELPWSKS 325
Query: 359 SEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSL 418
+ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK +GP+LCFVGPPGVGKTS+
Sbjct: 326 TTDC-LDIRAARVLLDNDHYAMEKLKKRVLEYLAVRQLKSTLKGPILCFVGPPGVGKTSV 384
Query: 419 ASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEI 478
SIA L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE+
Sbjct: 385 GRSIARTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDEV 444
Query: 479 DKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDR 538
DK G ++GDPA+ALLEVLDPEQN +F DHYLNVPFDLS+V+F+ATAN IPP LLDR
Sbjct: 445 DKLGKSLQGDPAAALLEVLDPEQNHSFTDHYLNVPFDLSQVLFIATANTTATIPPALLDR 504
Query: 539 MEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNL 598
MEV+++PGYT EEK++IA +HLIP L+QHGL+ + LQIP+ +I +YTREAGVR+L
Sbjct: 505 MEVLQVPGYTQEEKVEIAHRHLIPHQLEQHGLTPQQLQIPQDTTLQIISKYTREAGVRSL 564
Query: 599 ERNLASLARAAAVRVAEQEQV----VPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVN 654
ER + ++ RA AV+VAE ++V P + + T +E+ G E+
Sbjct: 565 ERKIGAVCRAVAVKVAEGQKVSRSEAPTEQHAEQNTDSKVED---SGIAAPPEM------ 615
Query: 655 SRDISNTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFV 714
P+V++ L+ +LGPP F+ E +ER+ PG+ +GL WT GGE+ FV
Sbjct: 616 ------------PIVIDHVALKDILGPPLFE-MEVSERLTLPGVAIGLAWTPMGGEIMFV 662
Query: 715 EATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFP 774
EA+ M G+G L LTGQLGDV+KESA +A++W+R+ A L +LLEG DIH+HFP
Sbjct: 663 EASRMEGEGQLTLTGQLGDVMKESAHLAISWLRSNAKTYLLNDGSA-DLLEGTDIHLHFP 721
Query: 775 AGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRC 834
AGA+ KDGPSAGVT+VT L SL S + VRSD AMTGE+TLRGLVLPVGGIKDK+LAAHR
Sbjct: 722 AGAVTKDGPSAGVTIVTCLASLLSGRLVRSDVAMTGEITLRGLVLPVGGIKDKVLAAHRA 781
Query: 835 GIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCP 881
+KR+IIP+RN KDL E+P++V +L+ +LA +++VL A +GG P
Sbjct: 782 NLKRIIIPKRNEKDLEEIPANVRADLDFVLAGTLDEVLNAAFDGGFP 828
>ref|XP_614496.1| PREDICTED: similar to peroxisomal lon protease [Bos taurus]
Length = 852
Score = 692 bits (1786), Expect = 0.0
Identities = 386/889 (43%), Positives = 560/889 (62%), Gaps = 71/889 (7%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRED--KGLIGILPVRDAA 62
+++PSRL +L VLLPG+ +R S +++LV L + ++G++P
Sbjct: 7 IQIPSRLPLLLTHEGVLLPGSTMRTSVDSARNLQLVRSRLLKGTSLQSTILGVIP----- 61
Query: 63 ETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+T D +S DA+ D+ H G AA A+ + G
Sbjct: 62 ---------------NTPDPAS------------DAQ---DLPPLHRIGTAALAVQVV-G 90
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
P Y +++ GLCRF + ++ Y A + L+ + + + LS
Sbjct: 91 SNWPKPH--YTLLITGLCRFQITQVVREKPYPVAEVEQLDRLEEFPNTCKTREELGELSE 148
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QF A++L+ +L+ + + LL+++P L DI + S +E+L +LD+V +
Sbjct: 149 QFYKYAVQLVEMLDMSVPAVAKLRRLLDSLPREALPDILTSIIRTSNKEKLQILDAVSLE 208
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG-----DND 297
R L+ R ++ + K+ QK K + + MR I G D D
Sbjct: 209 ERFKMTIPLLVRQIEGL----KLLQKTRKHKQDDDKRVIAIRPMRRITHVPGALADEDED 264
Query: 298 DDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQK 357
+D DD+ LE+K++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K
Sbjct: 265 EDNDDIVMLEKKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNK 324
Query: 358 ASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTS 417
++ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVGKTS
Sbjct: 325 STTDR-LDIRAARVLLDNDHYAMEKLKKRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTS 383
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDE 477
+ S+A L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE
Sbjct: 384 VGRSVAKTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDE 443
Query: 478 IDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLD 537
+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IPP LLD
Sbjct: 444 VDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANTTASIPPALLD 503
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRN 597
RME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAGVR+
Sbjct: 504 RMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQVTTLDIITRYTREAGVRS 563
Query: 598 LERNLASLARAAAVRVAE-QEQVVPLNK----EVQGLTTPLLENRLTDGAEVEMEV-IPM 651
L+R L ++ RA AV+VAE Q + L++ E +G LLE+ +D ++ +P
Sbjct: 564 LDRKLGAICRAVAVKVAEGQHREAKLDRPDVAEGEGCKEHLLEDGKSDPVSDTTDLALPP 623
Query: 652 DVNSRDISNTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEV 711
++ P++++ L+ +LGPP ++ E +ER++ PG+ +GL WT GGE+
Sbjct: 624 EM-------------PILIDFHALKDILGPPMYE-MEVSERLSQPGVAIGLAWTPLGGEI 669
Query: 712 QFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-INLLEGRDIH 770
FVEA+ M G+G L LTGQLG+V+KESA +A++W+R+ A L +A G +LLE DIH
Sbjct: 670 MFVEASRMDGEGQLTLTGQLGNVMKESAHLAISWLRSNAKKYHLTNASGSFDLLENTDIH 729
Query: 771 IHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILA 830
+HFPAGA+ KDGPSAGVT+ T L SLFS + VRSD AMTGE+TLRGLVLPVGGIKDK LA
Sbjct: 730 LHFPAGAVTKDGPSAGVTIATCLASLFSGRLVRSDVAMTGEITLRGLVLPVGGIKDKALA 789
Query: 831 AHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
AHR G+KRVIIP+RN KDL E+P++V +L + A +++VL A +GG
Sbjct: 790 AHRAGLKRVIIPQRNEKDLEEIPANVRQDLSFITASCLDEVLNAAFDGG 838
>ref|NP_080103.1| peroxisomal lon protease [Mus musculus] gi|29144996|gb|AAH49090.1|
RIKEN cDNA 1300002A08 [Mus musculus]
gi|26340950|dbj|BAC34137.1| unnamed protein product [Mus
musculus] gi|12836332|dbj|BAB23609.1| unnamed protein
product [Mus musculus]
Length = 852
Score = 688 bits (1776), Expect = 0.0
Identities = 380/889 (42%), Positives = 557/889 (61%), Gaps = 67/889 (7%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRED--KGLIGILPVRDAA 62
+++PSRL +L VLLPG+ +R + +++LV L + ++G++P
Sbjct: 7 IQIPSRLPLLLTHESVLLPGSTMRTSVDTARNLQLVRSRLLKGTSLQSTILGVIP----- 61
Query: 63 ETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+T D +S D+ H G AA A+ + G
Sbjct: 62 ---------------NTPDPASDTQ---------------DLPPLHRIGTAALAVQVV-G 90
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
P Y +++ GLCRF + ++ Y A + L+ + + + LS
Sbjct: 91 SNWPKPH--YTLLITGLCRFQIVQVLKEKPYPVAEVEQLDRLEEFPNICKSREELGELSE 148
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QF A++L+ +L+ + + LL+ +P L DI + S +E+L +LD+V +
Sbjct: 149 QFYRYAVQLVEMLDMSVPAVAKLRRLLDNLPREALPDILTSIIRTSNKEKLQILDAVSLE 208
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG-----DND 297
R L+ R ++ + K+ QK K + + +R I G + +
Sbjct: 209 DRFKMTIPLLVRQIEGL----KLLQKTRKPKQDDDKRVIAIRPIRRIPHIPGTLEDEEEE 264
Query: 298 DDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQK 357
+D DD+ LE+K++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K
Sbjct: 265 EDNDDIVMLEKKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNK 324
Query: 358 ASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTS 417
++ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVGKTS
Sbjct: 325 STTDR-LDIRAARILLDNDHYAMEKLKRRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTS 383
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDE 477
+ S+A L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE
Sbjct: 384 VGRSVAKTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDE 443
Query: 478 IDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLD 537
+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IPP LLD
Sbjct: 444 VDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANTTATIPPALLD 503
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRN 597
RME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAGVR+
Sbjct: 504 RMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQHTTLAIITRYTREAGVRS 563
Query: 598 LERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRD 657
L+R ++ RA AV+VAE + +KE + L + + DG + V+ D
Sbjct: 564 LDRKFGAICRAVAVKVAEGQ-----HKEAK-----LDRSDVADGEGCKEHVLE-DAKPES 612
Query: 658 ISNTFRIT----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQF 713
IS+T + P++++ L+ +LGPP ++ E +ER++ PG+ +GL WT GG++ F
Sbjct: 613 ISDTADLALPPEMPILIDSHALKDILGPPLYE-LEVSERLSQPGVAIGLAWTPLGGKIMF 671
Query: 714 VEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-INLLEGRDIHIH 772
VEA+ M G+G L LTGQLGDV+KESA +A++W+R+ A L +A G +LL+ DIH+H
Sbjct: 672 VEASRMDGEGQLTLTGQLGDVMKESAHLAISWLRSNAKKYHLTNAFGSFDLLDNTDIHLH 731
Query: 773 FPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAH 832
FPAGA+ KDGPSAGVT+VT L SLFS + VRSD AMTGE+TLRGLVLPVGGIKDK+LAAH
Sbjct: 732 FPAGAVTKDGPSAGVTIVTCLASLFSGRLVRSDVAMTGEITLRGLVLPVGGIKDKVLAAH 791
Query: 833 RCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCP 881
R G+K++IIP+RN KDL E+PS+V +L + A +++VL A +GG P
Sbjct: 792 RAGLKQIIIPQRNEKDLEEIPSNVRQDLSFVTASCLDEVLNAAFDGGFP 840
>ref|XP_535313.1| PREDICTED: similar to peroxisomal lon protease [Canis familiaris]
Length = 850
Score = 687 bits (1773), Expect = 0.0
Identities = 384/886 (43%), Positives = 554/886 (62%), Gaps = 67/886 (7%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRED--KGLIGILPVRDAA 62
+++P RL +L VLLPG+ +R S +++LV L + ++G++P
Sbjct: 7 IQIPRRLPLLLTNEGVLLPGSTMRTSVDSARNLQLVRSRLLKGTSLQSTILGVIP----- 61
Query: 63 ETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+T D +S DA+ D+ H G AA A+ + G
Sbjct: 62 ---------------NTPDPAS------------DAQ---DLPPLHRIGTAALAVQVV-G 90
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
P Y +++ GLCRF + ++ Y A + L+ + + + LS
Sbjct: 91 SNWPKPH--YTLLITGLCRFQIVQVLKEKPYPVAEVEQLDRLEEFPNTCKTREELGELSE 148
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QF A++L+ +L+ + + LL+++P L DI + S +E+L +LD+V +
Sbjct: 149 QFYKYAVQLVEMLDMSVPAVAKLRRLLDSLPREALPDILTSIIRTSNKEKLQILDAVSLE 208
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG-----DND 297
R L+ R ++ + K+ QK K + + +R I G D D
Sbjct: 209 ERFKMTIPLLVRQIEGL----KLLQKTRKHKQDDDKRVIAIRPIRRITHIPGTLEDDDED 264
Query: 298 DDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQK 357
+D DD+ LERK++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K
Sbjct: 265 EDNDDIVMLERKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNK 324
Query: 358 ASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTS 417
++ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVGKTS
Sbjct: 325 STTDR-LDIRAARILLDNDHYAMEKLKKRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTS 383
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDE 477
+ S+A L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE
Sbjct: 384 VGRSVAKTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDE 443
Query: 478 IDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLD 537
+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IPP LLD
Sbjct: 444 VDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANTTATIPPALLD 503
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRN 597
RME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAGVR+
Sbjct: 504 RMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQVTTLDIITRYTREAGVRS 563
Query: 598 LERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRD 657
L+R ++ RA AV+VAE + +KE + L+ E E I D
Sbjct: 564 LDRKFGAICRAVAVKVAEGQ-----HKEAK------LDRYDVAEGEGGREHILEDAKPES 612
Query: 658 ISNTFRIT----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQF 713
IS+T + P++++ L+ +LGPP ++ E +ER++ PG+ +GL WT GGE+ F
Sbjct: 613 ISDTTDLALPPEMPILIDFHALKDILGPPMYE-MEVSERLSQPGVAIGLAWTPLGGEIMF 671
Query: 714 VEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHF 773
VEA+ M G+G L LTGQLGDV+KESA +A++W+R+ A L + + +LL+ DIH+HF
Sbjct: 672 VEASRMDGEGQLTLTGQLGDVMKESAHLAISWLRSNAKKYHLTNGD-FDLLDNTDIHLHF 730
Query: 774 PAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHR 833
PAGA+ KDGPSAGVT+VT L SLFS + VRSD AMTGE+TLRGLVLPVGGIKDK+LAAHR
Sbjct: 731 PAGAVTKDGPSAGVTIVTCLASLFSGRLVRSDVAMTGEITLRGLVLPVGGIKDKVLAAHR 790
Query: 834 CGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
G+KRVIIP+RN KDL E+P +V +L + A +++VL A +GG
Sbjct: 791 AGLKRVIIPQRNEKDLEEIPGNVRQDLSFVTASCLDEVLNAAFDGG 836
>emb|CAD68987.1| peroxisomal lon protease [Homo sapiens] gi|62740210|gb|AAH93912.1|
Peroxisomal lon protease [Homo sapiens]
gi|62739604|gb|AAH93910.1| Peroxisomal lon protease
[Homo sapiens] gi|31377667|ref|NP_113678.2| peroxisomal
lon protease [Homo sapiens]
Length = 852
Score = 681 bits (1757), Expect = 0.0
Identities = 383/887 (43%), Positives = 555/887 (62%), Gaps = 67/887 (7%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRED--KGLIGILPVRDAA 62
+++PSRL +L VLLPG+ +R S +++LV L + ++G++P
Sbjct: 7 IQIPSRLPLLLTHEGVLLPGSTMRTSVDSARNLQLVRSRLLKGTSLQSTILGVIP----- 61
Query: 63 ETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+T D +S DA+ D+ H G AA A+ + G
Sbjct: 62 ---------------NTPDPAS------------DAQ---DLPPLHRIGTAALAVQVV-G 90
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
P Y +++ GLCRF + ++ Y A + L+ + + + LS
Sbjct: 91 SNWPKPH--YTLLITGLCRFQIVQVLKEKPYPIAEVEQLDRLEEFPNTCKMREELGELSE 148
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QF A++L+ +L+ + + LL+++P L DI + S +E+L +LD+V +
Sbjct: 149 QFYKYAVQLVEMLDMSVPAVAKLRRLLDSLPREALPDILTSIIRTSNKEKLQILDAVSLE 208
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG-----DND 297
R L+ R ++ + K+ QK K + + +R I G D D
Sbjct: 209 ERFKMTIPLLVRQIEGL----KLLQKTRKPKQDDDKRVIAIRPIRRITHISGTLEDEDED 264
Query: 298 DDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQK 357
+D DD+ LE+K++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K
Sbjct: 265 EDNDDIVMLEKKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNK 324
Query: 358 ASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTS 417
++ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVGKTS
Sbjct: 325 STTDR-LDIRAARILLDNDHYAMEKLKKRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTS 383
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDE 477
+ S+A L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE
Sbjct: 384 VGRSVAKTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDE 443
Query: 478 IDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLD 537
+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IP LLD
Sbjct: 444 VDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANTTATIPAALLD 503
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRN 597
RME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAGVR+
Sbjct: 504 RMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQVTTLDIITRYTREAGVRS 563
Query: 598 LERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRD 657
L+R L ++ RA AV+VAE + +KE + L+ E E I D
Sbjct: 564 LDRKLGAICRAVAVKVAEGQ-----HKEAK------LDRSDVTEREGCREHILEDEKPES 612
Query: 658 ISNTFRIT----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQF 713
IS+T + P++++ L+ +LGPP ++ E ++R++ PG+ +GL WT GGE+ F
Sbjct: 613 ISDTTDLALPPEMPILIDFHALKDILGPPMYE-MEVSQRLSQPGVAIGLAWTPLGGEIMF 671
Query: 714 VEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-INLLEGRDIHIH 772
VEA+ M G+G L LTGQLGDV+KESA +A++W+R+ A +L +A G +LL+ DIH+H
Sbjct: 672 VEASRMDGEGQLTLTGQLGDVMKESAHLAISWLRSNAKKYQLTNAFGSFDLLDNTDIHLH 731
Query: 773 FPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAH 832
FPAGA+ KDGPSAGVT+VT L SLFS + VRSD AMTGE+TLRGLVLPVGGIKDK+LAAH
Sbjct: 732 FPAGAVTKDGPSAGVTIVTCLASLFSGRLVRSDVAMTGEITLRGLVLPVGGIKDKVLAAH 791
Query: 833 RCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
R G+K+VIIP RN KDL +P +V +L + A +++VL A +GG
Sbjct: 792 RAGLKQVIIPRRNEKDLEGIPGNVRQDLSFVTASCLDEVLNAAFDGG 838
>dbj|BAC11201.1| unnamed protein product [Homo sapiens]
Length = 852
Score = 680 bits (1755), Expect = 0.0
Identities = 383/887 (43%), Positives = 554/887 (62%), Gaps = 67/887 (7%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRED--KGLIGILPVRDAA 62
+++PSRL +L VLLPG+ +R S +++LV L + ++G++P
Sbjct: 7 IQIPSRLPLLLTHEGVLLPGSTMRTSVDSARNLQLVRSRLLKGTSLQSTILGVIP----- 61
Query: 63 ETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+T D +S DA+ D+ H G AA A+ + G
Sbjct: 62 ---------------NTPDPAS------------DAQ---DLPPLHRIGTAALAVQVV-G 90
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
P Y +++ GLCRF + ++ Y A + L+ + + + LS
Sbjct: 91 SNWPKPH--YTLLITGLCRFQIVQVLKEKPYPIAEVEQLDRLEEFPNTCKMREELGELSE 148
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QF A++L+ +L+ + + LL+++P L DI + S +E+L +LD+V +
Sbjct: 149 QFYKYAVQLVEMLDMSVPAVAKLRRLLDSLPREALPDILTSIIRTSNKEKLQILDAVSLE 208
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG-----DND 297
R L+ R ++ + K+ QK K + + +R I G D D
Sbjct: 209 ERFKMTIPLLVRQIEGL----KLLQKTRKPKQDDDKRVIAIRPIRRITHISGTLEDEDED 264
Query: 298 DDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQK 357
+D DD+ LE+K++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K
Sbjct: 265 EDNDDIVMLEKKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNK 324
Query: 358 ASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTS 417
++ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVGKTS
Sbjct: 325 STTDR-LDIRAARILLDNDHYAMEKLKKRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTS 383
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDE 477
+ S+A L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE
Sbjct: 384 VGRSVAKTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDE 443
Query: 478 IDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLD 537
+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IP LLD
Sbjct: 444 VDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANTTATIPAALLD 503
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRN 597
RME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAGVR+
Sbjct: 504 RMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQVTTLDIITRYTREAGVRS 563
Query: 598 LERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRD 657
L+R L ++ RA AV+VAE + +KE + L+ E E I D
Sbjct: 564 LDRKLGAICRAVAVKVAEGQ-----HKEAK------LDRSDVTEREGCREHILEDEKPES 612
Query: 658 ISNTFRIT----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQF 713
IS+T + P+ ++ L+ +LGPP ++ E ++R++ PG+ +GL WT GGE+ F
Sbjct: 613 ISDTTDLALPPEMPIFIDFHALKDILGPPMYE-MEVSQRLSQPGVAIGLAWTPLGGEIMF 671
Query: 714 VEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-INLLEGRDIHIH 772
VEA+ M G+G L LTGQLGDV+KESA +A++W+R+ A +L +A G +LL+ DIH+H
Sbjct: 672 VEASRMDGEGQLTLTGQLGDVMKESAHLAISWLRSNAKKYQLTNAFGSFDLLDNTDIHLH 731
Query: 773 FPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAH 832
FPAGA+ KDGPSAGVT+VT L SLFS + VRSD AMTGE+TLRGLVLPVGGIKDK+LAAH
Sbjct: 732 FPAGAVTKDGPSAGVTIVTCLASLFSGRLVRSDVAMTGEITLRGLVLPVGGIKDKVLAAH 791
Query: 833 RCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
R G+K+VIIP RN KDL +P +V +L + A +++VL A +GG
Sbjct: 792 RAGLKQVIIPRRNEKDLEGIPGNVRQDLSFVTASCLDEVLNAAFDGG 838
>emb|CAH92585.1| hypothetical protein [Pongo pygmaeus]
Length = 852
Score = 677 bits (1747), Expect = 0.0
Identities = 380/887 (42%), Positives = 554/887 (61%), Gaps = 67/887 (7%)
Query: 5 VELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQRED--KGLIGILPVRDAA 62
+++PSRL +L VLLPG+ +R S +++LV L + ++G++P
Sbjct: 7 IQIPSRLPLLLTHEGVLLPGSTMRTSVDSARNLQLVRSRLLKGTSLQSTILGVIP----- 61
Query: 63 ETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLSRG 122
+T D +S DA+ D+ H G AA A+ + G
Sbjct: 62 ---------------NTPDPAS------------DAQ---DLPPLHRIGTAALAVQVV-G 90
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
P Y +++ GLCRF + ++ Y A + L+ + + + LS
Sbjct: 91 SNWPKPH--YTLLITGLCRFQIVQVLKEKPYPIAEVEQLDRLEEFPSTCKMREELGELSE 148
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
QF A++L+ +L+ + + LL+++P L DI + S +E+L +LD+V +
Sbjct: 149 QFYKYAVQLVEMLDMSVPAVAKLRRLLDSLPREALPDILTSIIRTSNKEKLQILDAVSLE 208
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG-----DND 297
R L+ R ++ + K+ QK K + + +R I G D D
Sbjct: 209 ERFKMTIPLLVRQIEGL----KLLQKTRKPKQDDDKRVIAIRPIRRITHISGTLEDEDED 264
Query: 298 DDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQK 357
+D DD+ LE++++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K
Sbjct: 265 EDNDDIVMLEKEIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNK 324
Query: 358 ASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTS 417
++ + +D+++A+ LD+DHY + K+K+R++EYL VR+LK + +GP+LCFVGPPGVGKTS
Sbjct: 325 STTDR-LDIRAARILLDNDHYAMEKLKKRVLEYLVVRQLKNNLKGPILCFVGPPGVGKTS 383
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDE 477
+ +A L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE
Sbjct: 384 VGRLVAKTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDE 443
Query: 478 IDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLD 537
+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IP LLD
Sbjct: 444 VDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSRVLFIATANTTATIPAALLD 503
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRN 597
RME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAGVR+
Sbjct: 504 RMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQVTTLDIITRYTREAGVRS 563
Query: 598 LERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRD 657
L+R L ++ RA AV+VAE + +KE + L+ E E I D
Sbjct: 564 LDRKLGAICRAVAVKVAEGQ-----HKEAK------LDRSDVTEREGCREHILEDEKPES 612
Query: 658 ISNTFRIT----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQF 713
IS+T + P++++ L+ +LGPP ++ E ++R++ PG+ +GL WT GGE+ F
Sbjct: 613 ISDTTDLALPPEMPILIDFHALKDILGPPMYE-MEVSQRLSQPGVAIGLAWTPLGGEIMF 671
Query: 714 VEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-INLLEGRDIHIH 772
VEA+ M G+G L LTGQLGDV+KESA +A++W+R+ A +L +A G +LL+ DIH+H
Sbjct: 672 VEASRMDGEGQLTLTGQLGDVMKESAHLAISWLRSNAKKYQLTNAFGSFDLLDNTDIHLH 731
Query: 773 FPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAH 832
FPAGA+ KDGPSAGVT+VT L SLFS++ VRSD AMTGE+TLRGLVLPVGGIKDK+LAAH
Sbjct: 732 FPAGAVTKDGPSAGVTIVTCLASLFSERLVRSDVAMTGEITLRGLVLPVGGIKDKVLAAH 791
Query: 833 RCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
R G+K+VIIP RN KDL +P +V +L + A +++VL A +GG
Sbjct: 792 RAGLKQVIIPRRNEKDLEGIPGNVRQDLSFVTASCLDEVLNAAFDGG 838
>emb|CAD38889.1| hypothetical protein [Homo sapiens]
Length = 715
Score = 655 bits (1691), Expect = 0.0
Identities = 344/710 (48%), Positives = 485/710 (67%), Gaps = 27/710 (3%)
Query: 180 LSRQFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSV 239
LS QF A++L+ +L+ + + LL+++P L DI + S +E+L +LD+V
Sbjct: 9 LSEQFYKYAVQLVEMLDMSVPAVAKLRRLLDSLPREALPDILTSIIRTSNKEKLQILDAV 68
Query: 240 DAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG----- 294
+ R L+ R ++ + K+ QK K + + +R I G
Sbjct: 69 SLEERFKMTIPLLVRQIEGL----KLLQKTRKPKQDDDKRVIAIRPIRRITHISGTLEDE 124
Query: 295 DNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLP 354
D D+D DD+ LE+K++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LP
Sbjct: 125 DEDEDNDDIVMLEKKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELP 184
Query: 355 WQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVG 414
W K++ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVG
Sbjct: 185 WNKSTTDR-LDIRAARILLDNDHYAMEKLKKRVLEYLAVRQLKNNLKGPILCFVGPPGVG 243
Query: 415 KTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVML 474
KTS+ S+A L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV L
Sbjct: 244 KTSVGRSVAKTLGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFL 303
Query: 475 LDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPP 534
LDE+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IP
Sbjct: 304 LDEVDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANTTATIPAA 363
Query: 535 LLDRMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAG 594
LLDRME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAG
Sbjct: 364 LLDRMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQVTTLDIITRYTREAG 423
Query: 595 VRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVN 654
VR+L+R L ++ RA AV+VAE + +KE + L+ E E I D
Sbjct: 424 VRSLDRKLGAICRAVAVKVAEGQ-----HKEAK------LDRSDVTEREGCREHILEDEK 472
Query: 655 SRDISNTFRIT----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGE 710
IS+T + P++++ L+ +LGPP ++ E ++R++ PG+ +GL WT GGE
Sbjct: 473 PESISDTTDLALPPEMPILIDFHALKDILGPPMYE-MEVSQRLSQPGVAIGLAWTPLGGE 531
Query: 711 VQFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-INLLEGRDI 769
+ FVEA+ M G+G L LTGQLGDV+KESA +A++W+R+ A +L +A G +LL+ DI
Sbjct: 532 IMFVEASRMDGEGQLTLTGQLGDVMKESAHLAISWLRSNAKKYQLTNAFGSFDLLDNTDI 591
Query: 770 HIHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKIL 829
H+HFPAGA+ KDGPSAGVT+VT L SLFS + VRSD AMTGE+TLRGLVLPVGGIKDK+L
Sbjct: 592 HLHFPAGAVTKDGPSAGVTIVTCLASLFSGRLVRSDVAMTGEITLRGLVLPVGGIKDKVL 651
Query: 830 AAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
AAHR G+K+VIIP RN KDL +P +V +L + A +++VL A +GG
Sbjct: 652 AAHRAGLKQVIIPRRNEKDLEGIPGNVRQDLSFVTASCLDEVLNAAFDGG 701
>ref|XP_214655.3| PREDICTED: similar to RIKEN cDNA 1300002A08 [Rattus norvegicus]
Length = 983
Score = 644 bits (1661), Expect = 0.0
Identities = 349/777 (44%), Positives = 500/777 (63%), Gaps = 44/777 (5%)
Query: 123 VEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVEHDPDFIMLSR 182
V+ P Y+V+L SV Y R S +E P L
Sbjct: 217 VQAPCAFNCYVVLLPLAWTCSV-------VYRHCRSSCVETVLQTCSPPRSHPVAAFLQE 269
Query: 183 QFKATAMELISVLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSVDAK 242
+ ++ L+ +L+ + + LL+++P L DI + S +E+L +LD+V +
Sbjct: 270 WQRVSSHRLVEMLDMSVPAVAKLRRLLDSLPREALPDILTSIIRTSNKEKLQILDAVSLE 329
Query: 243 LRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG-----DND 297
R L+ R ++ + K+ QK K + + +R I G + +
Sbjct: 330 DRFKMTIPLLVRQIEGL----KLLQKTRKPKQDDDKRVIAIRPIRRITHIPGALEDEEEE 385
Query: 298 DDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQK 357
+D DD+ LE+K++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K
Sbjct: 386 EDNDDIVMLEKKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNK 445
Query: 358 ASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTS 417
++ + +D+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVGKTS
Sbjct: 446 STTDR-LDIRAARILLDNDHYAMEKLKRRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTS 504
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGH----------RRTYIGSMPGRLIDGLKRVA 467
+ S+A L R+F RI+LGGV D++DIRGH RRTY+GSMPGR+I+GLK V
Sbjct: 505 VGRSVAKTLGREFHRIALGGVCDQSDIRGHSGLFTGVTDLRRTYVGSMPGRIINGLKTVG 564
Query: 468 VCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANR 527
V NPV LLDE+DK G ++GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN
Sbjct: 565 VNNPVFLLDEVDKLGKSLQGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANT 624
Query: 528 AQQIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQ 587
IPP LLDRME+I++PGYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I
Sbjct: 625 TATIPPALLDRMEIIQVPGYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQLTTLAIIT 684
Query: 588 RYTREAGVRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEME 647
RYTREAGVR+L+R ++ RA AV+VAE + +KE + L + + DG +
Sbjct: 685 RYTREAGVRSLDRKFGAICRAVAVKVAEGQ-----HKEAK-----LDRSDVADGEGCKEH 734
Query: 648 VIPMDVNSRDISNTFRIT----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLV 703
V+ D I + + P++++ L+ +LGPP ++ E +ER++ PG+ +GL
Sbjct: 735 VLE-DAKPESIGDAADLALPPEMPILIDSHALKDILGPPLYE-LEVSERLSQPGVAIGLA 792
Query: 704 WTTFGGEVQFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-IN 762
WT GG++ FVEA+ M G+G L LTGQLGDV+KESA +A++W+R+ A L +A G +
Sbjct: 793 WTPLGGKIMFVEASRMDGEGQLTLTGQLGDVMKESAHLAISWLRSNAKKYHLTNAFGSFD 852
Query: 763 LLEGRDIHIHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVG 822
LL+ DIH+HFPAGA+ KDGPSAGVT+VT L SLFS + VRSD AMTGE+TLRGLVLPVG
Sbjct: 853 LLDNTDIHLHFPAGAVTKDGPSAGVTIVTCLASLFSGRLVRSDVAMTGEITLRGLVLPVG 912
Query: 823 GIKDKILAAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
GIKDK+LAAHR G+K +IIP+RN KDL E+PS+V +L + A +++VL A +GG
Sbjct: 913 GIKDKVLAAHRAGLKHIIIPQRNEKDLEEIPSNVKQDLSFVTASCLDEVLNAAFDGG 969
>dbj|BAB55278.1| unnamed protein product [Homo sapiens]
Length = 581
Score = 610 bits (1573), Expect = e-173
Identities = 310/579 (53%), Positives = 425/579 (72%), Gaps = 18/579 (3%)
Query: 306 LERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHEMD 365
LE+K++++ MP+ K +E++RLKKM P Y+ +R YL+L+ +LPW K++ + +D
Sbjct: 2 LEKKIRTSSMPEQAHKVCVKEIKRLKKMPQSMPEYALTRNYLELMVELPWNKSTTDR-LD 60
Query: 366 LKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVGKTSLASSIAAA 425
+++A+ LD+DHY + K+K+R++EYLAVR+LK + +GP+LCFVGPPGVGKTS+ S+A
Sbjct: 61 IRAARILLDNDHYAMEKLKKRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTSVGRSVAKT 120
Query: 426 LDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDV 485
L R+F RI+LGGV D++DIRGHRRTY+GSMPGR+I+GLK V V NPV LLDE+DK G +
Sbjct: 121 LGREFHRIALGGVCDQSDIRGHRRTYVGSMPGRIINGLKTVGVNNPVFLLDEVDKLGKSL 180
Query: 486 RGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRMEVIELP 545
+GDPA+ALLEVLDPEQN F DHYLNV FDLS+V+F+ATAN IP LLDRME+I++P
Sbjct: 181 QGDPAAALLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATANTTATIPAALLDRMEIIQVP 240
Query: 546 GYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNLASL 605
GYT EEK++IA +HLIP+ L+QHGL+ + +QIP+ +I RYTREAGVR+L+R L ++
Sbjct: 241 GYTQEEKIEIAHRHLIPKQLEQHGLTPQQIQIPQVTTLDIITRYTREAGVRSLDRKLGAI 300
Query: 606 ARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRDISNTFRIT 665
RA AV+VAE + +KE + L+ E E I D IS+T +
Sbjct: 301 CRAVAVKVAEGQ-----HKEAK------LDRSDVTEREGCREHILEDEKPESISDTTDLA 349
Query: 666 ----SPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATEMVG 721
P++++ L+ +LGPP ++ E ++R++ PG+ +GL WT GGE+ FVEA+ M G
Sbjct: 350 LPPEMPILIDFHALKDILGPPMYE-MEVSQRLSQPGVAIGLAWTPLGGEIMFVEASRMDG 408
Query: 722 KGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEG-INLLEGRDIHIHFPAGAIPK 780
+G L LTGQLGDV+KESA +A++W+R+ A +L +A G +LL+ DIH+HFPAGA+ K
Sbjct: 409 EGQLTLTGQLGDVMKESAHLAISWLRSNAKKYQLTNAFGSFDLLDNTDIHLHFPAGAVTK 468
Query: 781 DGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRVI 840
DGPSAGVT+VT L SLFS + VRSD AMTGE+TLRGLVLPVGGIKDK+LAAHR G+K+VI
Sbjct: 469 DGPSAGVTIVTCLASLFSGRLVRSDVAMTGEITLRGLVLPVGGIKDKVLAAHRAGLKQVI 528
Query: 841 IPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGG 879
IP RN KDL +P +V +L + A +++VL A +GG
Sbjct: 529 IPRRNEKDLEGIPGNVRQDLSFVTASCLDEVLNAAFDGG 567
>ref|NP_636358.1| ATP-dependent serine proteinase La [Xanthomonas campestris pv.
campestris str. ATCC 33913] gi|21112003|gb|AAM40282.1|
ATP-dependent serine proteinase La [Xanthomonas
campestris pv. campestris str. ATCC 33913]
gi|66769565|ref|YP_244327.1| ATP-dependent serine
proteinase La [Xanthomonas campestris pv. campestris
str. 8004] gi|66574897|gb|AAY50307.1| ATP-dependent
serine proteinase La [Xanthomonas campestris pv.
campestris str. 8004]
Length = 823
Score = 585 bits (1509), Expect = e-165
Identities = 351/836 (41%), Positives = 509/836 (59%), Gaps = 74/836 (8%)
Query: 55 ILPVRDAAETEVKPVGPTISQDGD------TLDQSSKVHGASSDSHKLDAKAQNDVVHWH 108
+LP+RD + + +D ++ ++ + S + D A D+ +
Sbjct: 13 VLPLRDVVVFPHMVIPLFVGRDKSMRALEKAMEADKRILLVAQKSAETDDPAAGDL---Y 69
Query: 109 NRGVAARALHLSRGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEM 168
G A+ L L ++ P G + V++EGL R +V +++ + +L+ TE+
Sbjct: 70 TVGTLAQVLQL---LKLPDGTIK--VLVEGLSRVTVDKVAEQD-------GALQGRGTEV 117
Query: 169 EQVE-HDPDFIMLSRQFKATAMELISVLEQKQKTGGRTKV-LLETVP----VHKLADIFV 222
E + +P R+ +A A L+S+ EQ KT + LL+T+ +LAD
Sbjct: 118 EASDAREP------RELEAIARSLMSLFEQYVKTNRKLPPELLQTLAGIDEPGRLADTIA 171
Query: 223 ASFEISFEEQLSMLDSVDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLL 282
A + ++ +L+ + RL LVD + ++ ++I +V+ Q+ KSQ+E+ L
Sbjct: 172 AHIGVRLADKQRLLEITEIGDRLELLVGLVDGEIDVQQLEKRIRGRVKSQMEKSQREYYL 231
Query: 283 RQQMRAIKEELGDNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSS 342
+QM+AI++ELGD DD +L L RK+ AGMP+ + A EL +LK+M P +
Sbjct: 232 NEQMKAIQKELGDLDDAPGELEELARKIAEAGMPKPVETKAKAELNKLKQMSPMSAEAAV 291
Query: 343 SRAYLDLLADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARG 402
R YLD L +PW+K ++ + DLK AQ+ LD+DHYGL KVK+RI+EYLAV+ +G
Sbjct: 292 VRNYLDWLLGVPWKKRTKVRK-DLKVAQDTLDADHYGLDKVKERILEYLAVQSRVKQMKG 350
Query: 403 PVLCFVGPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDG 462
P+LC VGPPGVGKTSL SIA A +RKFVR+SLGGV+DEA+IRGHRRTY+GSMPGRL+
Sbjct: 351 PILCLVGPPGVGKTSLGQSIAKATNRKFVRMSLGGVRDEAEIRGHRRTYVGSMPGRLVQN 410
Query: 463 LKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFV 522
L +V NP+ LLDEIDK D RGDP+SALLEVLDPEQN TFNDHYL V DLS+V+FV
Sbjct: 411 LNKVGSKNPLFLLDEIDKMSMDFRGDPSSALLEVLDPEQNHTFNDHYLEVDLDLSEVMFV 470
Query: 523 ATANRAQQIPPPLLDRMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMV 582
AT+N + IP PLLDRMEVI +PGYT +EKL IAM++L+P+ + +GL E ++I +
Sbjct: 471 ATSN-SLNIPGPLLDRMEVIRIPGYTEDEKLNIAMRYLVPKQIKANGLKPEEIEIGSDAI 529
Query: 583 QLVIQRYTREAGVRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGA 642
Q V++ YTRE+GVRNLER +A + R +A + G
Sbjct: 530 QDVVRYYTRESGVRNLEREVAKICRKVVKEIA-----------LAG-------------- 564
Query: 643 EVEMEVIPMDVNSRDISNTFRITSPLVVNEAILEKVLGPPKFD-GREAAERVAAPGICVG 701
P + ++ + + + V L+K LG +FD GR AE G+ G
Sbjct: 565 -------PQPAAKKAVAKKAKPKALVTVTSKNLDKYLGVRRFDFGR--AEEENEIGLVTG 615
Query: 702 LVWTTFGGEVQFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGI 761
L WT GGE+ +E+T + GKG+L LTGQLG+V+KESA AL+ VR+RA L + +
Sbjct: 616 LAWTEVGGELLQIESTLVPGKGNLILTGQLGNVMKESASAALSVVRSRAERLGI----DV 671
Query: 762 NLLEGRDIHIHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPV 821
+ L+ +D+H+H P GA PKDGPSAG+ +VT+LVS+ +K +R+D AMTGE+TLRG V +
Sbjct: 672 DFLQKQDVHVHVPDGATPKDGPSAGIAMVTSLVSILTKVPIRADVAMTGEITLRGRVSAI 731
Query: 822 GGIKDKILAAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALE 877
GG+K+K+LAA R GI+ V+IP+ N KDL ++P++V +L+I+ K +++VL+ ALE
Sbjct: 732 GGLKEKLLAALRGGIRTVLIPDENRKDLADIPANVTRDLKIVPVKWIDEVLDLALE 787
>ref|YP_199674.1| ATP-dependent serine proteinase La [Xanthomonas oryzae pv. oryzae
KACC10331] gi|58425252|gb|AAW74289.1| ATP-dependent
serine proteinase La [Xanthomonas oryzae pv. oryzae
KACC10331]
Length = 850
Score = 581 bits (1498), Expect = e-164
Identities = 362/884 (40%), Positives = 523/884 (58%), Gaps = 104/884 (11%)
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRD 60
MA+S L +LP R+ V+ P +I + S++ +E+ + DK ++ L +
Sbjct: 28 MAQSQPEVLDLPVLPLRDVVVFPHMVIPLFVGRDKSMRALEKAM--EADKRIL--LVAQK 83
Query: 61 AAETEVKPVGPTISQDGDTLDQSSKVHGASSDSHKLDAKAQNDVVHWHNRGVAARALHLS 120
+AET+ D A V H G A+ L L
Sbjct: 84 SAETD-------------------------------DPAA----VDLHTVGTLAQVLQL- 107
Query: 121 RGVEKPSGRVTYIVVLEGLCRFSVQELSTRGTYHTARISSLEMNKTEMEQVE-HDPDFIM 179
++ P G + V++EGL R +V ++ + +L+ TE+E + +P
Sbjct: 108 --LKLPDGTIK--VLVEGLSRVTVDKVVEQD-------GALQGQGTEVEASDAREP---- 152
Query: 180 LSRQFKATAMELISVLEQKQKTGGRTKV-LLETVP----VHKLADIFVASFEISFEEQLS 234
R+ +A A L+S+ EQ KT + LL+T+ +LAD + ++
Sbjct: 153 --REVEAIARSLMSLFEQYVKTNRKLPPELLQTLAGIDEPGRLADTIAPHIGVRLADKQR 210
Query: 235 MLDSVDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELG 294
+L+ D RL LVD + ++ ++I +V+ Q+ KSQ+E+ L +QM+AI++ELG
Sbjct: 211 LLEITDIGERLELLVGLVDGEIDVQQLEKRIRGRVKSQMEKSQREYYLNEQMKAIQKELG 270
Query: 295 DNDDDEDDLAALERKMQSAGMPQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLP 354
D DD +L L RK+ AGMP+ + A EL +LK+M P + R YLD L +P
Sbjct: 271 DLDDVPGELEELARKIAEAGMPKPVETKAKAELNKLKQMSPMSAEAAVVRNYLDWLLGVP 330
Query: 355 WQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAVRKLKPDARGPVLCFVGPPGVG 414
W+K ++ + DLK A++ LD+DHYGL KVK+RI+EYLAV+ +GP+LC VGPPGVG
Sbjct: 331 WKKRTKVRK-DLKVAEDTLDADHYGLDKVKERILEYLAVQSRVKQMKGPILCLVGPPGVG 389
Query: 415 KTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVML 474
KTSL SIA A +RKFVR+SLGG++DEA+IRGHRRTY+GSMPGRL+ L +V NP+ L
Sbjct: 390 KTSLGQSIAKATNRKFVRMSLGGIRDEAEIRGHRRTYVGSMPGRLVQNLNKVGSKNPLFL 449
Query: 475 LDEIDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPP 534
LDEIDK D RGDP+SALLEVLDPEQN +FNDHYL V DLS+V+FVAT+N + IP P
Sbjct: 450 LDEIDKMSMDFRGDPSSALLEVLDPEQNNSFNDHYLEVDLDLSEVMFVATSN-SLNIPGP 508
Query: 535 LLDRMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAG 594
LLDRMEVI +PGYT +EKL IAM++L+P+ + +GL E ++I +Q +++ YTRE+G
Sbjct: 509 LLDRMEVIRIPGYTEDEKLNIAMRYLVPKQIKANGLKPEEIEIGGDAIQDIVRYYTRESG 568
Query: 595 VRNLERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVN 654
VRNLER +A + R +A + G P
Sbjct: 569 VRNLEREVAKICRKVVKEIA-----------LAG---------------------PQPAA 596
Query: 655 SRDISNTFRITSPLVVNEAILEKVLGPPKFD-GREAAERVAAPGICVGLVWTTFGGEVQF 713
+ ++ + + + VN L+K LG +FD GR AE G+ GL WT GGE+
Sbjct: 597 KKAVAKKGKPKALVTVNAKNLDKYLGVRRFDFGR--AEEENEIGLVTGLAWTEVGGELLQ 654
Query: 714 VEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHF 773
VE+T + GKG+L LTGQLG+V+KESA AL+ VR+RA L + ++ L+ +D+H+H
Sbjct: 655 VESTLVPGKGNLILTGQLGNVMKESASAALSVVRSRAERLGI----DVDFLQKQDVHVHV 710
Query: 774 PAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHR 833
P GA PKDGPSAG+ +VT+LVS+ +K +R+D AMTGE+TLRG V +GG+K+K+LAA R
Sbjct: 711 PDGATPKDGPSAGIAMVTSLVSVLTKVPIRADVAMTGEITLRGRVSAIGGLKEKLLAALR 770
Query: 834 CGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALE 877
GI+ V+IP N KDL ++P++V +L+I+ K +++VL+ ALE
Sbjct: 771 GGIRTVLIPGENRKDLADIPANVTRDLKIVPVKWIDEVLDLALE 814
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,425,159,552
Number of Sequences: 2540612
Number of extensions: 59195349
Number of successful extensions: 210065
Number of sequences better than 10.0: 3421
Number of HSP's better than 10.0 without gapping: 731
Number of HSP's successfully gapped in prelim test: 2693
Number of HSP's that attempted gapping in prelim test: 205392
Number of HSP's gapped (non-prelim): 5770
length of query: 888
length of database: 863,360,394
effective HSP length: 137
effective length of query: 751
effective length of database: 515,296,550
effective search space: 386987709050
effective search space used: 386987709050
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0550.4


