

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0546.8
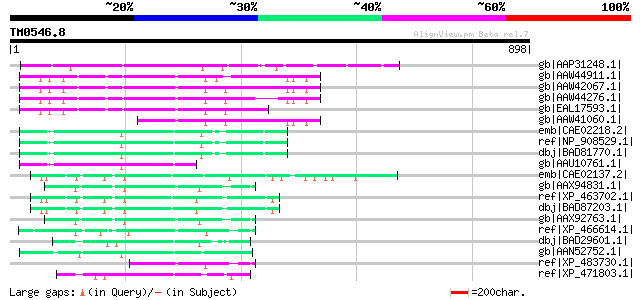
(898 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP31248.1| transposase [Fusarium oxysporum f. sp. melonis] 190 2e-46
gb|AAW44911.1| conserved hypothetical protein [Cryptococcus neof... 147 1e-33
gb|AAW42067.1| conserved hypothetical protein [Cryptococcus neof... 147 2e-33
gb|AAW44276.1| conserved hypothetical protein [Cryptococcus neof... 133 3e-29
gb|EAL17593.1| hypothetical protein CNBM0450 [Cryptococcus neofo... 122 4e-26
gb|AAW41060.1| conserved hypothetical protein [Cryptococcus neof... 122 5e-26
emb|CAE02218.2| OSJNBb0002N06.8 [Oryza sativa (japonica cultivar... 113 3e-23
ref|NP_908529.1| P0702D12.15 [Oryza sativa (japonica cultivar-gr... 109 4e-22
dbj|BAD81770.1| putative far-red impaired response protein [Oryz... 108 6e-22
gb|AAU10761.1| unknown protein [Oryza sativa (japonica cultivar-... 98 1e-18
emb|CAE02137.2| OSJNBa0074L08.5 [Oryza sativa (japonica cultivar... 90 3e-16
gb|AAX94831.1| transposon protein, putative, unclassified [Oryza... 90 4e-16
ref|XP_463702.1| putative far-red impaired response protein [Ory... 90 4e-16
dbj|BAD87203.1| far-red impaired response-like [Oryza sativa (ja... 90 4e-16
gb|AAX92763.1| transposon protein, putative, unclassified [Oryza... 90 4e-16
ref|XP_466614.1| putative far-red impaired response protein [Ory... 89 8e-16
dbj|BAD29601.1| putative far-red impaired response protein [Oryz... 89 8e-16
gb|AAN52752.1| Unknown protein [Oryza sativa (japonica cultivar-... 86 4e-15
ref|XP_483730.1| putative far-red impaired response protein [Ory... 86 4e-15
ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultiva... 86 4e-15
>gb|AAP31248.1| transposase [Fusarium oxysporum f. sp. melonis]
Length = 836
Score = 190 bits (483), Expect = 2e-46
Identities = 184/695 (26%), Positives = 311/695 (44%), Gaps = 67/695 (9%)
Query: 19 FESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMTLGCERGGKYKPSTSVLKRNRT 78
+ S ++L + A GY +I++S ++ R V+ C+RG PS S R +T
Sbjct: 21 YSSREELRSAINAWAAPRGYAFVIKRSSKTANGRTHVI-FNCDRGAGRIPSLS--DRRQT 77
Query: 79 GTKKCNCPFRLRARRSNKDKMWTVL----VHSGIHNHDTAEVLQGHSYVGRLNPEEKAMV 134
T++ C F + A+ S +W++ H HNH+ + H + +L+ +E+ V
Sbjct: 78 TTRRTGCLFSVLAKESLCKTIWSLRHRPGPHFSQHNHEPSFSEVAHPTLRQLSRQEEITV 137
Query: 135 GEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMKLLEEHK 194
++ + +I +R + T T+ + IYN R + + L L E
Sbjct: 138 NQLTNAGIAPKEIGSFLRITSNTLATQ-QDIYNCIAKGRRDLSKGQSNIHALADQLNEEG 196
Query: 195 YAHWSRV-QDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTST 253
+ W+R+ D + V ++ +AHP S+ +P V+ILDSTYKTNR+++PLL++VGV +
Sbjct: 197 F--WNRICLDESSRVTAVLFAHPKSLEYLKTYPEVLILDSTYKTNRFKMPLLDIVGVDAC 254
Query: 254 NLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINED-NLPKVFVTDKDQVLMEALETVFPAA 312
T+ IAFA++ E++ + WA+ L+ + + + LP V +TD+ M A+ + FP +
Sbjct: 255 QRTFCIAFAFLSGEEEGDFTWALQALRSVYEDHNIGLPSVILTDRCLACMNAVSSCFPGS 314
Query: 313 RCLLCQFHVLKSVTGEMKK-----------LVKDIETREDITDRWNRLMYAANEVEYDKA 361
LC +H+ K+V + L + E ++ + W+ ++ + E Y++
Sbjct: 315 ALFLCLWHINKAVQSYCRPAFTRGKDNPQGLGGESEEWKEFFNFWHEIVASTTEDIYNER 374
Query: 362 VGAMSN------TGELWTYIETNWLPLRSK-FVKFEIDTCMHMGCTTTNRVEGAHSKLKK 414
+ E+ +ET WL L K FVK ++T +H T+RVEG HS +K
Sbjct: 375 LEKFKKRYIPDYINEVGYILET-WLDLYKKSFVKAWVNTHLHFEQYATSRVEGIHSLIKL 433
Query: 415 LLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDP------FYKHLRGFV 468
L+ S+ DL AW + KL++M + QL +++P Y ++RG++
Sbjct: 434 HLNHSQVDLFEAWRVI-KLVLM-----NQLSQLEANQARQHISNPIRESRVLYSNIRGWI 487
Query: 469 SRAALHIIVKEKTQIGMIGVGGIYCECVLRRTHGLPCACELMKF--ESVPLLA--IHPHW 524
S AL K +TQ + C V RT GLPCA L ++ PLL H HW
Sbjct: 488 SHEALR---KVETQRERLLKEVPVCTGVFTRTLGLPCAHSLQPLLKQNQPLLLNHFHSHW 544
Query: 525 RKLTWDSSDRDMDPTLGLSFESEFDKL--RTKFEESSHGVRMSIIESLRMITYPETTSMC 582
R P + +FD+L + +S S E + P+ C
Sbjct: 545 H------LRRPGSPRFLIEPRKQFDRLTASSTLPPTSTQREPSTFERIEKALQPKAPPKC 598
Query: 583 APTKKKTRGRPMGSTNKP---KKFAKGCGETSTKRIPCRWETIDKEYPGSWRDSNTTPLQ 639
++ +G M S P K + +TST + P + T + S T L
Sbjct: 599 --SRCHQQGHMMTSKACPLRYKHLLQAPTQTSTIQAPTQDSTTTHIITHTTTRSITRSLS 656
Query: 640 EPSPKVRSQKASKASKASKASKVSTASKASKVQSP 674
S S +S S+ + T ++V SP
Sbjct: 657 PSS----SSGSSIVSEIVAYTTTHTTHTTTRVLSP 687
>gb|AAW44911.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58270124|ref|XP_572218.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 920
Score = 147 bits (372), Expect = 1e-33
Identities = 145/567 (25%), Positives = 245/567 (42%), Gaps = 65/567 (11%)
Query: 17 QAFESSKDLVNWA-KAVGKEHGYVIIIQKSDYGSDKR-----RAVMTLGCERGGKYK--- 67
QA+ SKD A +A G+ +I++S + R ++ C G +Y+
Sbjct: 94 QAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYRCTR 153
Query: 68 -PSTSVLKRNRTGTKKCNCPFRLRA--------RRSNKDKMWTVLVHSGIHNHDTAEVLQ 118
S ++ T K CPF A + DK WT V + HNH +
Sbjct: 154 GDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVANI- 212
Query: 119 GHSYVGRLN--PEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNR-- 174
G + + R + PE K ++ +I+ R I+ + + PT + + ++ + A+ R
Sbjct: 213 GIALLRRASRTPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLH--RLAFRRRA 270
Query: 175 -LKRGSLTEMQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDS 233
L+ GS + +L+ + H V +++ L + + L + FP V+ D
Sbjct: 271 DLRAGSSASAACIRQLVSNDEL-HRPFVNTRDELI-GLVYTKRTARALIHRFPEVLFFDC 328
Query: 234 TYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVF 293
TYKTN YR+P+L +VG TST +TY+ M E + A+N +L+ D KV
Sbjct: 329 TYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVV 386
Query: 294 VTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIE----TREDITDRWNRL 349
+TD+D L+ AL +V P A C +H+ ++V ++ D E + W
Sbjct: 387 ITDRDPALINALMSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMAVSNFCKAWLEE 446
Query: 350 MYAANE-----VEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTCMHMGCTTTNR 404
Y E +Y +A+ + E ++ +FV I+ +H G T +R
Sbjct: 447 GYRKMEELYPGQKYARAISYIRGLDE-----------IKERFVHAYINKQLHFGQTGNSR 495
Query: 405 VEGAHSKLKKLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDPFYKHL 464
+EG H+ LKK + GDL++ +++ Q KI + +L + F + L
Sbjct: 496 LEGQHATLKKSIDTKYGDLLLVIGSLSTYFDQQWLKILKRIELERTRCATHIPSTFVR-L 554
Query: 465 RGFVSRAALHIIVKE----KTQIGMIGVG------GIYCECVLRRTHGLPCACELMKF-- 512
+G +SRAA+ ++ ++ K +G G C ++HGLPCA L+ F
Sbjct: 555 KGSISRAAMKLLAEQLTLAKRYLGDYDEGVDDFEESHPCSGSFTKSHGLPCAHRLISFVR 614
Query: 513 --ESVPLLAIHPHWRKLTWDSSDRDMD 537
+ IHPHWR + S R +D
Sbjct: 615 ERRHLEKDHIHPHWRLGSAASLKRYLD 641
>gb|AAW42067.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|50260537|gb|EAL23192.1|
hypothetical protein CNBA5360 [Cryptococcus neoformans
var. neoformans B-3501A] gi|50259285|gb|EAL21958.1|
hypothetical protein CNBC0980 [Cryptococcus neoformans
var. neoformans B-3501A] gi|50256296|gb|EAL19021.1|
hypothetical protein CNBH1230 [Cryptococcus neoformans
var. neoformans B-3501A] gi|57229113|gb|AAW45547.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21] gi|58271396|ref|XP_572854.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21] gi|58264436|ref|XP_569374.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21]
Length = 932
Score = 147 bits (370), Expect = 2e-33
Identities = 146/569 (25%), Positives = 252/569 (43%), Gaps = 57/569 (10%)
Query: 17 QAFESSKDLVNWA-KAVGKEHGYVIIIQKSDYGSDKR-----RAVMTLGCERGGKYK--- 67
QA+ SKD A +A G+ +I++S + R ++ C G +Y+
Sbjct: 94 QAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYRCTR 153
Query: 68 -PSTSVLKRNRTGTKKCNCPFRLRA--------RRSNKDKMWTVLVHSGIHNHDTAEVLQ 118
S ++ T K CPF A + DK WT V + HNH +
Sbjct: 154 GDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVANI- 212
Query: 119 GHSYVGRLN--PEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNR-- 174
G + + R + PE K ++ +I+ R I+ + + PT + + ++ + A+ R
Sbjct: 213 GIALLRRASRTPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLH--RLAFRRRA 270
Query: 175 -LKRGSLTEMQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDS 233
L+ GS + +L+ + H V +++ L + + L + FP V+ D
Sbjct: 271 DLRAGSSASAACIRQLVSNDEL-HRPFVNTRDELI-GLVYTKRTARALIHRFPEVLFFDC 328
Query: 234 TYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVF 293
TYKTN YR+P+L +VG TST +TY+ M E + A+N +L+ D KV
Sbjct: 329 TYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVV 386
Query: 294 VTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIE----TREDITDRWNR- 348
+TD+D L+ AL +V P A C +H+ ++V ++ D E + W R
Sbjct: 387 ITDRDPALINALMSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMAVSNFCKAWVRY 446
Query: 349 LMYAANEVEYDKAVGAMSN--TGELW----TYIETNWLPLRSKFVKFEIDTCMHMGCTTT 402
++A ++ + ++ M G+ + +YI ++ +FV I+ +H G T
Sbjct: 447 CVHAKSDEQLEEGYRKMEELYPGQKYARAISYIR-GLDEIKERFVHAYINKQLHFGQTGN 505
Query: 403 NRVEGAHSKLKKLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDPFYK 462
+R+EG H+ LKK + GDL++ +++ Q KI + +L + F +
Sbjct: 506 SRLEGQHATLKKSIDTKYGDLLLVIGSLSTYFDQQWLKILKRIELERTRCATHIPSTFVR 565
Query: 463 HLRGFVSRAALHIIVKE----KTQIGMIGVG------GIYCECVLRRTHGLPCACELMKF 512
L+G +SRAA+ ++ ++ K +G G C ++HGLPCA L+ F
Sbjct: 566 -LKGSISRAAMKLLAEQLTLAKRYLGDYDEGVDDFEESHPCSGSFTKSHGLPCAHRLISF 624
Query: 513 ----ESVPLLAIHPHWRKLTWDSSDRDMD 537
+ IHPHWR + S R +D
Sbjct: 625 VRERRHLEKDHIHPHWRLGSAASLKRYLD 653
>gb|AAW44276.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58268854|ref|XP_571583.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 895
Score = 133 bits (334), Expect = 3e-29
Identities = 141/569 (24%), Positives = 240/569 (41%), Gaps = 94/569 (16%)
Query: 17 QAFESSKDLVNWA-KAVGKEHGYVIIIQKSDYGSDKR-----RAVMTLGCERGGKYK--- 67
QA+ SKD A +A G+ +I++S + R ++ C G +Y+
Sbjct: 94 QAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYRCTR 153
Query: 68 -PSTSVLKRNRTGTKKCNCPFRLRA--------RRSNKDKMWTVLVHSGIHNHDTAEVLQ 118
S ++ T K CPF A + DK WT V + HNH +
Sbjct: 154 GDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVANI- 212
Query: 119 GHSYVGRLN--PEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNR-- 174
G + + R + PE K ++ +I+ R I+ + + PT + + ++ + A+ R
Sbjct: 213 GIALLRRASRTPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLH--RLAFRRRA 270
Query: 175 -LKRGSLTEMQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDS 233
L+ GS + +L+ + H V +++ L + + L + FP V+ D
Sbjct: 271 DLRAGSSASAACIRQLVSNDEL-HRPFVNTRDELI-GLVYTKRTARALIHRFPEVLFFDC 328
Query: 234 TYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVF 293
TYKTN YR+P+L +VG TST +TY+ M E + A+N +L+ D KV
Sbjct: 329 TYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVV 386
Query: 294 VTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIE----TREDITDRWNR- 348
+TD+D L+ AL +V P A C +H+ ++V ++ D E + W R
Sbjct: 387 ITDRDPALINALMSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMAVSNFCKAWVRY 446
Query: 349 LMYAANEVEYDKAVGAMSN--TGELW----TYIETNWLPLRSKFVKFEIDTCMHMGCTTT 402
++A ++ + ++ M G+ + +YI ++ +FV I+ +H G T
Sbjct: 447 CVHAKSDEQLEEGYRKMEELYPGQKYARAISYIR-GLDEIKERFVHAYINKQLHFGQTGN 505
Query: 403 NRVEGAHSKLKKLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDPFYK 462
+R+EG H+ LKK + GDL++
Sbjct: 506 SRLEGQHATLKKSIDTKYGDLLL------------------------------------- 528
Query: 463 HLRGFVSRAALHIIVKE----KTQIGMIGVG------GIYCECVLRRTHGLPCACELMKF 512
L+G +SRAA+ ++ ++ K +G G C ++HGLPCA L+ F
Sbjct: 529 -LKGSISRAAMKLLAEQLTLAKRYLGDYDEGVDDFEESHPCSGSFTKSHGLPCAHRLISF 587
Query: 513 ----ESVPLLAIHPHWRKLTWDSSDRDMD 537
+ IHPHWR + S R +D
Sbjct: 588 VRERRHLEKDHIHPHWRLGSAASLKRYLD 616
>gb|EAL17593.1| hypothetical protein CNBM0450 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57226908|gb|AAW43368.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58267038|ref|XP_570675.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 580
Score = 122 bits (307), Expect = 4e-26
Identities = 118/465 (25%), Positives = 208/465 (44%), Gaps = 42/465 (9%)
Query: 17 QAFESSKDLVNWA-KAVGKEHGYVIIIQKSDYGSDKR-----RAVMTLGCERGGKYK--- 67
QA+ SKD A +A G+ +I++S + R ++ C G +Y+
Sbjct: 94 QAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYRCTR 153
Query: 68 -PSTSVLKRNRTGTKKCNCPFRLRA--------RRSNKDKMWTVLVHSGIHNHDTAEVLQ 118
S ++ T K CPF A + DK WT V + HNH +
Sbjct: 154 GDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVANI- 212
Query: 119 GHSYVGRLN--PEEKAMVGEMIEERVKASDILIAIRKHNPTNLTRIKQIYNEKQAYNR-- 174
G + + R + PE K ++ +I+ R I+ + + PT + + ++ + A+ R
Sbjct: 213 GIALLRRASRTPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLH--RLAFRRRA 270
Query: 175 -LKRGSLTEMQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDS 233
L+ GS + +L+ + H V +++ L + + L + FP V+ D
Sbjct: 271 DLRAGSSASAACIRQLVSNDEL-HRPFVNTRDELI-GLVYTKRTARALIHRFPEVLFFDC 328
Query: 234 TYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVF 293
TYKTN YR+P+L +VG TST +TY+ M E + A+N +L+ D KV
Sbjct: 329 TYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVV 386
Query: 294 VTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIE----TREDITDRWNR- 348
+TD+D L+ AL +V P A C +H+ ++V ++ D E + W R
Sbjct: 387 ITDRDPALINALMSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMAVSNFCKAWVRY 446
Query: 349 LMYAANEVEYDKAVGAMSN--TGELW----TYIETNWLPLRSKFVKFEIDTCMHMGCTTT 402
++A ++ + ++ M G+ + +YI ++ +FV I+ +H G T
Sbjct: 447 CVHAKSDEQLEEGYRKMEELYPGQKYARAISYIR-GLDEIKERFVHAYINKQLHFGQTGN 505
Query: 403 NRVEGAHSKLKKLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQL 447
+R+EG H+ LKK + GDL++ +++ Q KI + +L
Sbjct: 506 SRLEGQHATLKKSIDTKYGDLLLVIGSLSTYFDQQWLKILKRIEL 550
>gb|AAW41060.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58258933|ref|XP_566879.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 678
Score = 122 bits (306), Expect = 5e-26
Identities = 98/342 (28%), Positives = 162/342 (46%), Gaps = 29/342 (8%)
Query: 221 LFNEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLK 280
L + FP V+ D TYKTN YR+P+L +VG TST +TY+ M E + A+N
Sbjct: 62 LIHRFPEVLFFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFF 121
Query: 281 DLIINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIE--- 337
+L+ D KV +TD+D L+ AL +V P A C +H+ ++V ++ D E
Sbjct: 122 ELVGKPD--VKVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIV 179
Query: 338 -TREDITDRWNR-LMYAANEVEYDKAVGAMSN--TGELW----TYIETNWLPLRSKFVKF 389
+ W R ++A ++ + ++ M G+ + +YI ++ +FV
Sbjct: 180 MAVSNFCKAWVRYCVHAKSDEQLEEGYRKMEELYPGQKYARAISYIR-GLDEIKERFVHA 238
Query: 390 EIDTCMHMGCTTTNRVEGAHSKLKKLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSI 449
I+ +H G T +R+EG H+ LKK + GDL++ +++ Q KI + +L
Sbjct: 239 YINKQLHFGQTGNSRLEGQHATLKKSIDTKYGDLLLVIGSLSTYFDQQWLKILKRIELER 298
Query: 450 FVTEHSLNDPFYKHLRGFVSRAALHIIVKE----KTQIGMIGVG------GIYCECVLRR 499
+ F + L+G +SRAA+ ++ ++ K +G G C +
Sbjct: 299 TRCATHIPSTFVR-LKGSISRAAMKLLAEQLTLAKRYLGDYDEGVDDFEESHPCSGSFTK 357
Query: 500 THGLPCACELMKF----ESVPLLAIHPHWRKLTWDSSDRDMD 537
+HGLPCA L+ F + IHPHWR + S R +D
Sbjct: 358 SHGLPCAHRLISFVRERRHLEKDHIHPHWRLGSAASLKRYLD 399
>emb|CAE02218.2| OSJNBb0002N06.8 [Oryza sativa (japonica cultivar-group)]
gi|50923349|ref|XP_472035.1| OSJNBb0002N06.8 [Oryza
sativa (japonica cultivar-group)]
Length = 885
Score = 113 bits (282), Expect = 3e-23
Identities = 107/489 (21%), Positives = 196/489 (39%), Gaps = 50/489 (10%)
Query: 17 QAFESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMTLGCERGGKYKPSTSVLKRN 76
Q F++ +D N+ G+ I + K ++K+R + + C G+ P T
Sbjct: 128 QRFKTERDAYNFYNVYAVSKGFGIRLDKDRMNTEKQRTMRQICCSHQGR-NPKT------ 180
Query: 77 RTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAE---VLQGHSYVGRLNPEEKAM 133
+ + + CP ++ RS W+V HNH + V + + +++ + +
Sbjct: 181 KKPSVRIGCPAMMKINRSGAGSGWSVTKVVSTHNHPMKKSVGVTKNYQSHNQIDEGTRGI 240
Query: 134 VGEMIEERVKASDILIAIR-KHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMKLLEE 192
+ EM++ + +++ + H ++ + ++ AY + S +MQ + +L++
Sbjct: 241 IEEMVDSSMSLTNMYGMLSGMHGGPSMVPFTRKAMDRLAYAIRRDESSDDMQKTLDVLKD 300
Query: 193 ----HKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMV 248
K +S D V+++FW+H S F F V+ D+TYKTN+Y +P V
Sbjct: 301 LQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFV 360
Query: 249 GVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETV 308
GV + + A ++ E ++ W N K+ + +P +TD + A+ TV
Sbjct: 361 GVNNHFQSTFFGCALLREETEESFTWLFNTFKECM--NGKVPIGILTDNCPSMAAAIRTV 418
Query: 309 FPAARCLLCQFHVLKSVTGEM--------------KKLVKDIETREDITDRWNRLMYAAN 354
FP +C++HVLK M K++ T E+ W++L+
Sbjct: 419 FPNTIHRVCKWHVLKKAKEFMGNIYSKRHTFKKAFHKVLTQTLTEEEFVAAWHKLI---R 475
Query: 355 EVEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTCMHMGCTTTNRVEGAHSKLKK 414
+ +K+V Y+ W +R K+ G TTT R E A+ K
Sbjct: 476 DYNLEKSV-----------YLRHIW-DIRRKWAFVYFSHRFFAGMTTTQRSESANHVFKM 523
Query: 415 LL--SDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDPFYKHLRGFVSRAA 472
+ S S V + + + D E FQ S E P H +RA
Sbjct: 524 FVSPSSSMNGFVKRYDRFFNEKLQKED--SEEFQTSNDKVEIKTRSPIEIHASQVYTRAV 581
Query: 473 LHIIVKEKT 481
+ +E T
Sbjct: 582 FQLFSEELT 590
>ref|NP_908529.1| P0702D12.15 [Oryza sativa (japonica cultivar-group)]
Length = 832
Score = 109 bits (273), Expect = 4e-22
Identities = 106/489 (21%), Positives = 194/489 (38%), Gaps = 50/489 (10%)
Query: 17 QAFESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMTLGCERGGKYKPSTSVLKRN 76
Q F++ +D N+ G+ I + K + +R + + C G+ P T
Sbjct: 75 QRFKTERDAYNFYNVYAVSKGFGIRLDKDRKNTKNQRTMRQICCSHQGR-NPKT------ 127
Query: 77 RTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAE---VLQGHSYVGRLNPEEKAM 133
+ + + CP ++ RS W+V HNH + V + + +++ + +
Sbjct: 128 KKPSVRIGCPAMMKINRSRAGSGWSVTKVVSAHNHPMKKSVGVTKNYQSHNQIDEGTRGI 187
Query: 134 VGEMIEERVKASDILIAIR-KHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMKLLEE 192
+ EM++ + +++ + H ++ + ++ AY + S +MQ + +L++
Sbjct: 188 IEEMVDSSMSLTNMYGMLSGMHGGPSMVPFTRKAMDRLAYAIRRDESSDDMQKTLDVLKD 247
Query: 193 ----HKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMV 248
K +S D V+++FW+H S F F V+ D+TYKTN+Y +P V
Sbjct: 248 LQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFV 307
Query: 249 GVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETV 308
GV + + A ++ E ++ W N K+ + +P +TD + A+ TV
Sbjct: 308 GVNNHFQSTFFGCALLREETEESFTWLFNTFKECM--NGKVPIGILTDNCPSMAAAIRTV 365
Query: 309 FPAARCLLCQFHVLKSVTGEM--------------KKLVKDIETREDITDRWNRLMYAAN 354
FP +C++HVLK M K++ T E+ W++L+
Sbjct: 366 FPNTIHRVCKWHVLKKAKEFMGNIYSKRHTFKKAFHKVLTQTLTEEEFVAAWHKLI---R 422
Query: 355 EVEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTCMHMGCTTTNRVEGAHSKLKK 414
+ +K+V Y+ W +R K+ G TTT R E A+ K
Sbjct: 423 DYNLEKSV-----------YLRHIW-DIRRKWAFVYFSHRFFAGMTTTQRSESANHVFKM 470
Query: 415 LL--SDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDPFYKHLRGFVSRAA 472
+ S S V + + + D E FQ S E P H +RA
Sbjct: 471 FVSPSSSMNGFVKRYDRFFNEKLQKED--SEEFQTSNDKVEIKTRSPIEIHASQVYTRAV 528
Query: 473 LHIIVKEKT 481
+ +E T
Sbjct: 529 FQLFSEELT 537
>dbj|BAD81770.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)]
Length = 883
Score = 108 bits (271), Expect = 6e-22
Identities = 106/489 (21%), Positives = 193/489 (38%), Gaps = 50/489 (10%)
Query: 17 QAFESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMTLGCERGGKYKPSTSVLKRN 76
Q F++ +D N+ G+ I + K + K+R + + C G+ P T
Sbjct: 126 QRFKTERDAYNFYNVYAVSKGFGIRLDKDHMNTKKQRTMRQICCSHQGR-NPKT------ 178
Query: 77 RTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAE---VLQGHSYVGRLNPEEKAM 133
+ + + CP ++ S W+V HNH + V + + +++ + +
Sbjct: 179 KKPSVRIGCPAMMKINMSGAGSGWSVTKVVSAHNHPMKKSVGVTKNYQSHNQIDEGTRGI 238
Query: 134 VGEMIEERVKASDILIAIR-KHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMKLLEE 192
+ EM++ + +++ + H ++ + ++ AY + S +MQ + +L++
Sbjct: 239 IEEMVDSSMSLTNMYGMLSGMHGGPSMVPFTRKAMDRVAYAIRRDESSDDMQKTLDVLKD 298
Query: 193 ----HKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMV 248
K +S D V+++FW+H S F F V+ D+TYKTN+Y +P V
Sbjct: 299 LQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFV 358
Query: 249 GVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETV 308
GV + + A ++ E ++ W N K+ + +P +TD + A+ TV
Sbjct: 359 GVNNHFQSTFFGCALLREETEESFTWLFNTFKECM--NGKVPIGILTDNCPSMAAAIRTV 416
Query: 309 FPAARCLLCQFHVLKSVTGEM--------------KKLVKDIETREDITDRWNRLMYAAN 354
FP +C+ HVLK M K++ T E+ W++L+
Sbjct: 417 FPNTIHRVCKSHVLKKAKEFMGNIYSKCHTFKKAFHKVLTQTLTEEEFVAAWHKLI---R 473
Query: 355 EVEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTCMHMGCTTTNRVEGAHSKLKK 414
+ +K+V Y+ W +R K+ G TTT R E A+ K
Sbjct: 474 DYNLEKSV-----------YLRHIW-DIRRKWAFVYFSHRFFAGMTTTQRSESANHVFKM 521
Query: 415 LL--SDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTEHSLNDPFYKHLRGFVSRAA 472
+ S S V + + + D E FQ S E P H +RA
Sbjct: 522 FVSPSSSMNGFVKRYDRFFNEKLQKED--SEEFQTSNDKVEIKTRSPIEIHASQVYTRAV 579
Query: 473 LHIIVKEKT 481
+ +E T
Sbjct: 580 FQLFSEELT 588
>gb|AAU10761.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 822
Score = 98.2 bits (243), Expect = 1e-18
Identities = 71/315 (22%), Positives = 139/315 (43%), Gaps = 17/315 (5%)
Query: 17 QAFESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMTLGCERGGKYKPSTSVLKRN 76
Q F++ +D N+ G+ I + K + K+R + + C G+ P T
Sbjct: 128 QRFKTERDAYNFYNVYAVSKGFGIRLDKDRMNTKKQRTMRQICCSHQGR-NPKT------ 180
Query: 77 RTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAE---VLQGHSYVGRLNPEEKAM 133
+ + + CP ++ S W+V HNH + V + + +++ + +
Sbjct: 181 KKPSVRIGCPAMMKINMSGAGSGWSVTKVVSAHNHPMKKSVGVTKNYQSHNQIDEGTRGI 240
Query: 134 VGEMIEERVKASDILIAIR-KHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMKLLEE 192
+ EM++ + +++ + H ++ + ++ AY + S +MQ + +L++
Sbjct: 241 IEEMVDSSMSLTNMYGMLSGMHGGPSMVPFTRKAMDRVAYAIRRDESSDDMQKTLDVLKD 300
Query: 193 ----HKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMV 248
K +S D V+++FW+H S F F V+ D+TYKTN+Y +P V
Sbjct: 301 LQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFV 360
Query: 249 GVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETV 308
GV + + A ++ E ++ W N K+ + +P +TD + A+ TV
Sbjct: 361 GVNNHFQSTFFGCALLREETEESFTWLFNTFKECM--NGKVPIGILTDNCPSMAAAIRTV 418
Query: 309 FPAARCLLCQFHVLK 323
FP +C++HVLK
Sbjct: 419 FPNTIHRVCKWHVLK 433
>emb|CAE02137.2| OSJNBa0074L08.5 [Oryza sativa (japonica cultivar-group)]
gi|50926620|ref|XP_473257.1| OSJNBa0074L08.5 [Oryza
sativa (japonica cultivar-group)]
Length = 1061
Score = 90.1 bits (222), Expect = 3e-16
Identities = 156/759 (20%), Positives = 277/759 (35%), Gaps = 145/759 (19%)
Query: 37 GYVIIIQKSDYGSDKRR----AVMTLGCER------GGKYKPSTSVLKRNRTGTKKCNCP 86
G+ ++I + K+R T C R G K K + +RN + +C
Sbjct: 321 GFSVVITHHYKSTSKKRFGQITKYTYQCYRYGKNDDGQKKKKKKTQQQRNTQVIDRTDCK 380
Query: 87 FRLRARRSNKDKMWTVLVHSGIHNHDT-----AEVLQGHSYVGRLNPEEKAMVGEMIEER 141
L R +W +L + HNH A+ L+ H + + EEK ++ + +
Sbjct: 381 CMLTVRLDGD--VWRILSMNLNHNHTLTPNREAKFLRSHKH---MTDEEKKLIRTLKQCN 435
Query: 142 VKASDILIAIRKHNPTNLTRI----KQIYNEKQAYNRLKRGSLTEMQHLMKLLEEHKYA- 196
+ + +I+I + + K + N A N R T+M +M+ L + +
Sbjct: 436 IPTRN-MISILSFLRGGIAALPYTGKDVSNVGTAINSETRN--TDMNQVMEYLRQKEAKD 492
Query: 197 ---HWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTST 253
++ D + V+S+FW S L+ ++ V D+TYKTNRY +P +VGVT
Sbjct: 493 PGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYKTNRYNMPFAPIVGVTGH 552
Query: 254 NLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAAR 313
A A++ +E + W L P+ +TD+D + A+ VFP ++
Sbjct: 553 GNICIFACAFLGDETTETFKWVFETF--LTAMGGKHPETIITDQDLAMRAAIRQVFPNSK 610
Query: 314 CLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLMY-AANEVEYDK---AVGAMSNTG 369
C FH+LK D + R+D+ +N +++ + E++ + A N
Sbjct: 611 HRNCLFHILKKCRERSGNTFSD-KRRKDLYAEFNDIVHNSLTRAEFESLWLQMIAQYNLK 669
Query: 370 ELWTYIETNW------LPLRSK--FVKFEIDTCMHMGCTTTNR--VEGAHSKLKKLLSDS 419
+ Y+E W +P+ SK F F T + G + V HS + LL
Sbjct: 670 NI-KYLEIMWRTRKNFIPVYSKTDFCPFIHSTALSEGTNARFKRGVGPTHSVMTFLLEYE 728
Query: 420 KGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTE-------HSLNDPFYKHLRGFVS--- 469
+ +I T K ++ K K SF + + E H + D F L+G ++
Sbjct: 729 TINDIIFDTEYGKDYESRNKKPK-SFWSNYMIEEQAADLYNHGIFDKFQDELKGTLNLEV 787
Query: 470 ----------------------RAALHIIVKEKTQIGMIGVGGIYCECVLRRTHGLPCAC 507
R ++++ + Q + C C G+ C+
Sbjct: 788 AAIQQGKAYEVYAAPNLKQQEFRPRRYVVMTDLPQENFV------CICAKFSKDGILCSH 841
Query: 508 EL-----MKFESVPLLAIHPHWRK------LTWDSSDRDMDPTLGLSFE----------- 545
L +K +P I WRK ++ + D + ++ L F
Sbjct: 842 VLKVMLYLKMSKIPDKYIIERWRKKERKLTVSMQLATNDENSSV-LRFNVLSRRICNMAS 900
Query: 546 -------------SEFDKLRTKFE-------------ESSHGVRMSIIESLRMITYPETT 579
E D L + + + S+ V + + T E
Sbjct: 901 KASKSKETCEYLLGEIDNLDSNLDLIIEDAAQNQASIDESNTVENTSEDQAEKETTNEQE 960
Query: 580 SMCAPTKKKTRGRPMGST--------NKPKKFAKGCGETSTKRIPCRWETIDKEYPGSWR 631
+ P T+G+ T +PKK CG TS C I + +
Sbjct: 961 DIQDPDIANTKGKKSARTKGMVEKIKEQPKKHCTRCGRTSHTIENCPLMQIARSTKKIQK 1020
Query: 632 DSNTTPLQEPSPKVRSQKASKASKASKASKVSTASKASK 670
+ T +++ + R +K + K SK K ++K +K
Sbjct: 1021 STTTRTIEKQAMPARKKKHEENDKGSKLPKDQGSTKKNK 1059
>gb|AAX94831.1| transposon protein, putative, unclassified [Oryza sativa (japonica
cultivar-group)]
Length = 1023
Score = 89.7 bits (221), Expect = 4e-16
Identities = 91/391 (23%), Positives = 157/391 (39%), Gaps = 48/391 (12%)
Query: 60 CERGGKYKP---STSVLKRNRTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNH----- 111
C R G K S R +C C + + K+ W V+ HNH
Sbjct: 625 CSREGFRKELYMDVSNRSREPQALTRCGCKAQFEIKLYQKEGNWYVVRFVSKHNHPLCKG 684
Query: 112 DTAEVLQGHSYVGRLNPEEKAMVGEMIEERVKASDIL-IAIRKHNPTNLTRI--KQIYN- 167
D L+ H R+ P E+A + E+ + + ++ I R H T + +YN
Sbjct: 685 DQVPFLRSHR---RITPAEQAKMVELRDVGLHQHQVMDIMERDHGGYEGTGFTSRDMYNF 741
Query: 168 -EKQAYNRLKRGSLTEMQHLMKLLEEHKYA----HWSRVQDGTDVVRSLFWAHPDSIHLF 222
K R+K G + H++K ++ + ++ D ++ LFW+ P S +
Sbjct: 742 FVKLKKKRIKGG---DADHVLKYMQARQKDDMEFYYDYEIDQAGCLKRLFWSDPQSRIDY 798
Query: 223 NEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDL 282
+ F VV+ DSTY+ N+Y +P + VGV T A A + +E+ + W + +
Sbjct: 799 DAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTVIFACAVVSDERVETYEWVLRQFLTC 858
Query: 283 IINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSVT--------GEMKKLVK 334
+ + PK +TD D + A+ VFP + LC +H+ +++ + + LV
Sbjct: 859 MCQKH--PKSVITDGDNAMRRAILHVFPNSDHRLCTWHIEQNMARNLSPAMLSDFRTLVH 916
Query: 335 DIETREDITDRWNRLMYAANEVEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTC 394
++ +W + +K + M N LR K+
Sbjct: 917 SEFDEDEFERKWVEFKIKHKVSDDNKWLKRMYN--------------LRKKWAATYTKGR 962
Query: 395 MHMGCTTTNRVEGAHSKLKKLLSDSKGDLVI 425
+ + + R E +SKL +LL D K LVI
Sbjct: 963 VFLSMKSNQRSESLNSKLHRLL-DRKMSLVI 992
>ref|XP_463702.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)]
Length = 1114
Score = 89.7 bits (221), Expect = 4e-16
Identities = 113/480 (23%), Positives = 189/480 (38%), Gaps = 69/480 (14%)
Query: 37 GYVIIIQKSDYGSDKRR----AVMTLGCER------GGKYKPSTSVLKRNRTGTKKCNCP 86
G+ ++I + K+R T C R G K K + +RN + +C
Sbjct: 374 GFSVVITHHYKSTSKKRFGQITKYTYQCYRYGKNDDGQKKKKKKTQQQRNTQVIDRTDCK 433
Query: 87 FRLRARRSNKDKMWTVLVHSGIHNHDT-----AEVLQGHSYVGRLNPEEKAMVGEMIEER 141
L R +W +L + HNH A+ L+ H + + EEK ++ + +
Sbjct: 434 CMLTVRLEGD--VWRILSMNLNHNHTLTPNREAKFLRSHKH---MTDEEKKLIRTLKQCN 488
Query: 142 VKASDILIAIRKHNPTNLTRI----KQIYNEKQAYNRLKRGSLTEMQHLMKLLEEHKYA- 196
+ + +I+I + + K + N A N R T+M +M+ L + +
Sbjct: 489 IPTRN-MISILSFLRGGIAALPYTRKDVSNVGTAINSETRN--TDMNQVMEYLRQKEAKD 545
Query: 197 ---HWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTST 253
++ D + V+S+FW S L+ ++ V D+TYKTNRY +P +VGVT
Sbjct: 546 PGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYKTNRYNMPFAPIVGVTGH 605
Query: 254 NLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAAR 313
A A++ +E + W L P+ +TD+D + A+ VFP ++
Sbjct: 606 GNICIFACAFLGDETTETFKWVFETF--LTAMGGKHPETIITDQDLAMRAAIRQVFPNSK 663
Query: 314 CLLCQFHVL----------------KSVTGEMKKLVKDIETREDITDRWNRLMYAANEVE 357
C FH+L K + E +V + TR + W + M A +E
Sbjct: 664 HRNCLFHILKKCRERSGNTFSDKRRKDLYAEFNDIVHNSLTRAEFESLWLQ-MIAQYNLE 722
Query: 358 YDKAVGAMSNTGELWTYIETNWLPLRSK--FVKFEIDTCMHMGCTTTNR--VEGAHSKLK 413
K + M T N++P+ SK F F T + G + V HS +
Sbjct: 723 NIKYLEIMWRT-------RKNFIPVYSKTDFCPFIHSTALSEGTNARFKRGVGPTHSVMT 775
Query: 414 KLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTE-------HSLNDPFYKHLRG 466
LL + +I T K ++ K K SF + + E H + D F L+G
Sbjct: 776 FLLEYETINDIIFDTEYGKDYESRNKKPK-SFWSNYMIEEQAADLYNHGIFDKFQDELKG 834
>dbj|BAD87203.1| far-red impaired response-like [Oryza sativa (japonica
cultivar-group)]
Length = 1130
Score = 89.7 bits (221), Expect = 4e-16
Identities = 113/480 (23%), Positives = 189/480 (38%), Gaps = 69/480 (14%)
Query: 37 GYVIIIQKSDYGSDKRR----AVMTLGCER------GGKYKPSTSVLKRNRTGTKKCNCP 86
G+ ++I + K+R T C R G K K + +RN + +C
Sbjct: 390 GFSVVITHHYKSTSKKRFGQITKYTYQCYRYGKNDDGQKKKKKKTQQQRNTQVIDRTDCK 449
Query: 87 FRLRARRSNKDKMWTVLVHSGIHNHDT-----AEVLQGHSYVGRLNPEEKAMVGEMIEER 141
L R +W +L + HNH A+ L+ H + + EEK ++ + +
Sbjct: 450 CMLTVRLEGD--VWRILSMNLNHNHTLTPNREAKFLRSHKH---MTDEEKKLIRTLKQCN 504
Query: 142 VKASDILIAIRKHNPTNLTRI----KQIYNEKQAYNRLKRGSLTEMQHLMKLLEEHKYA- 196
+ + +I+I + + K + N A N R T+M +M+ L + +
Sbjct: 505 IPTRN-MISILSFLRGGIAALPYTRKDVSNVGTAINSETRN--TDMNQVMEYLRQKEAKD 561
Query: 197 ---HWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTST 253
++ D + V+S+FW S L+ ++ V D+TYKTNRY +P +VGVT
Sbjct: 562 PGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYKTNRYNMPFAPIVGVTGH 621
Query: 254 NLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAAR 313
A A++ +E + W L P+ +TD+D + A+ VFP ++
Sbjct: 622 GNICIFACAFLGDETTETFKWVFETF--LTAMGGKHPETIITDQDLAMRAAIRQVFPNSK 679
Query: 314 CLLCQFHVL----------------KSVTGEMKKLVKDIETREDITDRWNRLMYAANEVE 357
C FH+L K + E +V + TR + W + M A +E
Sbjct: 680 HRNCLFHILKKCRERSGNTFSDKRRKDLYAEFNDIVHNSLTRAEFESLWLQ-MIAQYNLE 738
Query: 358 YDKAVGAMSNTGELWTYIETNWLPLRSK--FVKFEIDTCMHMGCTTTNR--VEGAHSKLK 413
K + M T N++P+ SK F F T + G + V HS +
Sbjct: 739 NIKYLEIMWRT-------RKNFIPVYSKTDFCPFIHSTALSEGTNARFKRGVGPTHSVMT 791
Query: 414 KLLSDSKGDLVIAWTAMNKLIIMQHDKIKESFQLSIFVTE-------HSLNDPFYKHLRG 466
LL + +I T K ++ K K SF + + E H + D F L+G
Sbjct: 792 FLLEYETINDIIFDTEYGKDYESRNKKPK-SFWSNYMIEEQAADLYNHGIFDKFQDELKG 850
>gb|AAX92763.1| transposon protein, putative, unclassified [Oryza sativa (japonica
cultivar-group)]
Length = 1185
Score = 89.7 bits (221), Expect = 4e-16
Identities = 91/391 (23%), Positives = 157/391 (39%), Gaps = 48/391 (12%)
Query: 60 CERGGKYKP---STSVLKRNRTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNH----- 111
C R G K S R +C C + + K+ W V+ HNH
Sbjct: 625 CSREGFRKELYMDVSNRSREPQALTRCGCKAQFEIKLYQKEGNWYVVRFVSKHNHPLCKG 684
Query: 112 DTAEVLQGHSYVGRLNPEEKAMVGEMIEERVKASDIL-IAIRKHNPTNLTRI--KQIYN- 167
D L+ H R+ P E+A + E+ + + ++ I R H T + +YN
Sbjct: 685 DQVPFLRSHR---RITPAEQAKMVELRDVGLHQHQVMDIMERDHGGYEGTGFTSRDMYNF 741
Query: 168 -EKQAYNRLKRGSLTEMQHLMKLLEEHKYA----HWSRVQDGTDVVRSLFWAHPDSIHLF 222
K R+K G + H++K ++ + ++ D ++ LFW+ P S +
Sbjct: 742 FVKLKKKRIKGG---DADHVLKYMQARQKDDMEFYYDYEIDQAGCLKRLFWSDPQSRIDY 798
Query: 223 NEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDL 282
+ F VV+ DSTY+ N+Y +P + VGV T A A + +E+ + W + +
Sbjct: 799 DAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTVIFACAVVSDERVETYEWVLRQFLTC 858
Query: 283 IINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSVT--------GEMKKLVK 334
+ + PK +TD D + A+ VFP + LC +H+ +++ + + LV
Sbjct: 859 MCQKH--PKSVITDGDNAMRRAILHVFPNSDHRLCTWHIEQNMARNLSPAMLSDFRTLVH 916
Query: 335 DIETREDITDRWNRLMYAANEVEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTC 394
++ +W + +K + M N LR K+
Sbjct: 917 SEFDEDEFERKWVEFKIKHKVSDDNKWLKRMYN--------------LRKKWAATYTKGR 962
Query: 395 MHMGCTTTNRVEGAHSKLKKLLSDSKGDLVI 425
+ + + R E +SKL +LL D K LVI
Sbjct: 963 VFLSMKSNQRSESLNSKLHRLL-DRKMSLVI 992
>ref|XP_466614.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|47497275|dbj|BAD19318.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 880
Score = 88.6 bits (218), Expect = 8e-16
Identities = 98/446 (21%), Positives = 175/446 (38%), Gaps = 67/446 (15%)
Query: 15 TSQAFESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMT---LGCERGGKYKP--- 68
T+ F++ K+ + KE G+ + +K+ D +T C R G K
Sbjct: 59 TAMRFKTEKEGFLFYNRYAKEKGFSV--RKNYIRRDPITVAVTHRQYQCSREGHRKEVYM 116
Query: 69 STSVLKRNRTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNH-----DTAEVLQGHSYV 123
+ R +C C + K W V+ + HNH D L+ H +
Sbjct: 117 EAANRSREPKALTRCGCNALFEIKLDKKKGDWFVVKYVAKHNHPLAKSDEVAFLRSHRTI 176
Query: 124 GRLNPEEKAMVGEMIEERVKASDILIAIRKHN----PTNLTRIKQIYNEKQAYNRLKRGS 179
+KA + E+ E ++ ++ + +H+ T + +YN + RL++
Sbjct: 177 SNA---QKANILELKEVGLRQHQVMDVMERHHGGFDATGFVS-RDLYNY---FTRLRK-- 227
Query: 180 LTEMQHLMKLLEEH--KYAHWSRVQD-------GTDV---VRSLFWAHPDSIHLFNEFPH 227
+H++ E KY W + D TD ++ LFW+ P S ++ F
Sbjct: 228 ----KHILGGDAERVIKYFQWRQKHDMEFFFEYETDEAGRLKRLFWSDPQSRIDYDAFGD 283
Query: 228 VVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINED 287
VV+ DSTY+ NRY +P + VGV T A + +E+ W + + D + +
Sbjct: 284 VVVFDSTYRVNRYNLPFIPFVGVNHHGSTIIFGCAIVADEKVATYEWILKQFLDCMYQKH 343
Query: 288 NLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSVTGEMK-KLVKDIET-------R 339
P+ +TD D + A+ V P + LC +H+ +++ ++ ++ D T
Sbjct: 344 --PRALITDGDNAMRRAIAAVMPDSEHWLCTWHIEQNMARHLRPDMLSDFRTLVHAPYDH 401
Query: 340 EDITDRWNRLMYAANEVEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTCMHMGC 399
E+ +W E ++ + M N + W T L +G
Sbjct: 402 EEFERKWVEFKVKHKGCEDNQWLVRMYNLRKKWATAYTKGL--------------FFLGM 447
Query: 400 TTTNRVEGAHSKLKKLLSDSKGDLVI 425
+ R E +SKL + L D K LV+
Sbjct: 448 KSNQRSESLNSKLHRYL-DRKMSLVL 472
>dbj|BAD29601.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|50251625|dbj|BAD29488.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 688
Score = 88.6 bits (218), Expect = 8e-16
Identities = 83/371 (22%), Positives = 142/371 (37%), Gaps = 57/371 (15%)
Query: 74 KRNRTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVGRLNPEEKAM 133
KR C C +L R+ + +W V H H+ A NP+ A
Sbjct: 79 KREPRALTHCGCLAKLEIERNEEKGVWFVKEFDNQHTHELA------------NPDHVAF 126
Query: 134 VG--EMIEERVKASDILIAIRKHNPTNLTRIKQIYNE---------KQAYNRLKRGSLTE 182
+G ++ + KA + + + P + + + ++ K YN R +
Sbjct: 127 LGVHRVMSDSKKAQAVELRMSGLRPFQVMEVMENNHDELDEVGFVMKDLYNFFTRYEMKN 186
Query: 183 ---------MQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDS 233
+++L + EE + D +R++FWA +S + F VVI DS
Sbjct: 187 IKGHDAEDVLKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDS 246
Query: 234 TYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVF 293
TY+ N+Y +P + +GV T + NE + W + + + PK
Sbjct: 247 TYRVNKYNLPFIPFIGVNHHRSTTIFGCGILSNESVNSYCWLLETFLEAM--RQVHPKSL 304
Query: 294 VTDKDQVLMEALETVFPAARCLLCQFHVLKSVT--------GEMKKLVKDIETREDITDR 345
+TD D + +A+ V P A LC +H+ ++++ E++KL+ + E+ R
Sbjct: 305 ITDGDLAMAKAISKVMPGAYHRLCTWHIEENMSRHLRKPKLDELRKLIYESMDEEEFERR 364
Query: 346 WNRLMYAANEVEYDKAVGAMSNTGELWTYIETNWLPLRSKFVKFEIDTCMHMGCTTTNRV 405
W K G N E W + LR K+ D +G + R
Sbjct: 365 WADF----------KENGGTGN--EQWIAL---MYRLREKWAAAYTDGKYLLGMRSNQRS 409
Query: 406 EGAHSKLKKLL 416
E +SKL LL
Sbjct: 410 ESLNSKLHTLL 420
>gb|AAN52752.1| Unknown protein [Oryza sativa (japonica cultivar-group)]
gi|34902304|ref|NP_912498.1| Unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 909
Score = 86.3 bits (212), Expect = 4e-15
Identities = 89/417 (21%), Positives = 161/417 (38%), Gaps = 30/417 (7%)
Query: 17 QAFESSKDLVNWAKAVGKEHGYVIIIQKSDYGSDKRRAVMTLGCERGGKYKPSTSVLKRN 76
Q F D + + G+ I K+ Y +++ C R G+ T R
Sbjct: 140 QTFNEDSDGYAFYNLYARFTGFGIRRSKNRYKDGGVKSMQEFCCIREGRDNSVTGPPTR- 198
Query: 77 RTGTKKCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQ------GHSYVGRLNPEE 130
C +R RS++ + W V HNH+ LQ H+++ +
Sbjct: 199 ------IGCKAMVRLNRSSESQKWRVSAFVSEHNHEMKRDLQHTKHFRSHNFI---DEGT 249
Query: 131 KAMVGEMIEERVKASDIL-IAIRKHNPTNLTRIKQIYNEKQAYNRLKRGSLTEMQHLMKL 189
K + EM++ + + + + H +LT + + AY + ++Q +
Sbjct: 250 KRNIKEMVDNGMTPTAMYGLLSGMHGGPSLTPFTRRAVTRMAYAIRRDECSNDVQKTLNF 309
Query: 190 LEE----HKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLL 245
E K ++ DG ++++FW H S F+ F + D+TY+TNRY +P
Sbjct: 310 FREMQCRSKNFFYTIQIDGASRIKNIFWCHSLSRLSFDHFGDAITFDTTYQTNRYNMPFG 369
Query: 246 EMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEAL 305
V V + T A ++ E + W D + N P +TD + A+
Sbjct: 370 IFVDVNNHFQTAIFGCALLREETIEAFKWLFQTFTDAM--HGNRPAAILTDNCHQMEVAI 427
Query: 306 ETVFPAARCLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLM-YAANEVEYDKAVGA 364
+ V+P +C++HVLK+ + + R ++R++ E E+DKA
Sbjct: 428 KAVWPETIHRVCKWHVLKNAKENLGNI---YSKRSSFKQEFHRVLNEPQTEAEFDKAWSD 484
Query: 365 MSNTGELWT--YIETNWLPLRSKFVKFEIDTCMHMGCTTTNRVEGAHSKLKKLLSDS 419
+ L + Y+ W ++ K+ +TT R E + LKK + S
Sbjct: 485 LMEQYNLESSVYLRRMW-DMKKKWAPAYFREFFFARMSTTQRSESMNHVLKKYVKPS 540
>ref|XP_483730.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|42407926|dbj|BAD09065.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)] gi|42407459|dbj|BAD10392.1|
putative far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 690
Score = 86.3 bits (212), Expect = 4e-15
Identities = 59/226 (26%), Positives = 101/226 (44%), Gaps = 25/226 (11%)
Query: 208 VRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYRIPLLEMVGVTSTNLTYSIAFAYMQNE 267
++ LFWA P S ++ F VV+ DSTY+ N+Y +P + VGV T A A + +E
Sbjct: 198 LKRLFWADPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTIIFACAIVADE 257
Query: 268 QKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVLMEALETVFPAARCLLCQFHVLKSVTG 327
+ W + R D + + PK +TD D + A+ TV P + LC +H+ +++
Sbjct: 258 KIATYEWVLKRFLDCMCQKH--PKGLITDSDNSMRRAIATVMPNSEHRLCTWHIEQNMAR 315
Query: 328 EMK-KLVKDIE-------TREDITDRWNRLMYAANEVEYDKAVGAMSNTGELWTYIETNW 379
++ K++ D + E+ ++W E ++ +G M N + W T
Sbjct: 316 HLRPKMISDFRVLVHATYSAEEFEEKWVEFKLKRKVAEDNQWLGRMYNLRKKWAAAYTKG 375
Query: 380 LPLRSKFVKFEIDTCMHMGCTTTNRVEGAHSKLKKLLSDSKGDLVI 425
+ +G + R E +SKL + L D K LV+
Sbjct: 376 M--------------YFLGMKSNQRSESLNSKLHRHL-DRKMSLVL 406
>ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultivar-group)]
gi|38345091|emb|CAD40514.2| OSJNBa0050F15.2 [Oryza
sativa (japonica cultivar-group)]
Length = 688
Score = 86.3 bits (212), Expect = 4e-15
Identities = 81/360 (22%), Positives = 146/360 (40%), Gaps = 51/360 (14%)
Query: 82 KCNCPFRLRARRSNKDKMWTVLVHSGIHNHDTAEVLQGHSYVGRLNPEEKAMVG--EMIE 139
+C C +L R+ + +W V H H+ A NP+ A +G ++
Sbjct: 87 RCGCLAKLEIERNEEKGVWFVKEFDNQHTHELA------------NPDHVAFLGVHRVMS 134
Query: 140 ERVKASDI-----------LIAIRKHNPTNLTRI----KQIYNEKQAYNRLK-RGSLTE- 182
+ KA + ++ + ++N L + K +YN Y+ +G E
Sbjct: 135 DSKKAQAVELRMSGLRPFQVMEVMENNHDELDEVGFVMKDLYNFFTRYDMKNIKGHDAED 194
Query: 183 -MQHLMKLLEEHKYAHWSRVQDGTDVVRSLFWAHPDSIHLFNEFPHVVILDSTYKTNRYR 241
+++L + EE + D +R++FWA +S + F VVI DSTY+ N+Y
Sbjct: 195 VLKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYRVNKYN 254
Query: 242 IPLLEMVGVTSTNLTYSIAFAYMQNEQKDEVVWAVNRLKDLIINEDNLPKVFVTDKDQVL 301
+P + +GV T + NE + W + + + PK +TD D +
Sbjct: 255 LPFIPFIGVNHHRSTTIFGCGILSNESVNSYCWLLETFLEAM--RQVHPKSLITDGDLAM 312
Query: 302 MEALETVFPAARCLLCQFHVLKSVTGEMKKLVKDIETREDITDRWNRLMY-AANEVEYDK 360
+A+ V P A LC +H+ ++++ ++K D +L+Y + +E E+++
Sbjct: 313 AKAISKVMPGAYHRLCTWHIEENMSRHLRK---------PKLDELRKLIYESMDEEEFER 363
Query: 361 AVGAMSNTGELWTYIETNWLP----LRSKFVKFEIDTCMHMGCTTTNRVEGAHSKLKKLL 416
G W+ LR K+ D +G + R E +SKL LL
Sbjct: 364 RWADFKENGGTG---NGQWIALMYRLREKWAAAYTDGKYLLGMRSNQRSESLNSKLHTLL 420
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,519,787,076
Number of Sequences: 2540612
Number of extensions: 63736990
Number of successful extensions: 158812
Number of sequences better than 10.0: 441
Number of HSP's better than 10.0 without gapping: 254
Number of HSP's successfully gapped in prelim test: 187
Number of HSP's that attempted gapping in prelim test: 158060
Number of HSP's gapped (non-prelim): 721
length of query: 898
length of database: 863,360,394
effective HSP length: 137
effective length of query: 761
effective length of database: 515,296,550
effective search space: 392140674550
effective search space used: 392140674550
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0546.8


