

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
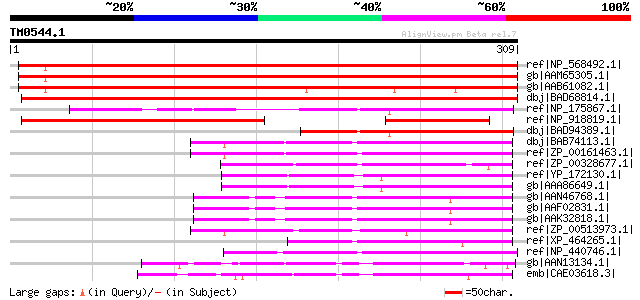
Query= TM0544.1
(309 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568492.1| expressed protein [Arabidopsis thaliana] 449 e-125
gb|AAM65305.1| unknown [Arabidopsis thaliana] 446 e-124
gb|AAB61082.1| contains similarity to Synechococcus PCC7942 chro... 405 e-112
dbj|BAD68814.1| ATP-dependent Zn proteases-like protein [Oryza s... 382 e-105
ref|NP_175867.1| expressed protein [Arabidopsis thaliana] gi|377... 148 2e-34
ref|NP_918819.1| similar to Oryza sativa chromosome 3, OSJNBa007... 145 1e-33
dbj|BAD94389.1| hypothetical protein [Arabidopsis thaliana] 127 3e-28
dbj|BAB74113.1| alr2414 [Nostoc sp. PCC 7120] gi|17229906|ref|NP... 112 1e-23
ref|ZP_00161463.1| COG0465: ATP-dependent Zn proteases [Anabaena... 108 2e-22
ref|ZP_00328677.1| COG0465: ATP-dependent Zn proteases [Trichode... 97 5e-19
ref|YP_172130.1| hypothetical protein syc1420_c [Synechococcus e... 91 6e-17
gb|AAA86649.1| orf5; Method: conceptual translation supplied by ... 91 6e-17
gb|AAN46768.1| At1g56180/F14G9_20 [Arabidopsis thaliana] gi|1840... 88 4e-16
gb|AAF02831.1| Hypothetical protein [Arabidopsis thaliana] 88 4e-16
gb|AAK32818.1| At1g56180/F14G9_20 [Arabidopsis thaliana] 88 4e-16
ref|ZP_00513973.1| conserved hypothetical protein [Crocosphaera ... 82 2e-14
ref|XP_464265.1| unknown protein [Oryza sativa (japonica cultiva... 76 1e-12
ref|NP_440746.1| hypothetical protein sll1738 [Synechocystis sp.... 70 6e-11
gb|AAN13134.1| unknown protein [Arabidopsis thaliana] gi|1433463... 64 7e-09
emb|CAE03618.3| OSJNBb0003B01.9 [Oryza sativa (japonica cultivar... 61 4e-08
>ref|NP_568492.1| expressed protein [Arabidopsis thaliana]
Length = 341
Score = 449 bits (1155), Expect = e-125
Identities = 221/311 (71%), Positives = 264/311 (84%), Gaps = 7/311 (2%)
Query: 6 LMHCVGVTSCPGQLK------LVRVRIQCSTTNVGVS-RRQLLEKLDKELLKGDDRAALA 58
L+H VG+ C Q K R R ++ G+S RRQ LE++D +L GD+RAAL+
Sbjct: 4 LLHSVGLIPCSNQQKSFLFHSYYRYRCIVCSSETGLSIRRQALEQVDSKLSSGDERAALS 63
Query: 59 LVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAA 118
LVKDLQGKPDGLRCFGAARQVPQRLYTL+ELKLNGI A SLLSP D+TLGSIER LQIAA
Sbjct: 64 LVKDLQGKPDGLRCFGAARQVPQRLYTLEELKLNGINAASLLSPTDTTLGSIERNLQIAA 123
Query: 119 IAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRV 178
++GG+ AW AF +S QQLF+++LG +FLWTLD VS+ GG+G+LV+DT GH+FS++YHNRV
Sbjct: 124 VSGGIVAWKAFDLSSQQLFFLTLGFMFLWTLDLVSFNGGIGSLVLDTTGHTFSQRYHNRV 183
Query: 179 IQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAK 238
+QHEAGHFL+AYLVGILP+GYTLSSL+AL+K+GSLN+QAGSAFVD+EFLEEVN+GKVSA
Sbjct: 184 VQHEAGHFLVAYLVGILPRGYTLSSLEALQKEGSLNIQAGSAFVDYEFLEEVNSGKVSAT 243
Query: 239 TLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVL 298
LN+FSCIALAGV TEYL+YG++EGGLDDI KLD L+ LGFTQKK DSQVRWSVLNT+L
Sbjct: 244 MLNRFSCIALAGVATEYLLYGYAEGGLDDISKLDGLVKSLGFTQKKADSQVRWSVLNTIL 303
Query: 299 LLRRHEAARAK 309
LLRRHE AR+K
Sbjct: 304 LLRRHEIARSK 314
>gb|AAM65305.1| unknown [Arabidopsis thaliana]
Length = 341
Score = 446 bits (1147), Expect = e-124
Identities = 220/311 (70%), Positives = 263/311 (83%), Gaps = 7/311 (2%)
Query: 6 LMHCVGVTSCPGQLK------LVRVRIQCSTTNVGVS-RRQLLEKLDKELLKGDDRAALA 58
L+H VG+ C Q K R R ++ G+S RRQ LE++D +L GD+RAAL+
Sbjct: 4 LLHSVGLIPCSNQQKSFLFHSYYRYRCIVCSSETGLSIRRQALEQVDSKLSSGDERAALS 63
Query: 59 LVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAA 118
LVKDLQGKPDGLRCFGAARQVPQRLYTL+ELKLNGI A SLLSP D+TLGSIER LQIAA
Sbjct: 64 LVKDLQGKPDGLRCFGAARQVPQRLYTLEELKLNGINAASLLSPTDTTLGSIERNLQIAA 123
Query: 119 IAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRV 178
++GG+ AW AF +S QQLF+++LG +FLWTLD VS+ GG+G+LV+DT GH+FS++YHNRV
Sbjct: 124 VSGGIVAWKAFDLSSQQLFFLTLGFMFLWTLDLVSFNGGIGSLVLDTTGHTFSQRYHNRV 183
Query: 179 IQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAK 238
+QHEAGHFL+AYLV ILP+GYTLSSL+AL+K+GSLN+QAGSAFVD+EFLEEVN+GKVSA
Sbjct: 184 VQHEAGHFLVAYLVEILPRGYTLSSLEALQKEGSLNIQAGSAFVDYEFLEEVNSGKVSAT 243
Query: 239 TLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVL 298
LN+FSCIALAGV TEYL+YG++EGGLDDI KLD L+ LGFTQKK DSQVRWSVLNT+L
Sbjct: 244 MLNRFSCIALAGVATEYLLYGYAEGGLDDISKLDGLVKSLGFTQKKADSQVRWSVLNTIL 303
Query: 299 LLRRHEAARAK 309
LLRRHE AR+K
Sbjct: 304 LLRRHEIARSK 314
>gb|AAB61082.1| contains similarity to Synechococcus PCC7942 chromosomal region
used as basis of neutral siteII recombinational cloning
vector (PID:g1174192) [Arabidopsis thaliana]
gi|7485298|pir||T01794 hypothetical protein
A_TM021B04.11 - Arabidopsis thaliana
Length = 386
Score = 405 bits (1041), Expect = e-112
Identities = 215/356 (60%), Positives = 258/356 (72%), Gaps = 52/356 (14%)
Query: 6 LMHCVGVTSCPGQLK------LVRVRIQCSTTNVGVS-RRQLLEKLDKELLKGDDRAALA 58
L+H VG+ C Q K R R ++ G+S RRQ LE++D +L GD+RAAL+
Sbjct: 4 LLHSVGLIPCSNQQKSFLFHSYYRYRCIVCSSETGLSIRRQALEQVDSKLSSGDERAALS 63
Query: 59 LVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAA 118
LVKDLQGKPDGLRCFGAARQVPQRLYTL+ELKLNGI A SLLSP D+TLGSIER LQIAA
Sbjct: 64 LVKDLQGKPDGLRCFGAARQVPQRLYTLEELKLNGINAASLLSPTDTTLGSIERNLQIAA 123
Query: 119 IAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRV 178
++GG+ AW AF +S QQLF+++LG +FLWTLD VS+ GG+G+LV+DT GH+FS++YHNRV
Sbjct: 124 VSGGIVAWKAFDLSSQQLFFLTLGFMFLWTLDLVSFNGGIGSLVLDTTGHTFSQRYHNRV 183
Query: 179 I----------------QHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFV 222
+ QHEAGHFL+AYLVGILP+GYTLSSL+AL+K+GSLN+QAGSAFV
Sbjct: 184 VQKHYIIFHWTYCELRSQHEAGHFLVAYLVGILPRGYTLSSLEALQKEGSLNIQAGSAFV 243
Query: 223 DFEFLEEVNAG---KVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRK--------- 270
D+EFLEE N + LN+FSCIALAGV TEYL+YG++EGGLDDI K
Sbjct: 244 DYEFLEEPNKKLCLLFQNQMLNRFSCIALAGVATEYLLYGYAEGGLDDISKVSFLLPLKN 303
Query: 271 -----------------LDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAARAK 309
LD L+ LGFTQKK DSQVRWSVLNT+LLLRRHE AR+K
Sbjct: 304 SSDYVNMLYGFVVLMEQLDGLVKSLGFTQKKADSQVRWSVLNTILLLRRHEIARSK 359
>dbj|BAD68814.1| ATP-dependent Zn proteases-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 346
Score = 382 bits (980), Expect = e-105
Identities = 195/303 (64%), Positives = 242/303 (79%), Gaps = 1/303 (0%)
Query: 8 HCVGVTSCPGQLKLVRVRIQCSTTNVGVSR-RQLLEKLDKELLKGDDRAALALVKDLQGK 66
H G + +L+L+ +T++ +R R +LE++D+EL KG+D AAL+LV+ QG
Sbjct: 17 HPAGGVAAAVRLRLLPPARAANTSSEPAARLRAVLEQVDEELRKGNDEAALSLVRGSQGA 76
Query: 67 PDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAAIAGGLTAW 126
GLR FGAARQVPQRLYTLDELKLNGI+ + LSPVD TLGSIER LQIAA+ GGL+
Sbjct: 77 DGGLRFFGAARQVPQRLYTLDELKLNGIDTSAFLSPVDLTLGSIERNLQIAAVLGGLSVS 136
Query: 127 NAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHF 186
AF +S Q+ ++ LGLL LW++D V +GGG+ NL++DTIGH+ S+KY NRVIQHEAGHF
Sbjct: 137 AAFELSKLQVLFLFLGLLSLWSVDLVYFGGGVRNLILDTIGHNLSQKYRNRVIQHEAGHF 196
Query: 187 LIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCI 246
LIAYL+G+LPKGYT++SLD K GSLNVQAG+AFVDFEFL+EVN+GK+SA LNKFSCI
Sbjct: 197 LIAYLLGVLPKGYTITSLDTFIKKGSLNVQAGTAFVDFEFLQEVNSGKLSATMLNKFSCI 256
Query: 247 ALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
ALAGV TEYL+YG++EGGL DI +LD LL GLGFTQKK DSQVRW+VLNTV LRRH+ A
Sbjct: 257 ALAGVATEYLLYGYAEGGLADIGQLDGLLKGLGFTQKKADSQVRWAVLNTVPALRRHKKA 316
Query: 307 RAK 309
R++
Sbjct: 317 RSQ 319
>ref|NP_175867.1| expressed protein [Arabidopsis thaliana] gi|3776580|gb|AAC64897.1|
Contains similarity to TM021B04.11 gi|2191197 from A.
thaliana BAC gb|AF007271. [Arabidopsis thaliana]
gi|25405756|pir||H96588 hypothetical protein T22H22.11
[imported] - Arabidopsis thaliana
Length = 289
Score = 148 bits (373), Expect = 2e-34
Identities = 96/286 (33%), Positives = 152/286 (52%), Gaps = 63/286 (22%)
Query: 37 RRQLLEKLDKELLKGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEA 96
RR+ LE++DKELL+G+ AL+LVK L+ K L FG+A+ +P+ KL+
Sbjct: 23 RRKSLERVDKELLRGNYETALSLVKQLKSKHGCLSAFGSAKLLPK--------KLDMSSK 74
Query: 97 ESLLSPVDSTLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGG 156
L S +DS SIE ++ + + + SP++ ++
Sbjct: 75 TDLRSLIDSVSRSIE-SVYVQEDSVRTSKEMEIKTSPEEDWF------------------ 115
Query: 157 GLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQ 216
V+QHE+GHFL+ YL+G+LP+ Y + +L+A++++ S NV
Sbjct: 116 --------------------SVVQHESGHFLVGYLLGVLPRHYEIPTLEAVRQNVS-NVT 154
Query: 217 AGSAFVDFEFLEEV---------------NAGKVSAKTLNKFSCIALAGVCTEYLIYGFS 261
FV FEFL++V N G +S+KTLN FSC+ L G+ TE++++G+S
Sbjct: 155 GRVEFVGFEFLKQVGAANQLMKDDVDGQMNQGNISSKTLNNFSCVILGGMVTEHILFGYS 214
Query: 262 EGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAAR 307
EG DI KL+ +L LGFT+ + ++ ++W+V NTV LL H+ AR
Sbjct: 215 EGLYSDIVKLNDVLRWLGFTESEKEAHIKWAVSNTVSLLHSHKEAR 260
>ref|NP_918819.1| similar to Oryza sativa chromosome 3, OSJNBa0077G22.24 [Oryza
sativa (japonica cultivar-group)]
Length = 516
Score = 145 bits (367), Expect = 1e-33
Identities = 80/149 (53%), Positives = 106/149 (70%), Gaps = 1/149 (0%)
Query: 8 HCVGVTSCPGQLKLVRVRIQCSTTNVGVSR-RQLLEKLDKELLKGDDRAALALVKDLQGK 66
H G + +L+L+ +T++ +R R +LE++D+EL KG+D AAL+LV+ QG
Sbjct: 264 HPAGGVAAAVRLRLLPPARAANTSSEPAARLRAVLEQVDEELRKGNDEAALSLVRGSQGA 323
Query: 67 PDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDSTLGSIERTLQIAAIAGGLTAW 126
GLR FGAARQVPQRLYTLDELKLNGI+ + LSPVD TLGSIER LQIAA+ GGL+
Sbjct: 324 DGGLRFFGAARQVPQRLYTLDELKLNGIDTSAFLSPVDLTLGSIERNLQIAAVLGGLSVS 383
Query: 127 NAFGISPQQLFYISLGLLFLWTLDTVSYG 155
AF +S Q+ ++ LGLL LW++D V+ G
Sbjct: 384 AAFELSKLQVLFLFLGLLSLWSVDLVNSG 412
Score = 97.1 bits (240), Expect = 6e-19
Identities = 47/63 (74%), Positives = 54/63 (85%)
Query: 230 VNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQV 289
VN+GK+SA LNKFSCIALAGV TEYL+YG++EGGL DI +LD LL GLGFTQKK DSQV
Sbjct: 409 VNSGKLSATMLNKFSCIALAGVATEYLLYGYAEGGLADIGQLDGLLKGLGFTQKKADSQV 468
Query: 290 RWS 292
+S
Sbjct: 469 LYS 471
>dbj|BAD94389.1| hypothetical protein [Arabidopsis thaliana]
Length = 223
Score = 127 bits (320), Expect = 3e-28
Identities = 63/145 (43%), Positives = 97/145 (66%), Gaps = 16/145 (11%)
Query: 178 VIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEV------- 230
V+QHE+GHFL+ YL+G+LP+ Y + +L+A++++ S NV FV FEFL++V
Sbjct: 51 VVQHESGHFLVGYLLGVLPRHYEIPTLEAVRQNVS-NVTGRVEFVGFEFLKQVGAANQLM 109
Query: 231 --------NAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQ 282
N G +S+KTLN FSC+ L G+ TE++++G+SEG DI KL+ +L LGFT+
Sbjct: 110 KDDVDGQMNQGNISSKTLNNFSCVILGGMVTEHILFGYSEGLYSDIVKLNDVLRWLGFTE 169
Query: 283 KKVDSQVRWSVLNTVLLLRRHEAAR 307
+ ++ ++W+V NTV LL H+ AR
Sbjct: 170 SEKEAHIKWAVSNTVSLLHSHKEAR 194
>dbj|BAB74113.1| alr2414 [Nostoc sp. PCC 7120] gi|17229906|ref|NP_486454.1|
hypothetical protein alr2414 [Nostoc sp. PCC 7120]
gi|25341220|pir||AG2107 hypothetical protein alr2414
[imported] - Nostoc sp. (strain PCC 7120)
Length = 228
Score = 112 bits (280), Expect = 1e-23
Identities = 68/201 (33%), Positives = 112/201 (54%), Gaps = 8/201 (3%)
Query: 111 ERTLQIAAIAGGLTAWNAF-----GISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDT 165
+ L + AI+ L +A +SP + +L + T D+ S G G +++D
Sbjct: 3 QTALNLVAISVFLITMSALLGPLINLSPAIPAIATFTILGIATFDSFSLQGKGGTILLDW 62
Query: 166 IGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFE 225
I FS ++ +R+I HEAGHFL+AYL+G+ GYTLS+ +A ++ L Q G F D E
Sbjct: 63 IA-GFSPQHRDRIIHHEAGHFLVAYLLGVPVTGYTLSAWEAWRQ--GLPGQGGVTFDDVE 119
Query: 226 FLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKV 285
+ +V GK+S + L ++ I +AG+ E L++ +EGG+DD KL ++ LGF++
Sbjct: 120 LMSQVQQGKISNQVLERYCTICMAGIAAETLVFERAEGGIDDKSKLATIFKVLGFSESVC 179
Query: 286 DSQVRWSVLNTVLLLRRHEAA 306
+ R+ VL LL+ + A+
Sbjct: 180 QQKQRFHVLQAKTLLQNNWAS 200
>ref|ZP_00161463.1| COG0465: ATP-dependent Zn proteases [Anabaena variabilis ATCC
29413]
Length = 231
Score = 108 bits (271), Expect = 2e-22
Identities = 67/201 (33%), Positives = 110/201 (54%), Gaps = 8/201 (3%)
Query: 111 ERTLQIAAIAGGLTAWNAF-----GISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDT 165
+ L + AI+ L +A +SP + +L + T D+ S G G +++D
Sbjct: 3 QTALNLVAISVFLMTMSALLGPLINLSPAVPAIATFTILGIATFDSFSLQGKGGTILLDW 62
Query: 166 IGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFE 225
I FS ++ +R+I HEAGHFL+AYL+G+ GYTLS+ +A ++ Q G F D E
Sbjct: 63 IA-GFSPQHRDRIIHHEAGHFLVAYLLGVPVTGYTLSAWEAWRQGQP--GQGGVTFDDVE 119
Query: 226 FLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKV 285
+ +V GK+S + L ++ I +AG+ E L++ +EGG DD KL ++ LGF++
Sbjct: 120 LVSQVEQGKISNQALERYCTICMAGIAAETLVFERAEGGTDDKSKLATIFKVLGFSESVC 179
Query: 286 DSQVRWSVLNTVLLLRRHEAA 306
+ R+ VL LL+ + A+
Sbjct: 180 QQKQRFHVLQAKTLLQNNWAS 200
>ref|ZP_00328677.1| COG0465: ATP-dependent Zn proteases [Trichodesmium erythraeum
IMS101]
Length = 229
Score = 97.4 bits (241), Expect = 5e-19
Identities = 58/180 (32%), Positives = 107/180 (59%), Gaps = 8/180 (4%)
Query: 129 FGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLI 188
F ISP + + +L L T+DT+ + G ++VD + + S+K +R+I HEAGHFL+
Sbjct: 26 FNISPFYIAIATFSVLVLATIDTLGWQGQGSMILVDLVAGTSSEK-RDRIICHEAGHFLV 84
Query: 189 AYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIAL 248
AYL+ I GY L++ +A ++ S Q G F D + ++ +G +S++ ++++ + +
Sbjct: 85 AYLLEIPISGYALNAWEAFRQGQS--SQGGVRFDDQKLAAQLYSGVISSQLVDRYCTVWM 142
Query: 249 AGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVR--WSVLNTVLLLRRHEAA 306
AG+ E L+YG +EGG +D K+ ++L L ++ +S+++ W+ L LL H++A
Sbjct: 143 AGIAAENLVYGNAEGGAEDRTKITAILRQL---KRPGESKLKQSWASLQARNLLENHQSA 199
>ref|YP_172130.1| hypothetical protein syc1420_c [Synechococcus elongatus PCC 6301]
gi|56686388|dbj|BAD79610.1| hypothetical protein
[Synechococcus elongatus PCC 6301]
Length = 212
Score = 90.5 bits (223), Expect = 6e-17
Identities = 59/180 (32%), Positives = 92/180 (50%), Gaps = 9/180 (5%)
Query: 130 GISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIA 189
G SP + LL L++LD V++ G L++D I S +Y R++ HEAGH+L+A
Sbjct: 6 GSSPLLPAGLGFSLLVLFSLDAVTWQGRGATLLLDGIQQR-SPEYRQRILHHEAGHYLVA 64
Query: 190 YLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFE---FLEEVNAGKVSAKTLNKFSCI 246
+G+ GYTLS+ +AL++ Q G V F+ E G++S ++L ++ +
Sbjct: 65 TALGLPVTGYTLSAWEALRQG-----QPGRGGVQFQAAALEAEAAQGQLSQRSLEQWCQV 119
Query: 247 ALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
+AG E L+YG EGG DD + L L + D + RW +L LL + A
Sbjct: 120 LMAGAAAEQLVYGNVEGGADDRAQWKQLWRQLDRNPAEADLRSRWGLLRAKTLLEQQRPA 179
>gb|AAA86649.1| orf5; Method: conceptual translation supplied by author
gi|53763205|ref|ZP_00163803.2| COG0620: Methionine
synthase II (cobalamin-independent) [Synechococcus
elongatus PCC 7942]
Length = 233
Score = 90.5 bits (223), Expect = 6e-17
Identities = 59/180 (32%), Positives = 92/180 (50%), Gaps = 9/180 (5%)
Query: 130 GISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIA 189
G SP + LL L++LD V++ G L++D I S +Y R++ HEAGH+L+A
Sbjct: 27 GSSPLLPAGLGFSLLVLFSLDAVTWQGRGATLLLDGIQQR-SPEYRQRILHHEAGHYLVA 85
Query: 190 YLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFE---FLEEVNAGKVSAKTLNKFSCI 246
+G+ GYTLS+ +AL++ Q G V F+ E G++S ++L ++ +
Sbjct: 86 TALGLPVTGYTLSAWEALRQG-----QPGRGGVQFQAAALEAEAAQGQLSQRSLEQWCQV 140
Query: 247 ALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
+AG E L+YG EGG DD + L L + D + RW +L LL + A
Sbjct: 141 LMAGAAAEQLVYGNVEGGADDRAQWKQLWRQLDRNPAEADLRSRWGLLRAKTLLEQQRPA 200
>gb|AAN46768.1| At1g56180/F14G9_20 [Arabidopsis thaliana]
gi|18405720|ref|NP_564711.1| expressed protein
[Arabidopsis thaliana] gi|12321763|gb|AAG50923.1|
hypothetical protein [Arabidopsis thaliana]
gi|25404102|pir||C96603 hypothetical protein F14G9.20
[imported] - Arabidopsis thaliana
Length = 389
Score = 87.8 bits (216), Expect = 4e-16
Identities = 60/196 (30%), Positives = 106/196 (53%), Gaps = 12/196 (6%)
Query: 113 TLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSK 172
++ +AA+ GG++ + I + + LGL +L D+V GG V +
Sbjct: 175 SIALAALLGGVSYLLSQEIDVRPNLAVILGLAYL---DSVFLGGTCLAQV-----SCYWP 226
Query: 173 KYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNA 232
+ R++ HEAGH L+AYL+G +G L + A++ + QAG+ F D + E+
Sbjct: 227 PHKRRIVVHEAGHLLVAYLMGCPIRGVILDPVVAMQM--GVQGQAGTQFWDQKMESEIAE 284
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDD--IRKLDSLLSGLGFTQKKVDSQVR 290
G++S + +++S + AG+ E L+YG +EGG +D + + S+L + ++ +Q R
Sbjct: 285 GRLSGSSFDRYSMVLFAGIAAEALVYGEAEGGENDENLFRSISVLLEPPLSVAQMSNQAR 344
Query: 291 WSVLNTVLLLRRHEAA 306
WSVL + LL+ H+AA
Sbjct: 345 WSVLQSYNLLKWHKAA 360
>gb|AAF02831.1| Hypothetical protein [Arabidopsis thaliana]
Length = 368
Score = 87.8 bits (216), Expect = 4e-16
Identities = 60/196 (30%), Positives = 106/196 (53%), Gaps = 12/196 (6%)
Query: 113 TLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSK 172
++ +AA+ GG++ + I + + LGL +L D+V GG V +
Sbjct: 154 SIALAALLGGVSYLLSQEIDVRPNLAVILGLAYL---DSVFLGGTCLAQV-----SCYWP 205
Query: 173 KYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNA 232
+ R++ HEAGH L+AYL+G +G L + A++ + QAG+ F D + E+
Sbjct: 206 PHKRRIVVHEAGHLLVAYLMGCPIRGVILDPVVAMQM--GVQGQAGTQFWDQKMESEIAE 263
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDD--IRKLDSLLSGLGFTQKKVDSQVR 290
G++S + +++S + AG+ E L+YG +EGG +D + + S+L + ++ +Q R
Sbjct: 264 GRLSGSSFDRYSMVLFAGIAAEALVYGEAEGGENDENLFRSISVLLEPPLSVAQMSNQAR 323
Query: 291 WSVLNTVLLLRRHEAA 306
WSVL + LL+ H+AA
Sbjct: 324 WSVLQSYNLLKWHKAA 339
>gb|AAK32818.1| At1g56180/F14G9_20 [Arabidopsis thaliana]
Length = 389
Score = 87.8 bits (216), Expect = 4e-16
Identities = 60/196 (30%), Positives = 106/196 (53%), Gaps = 12/196 (6%)
Query: 113 TLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSK 172
++ +AA+ GG++ + I + + LGL +L D+V GG V +
Sbjct: 175 SIALAALLGGVSYLLSQEIDVRPNLAVILGLAYL---DSVFLGGPCLAQV-----SCYWP 226
Query: 173 KYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNA 232
+ R++ HEAGH L+AYL+G +G L + A++ + QAG+ F D + E+
Sbjct: 227 PHKRRIVVHEAGHLLVAYLMGCPIRGVILDPVVAMQM--GVQGQAGTQFWDQKMESEIAE 284
Query: 233 GKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDD--IRKLDSLLSGLGFTQKKVDSQVR 290
G++S + +++S + AG+ E L+YG +EGG +D + + S+L + ++ +Q R
Sbjct: 285 GRLSGSSFDRYSMVLFAGIAAEALVYGEAEGGENDENLFRSISVLLEPPLSVAQMSNQAR 344
Query: 291 WSVLNTVLLLRRHEAA 306
WSVL + LL+ H+AA
Sbjct: 345 WSVLQSYNLLKWHKAA 360
>ref|ZP_00513973.1| conserved hypothetical protein [Crocosphaera watsonii WH 8501]
gi|67857937|gb|EAM53176.1| conserved hypothetical
protein [Crocosphaera watsonii WH 8501]
Length = 224
Score = 82.4 bits (202), Expect = 2e-14
Identities = 64/203 (31%), Positives = 100/203 (48%), Gaps = 14/203 (6%)
Query: 111 ERTLQIAAIAGGLTAWNAF-----GISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDT 165
+ +L + AIA + +A ISP + +L L T+D+ S+GG L +D
Sbjct: 3 QTSLNLVAIAVFVMTLSALLSPVLNISPFIPAATTFAVLGLATVDSFSWGGKGLTLFLDL 62
Query: 166 IGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFE 225
S +K R+I HEAGHFL AY +G+ GY+L++ + ++ +AG V F+
Sbjct: 63 FTSSEERK---RIIHHEAGHFLAAYCLGVPITGYSLTAWETFRQ----GEKAGIGGVQFD 115
Query: 226 FLEEVNAGKVSAKTL--NKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQK 283
F + KVS L + + +AG+ E +IY EGG +D + L LL LG +
Sbjct: 116 FSLLSDQEKVSRNPLIVERTFTVLMAGIAAEKVIYNNVEGGEEDKQNLRELLKILGLRAE 175
Query: 284 KVDSQVRWSVLNTVLLLRRHEAA 306
+ W++L LL RH+ A
Sbjct: 176 LYQQKENWALLQAKNLLIRHQTA 198
>ref|XP_464265.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|49388605|dbj|BAD25720.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 374
Score = 76.3 bits (186), Expect = 1e-12
Identities = 47/139 (33%), Positives = 75/139 (53%), Gaps = 4/139 (2%)
Query: 170 FSKKYHNRVIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEE 229
F Y R++ HEAGH L AYL+G +G L AL+ + QAG+ F D + +E
Sbjct: 209 FWPPYKRRILVHEAGHLLTAYLMGCPIRGVILDPFVALRM--GIQGQAGTQFWDEKMEKE 266
Query: 230 VNAGKVSAKTLNKFSCIALAGVCTEYLIYGFSEGGLDDIRKLDSL--LSGLGFTQKKVDS 287
+ G +S+ +++ I AG+ E L+YG +EGG +D SL L + ++ +
Sbjct: 267 LAEGHLSSTAFDRYCMILFAGIAAEALVYGEAEGGENDENLFRSLCILLDPPLSVAQMAN 326
Query: 288 QVRWSVLNTVLLLRRHEAA 306
+ RWSV+ + LL+ H+ A
Sbjct: 327 RARWSVMQSYNLLKWHKKA 345
>ref|NP_440746.1| hypothetical protein sll1738 [Synechocystis sp. PCC 6803]
gi|1652505|dbj|BAA17426.1| sll1738 [Synechocystis sp.
PCC 6803] gi|7470142|pir||S77323 hypothetical protein
sll1738 - Synechocystis sp. (strain PCC 6803)
Length = 231
Score = 70.5 bits (171), Expect = 6e-11
Identities = 49/179 (27%), Positives = 85/179 (47%), Gaps = 5/179 (2%)
Query: 131 ISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
+SP +LG+L + T D +S+ G + V G +K R++ HEAGHFL+A+
Sbjct: 28 LSPAIPAAATLGILGIITADQISWQGKGTDFFV---GLFQTKAEKERILCHEAGHFLVAH 84
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
+ I Y+LS + L++ AG F + + + L +++ + +AG
Sbjct: 85 CLQIPITNYSLSPWEVLRQGAG--GMAGIQFDTTNLENQCRDWHLRPQALERWATVWMAG 142
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAARAK 309
+ E +IYG S+GG D ++L G + K+ + W+ L LL +H A +
Sbjct: 143 IAAEKIIYGESQGGNGDRQQLRQAFRRAGLPEIKLQQKESWAFLQAKNLLEQHRQAHGQ 201
>gb|AAN13134.1| unknown protein [Arabidopsis thaliana] gi|14334630|gb|AAK59493.1|
unknown protein [Arabidopsis thaliana]
gi|20198006|gb|AAD20413.2| expressed protein
[Arabidopsis thaliana] gi|18399842|ref|NP_565523.1|
expressed protein [Arabidopsis thaliana]
Length = 332
Score = 63.5 bits (153), Expect = 7e-09
Identities = 73/247 (29%), Positives = 115/247 (46%), Gaps = 44/247 (17%)
Query: 81 QRLYTLDELKLNGIE-AESLLSP--------VDSTLGSIERTLQIAAIAGGLTA-WNAFG 130
+R +L EL GI+ AE+L P + + +GS T IA +AG L W F
Sbjct: 92 RRTTSLRELTTLGIKNAETLAIPSVRNDAAFLFTVVGS---TGFIAVLAGQLPGDWGFF- 147
Query: 131 ISPQQLFYISLGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
+ Y+ +G + L L S GL + +F Y R+ HEA HFL+AY
Sbjct: 148 -----VPYL-VGSISLVVLAVGSVSPGLLQAAISGFS-TFFPDYQERIAAHEAAHFLVAY 200
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
L+G+ GY SLD K+ +L +D + + +GK+ +K L++ + +A+AG
Sbjct: 201 LIGLPILGY---SLDIGKEHVNL--------IDERLAKLIYSGKLDSKELDRLAAVAMAG 249
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQ-----VRWSVLNTVLLLRR--- 302
+ E L Y G D+ L ++ +Q K+ ++ RW+VL + LL+
Sbjct: 250 LAAEGLKYDKVIGQSADLFSLQRFINR---SQPKISNEQQQNLTRWAVLYSASLLKNNKT 306
Query: 303 -HEAARA 308
HEA A
Sbjct: 307 IHEALMA 313
>emb|CAE03618.3| OSJNBb0003B01.9 [Oryza sativa (japonica cultivar-group)]
Length = 325
Score = 61.2 bits (147), Expect = 4e-08
Identities = 64/238 (26%), Positives = 109/238 (44%), Gaps = 30/238 (12%)
Query: 79 VPQRLYTLDELKLNGIE-AESLLSPVDSTLGSIERTLQIAAIAGGLTAWNAFGISPQQL- 136
V +R +L EL GI+ AE+L P S+ G T + G+ QL
Sbjct: 83 VSRRTTSLRELTTLGIKNAENLAIP------SVRNDAAFLFTVVGSTGF--LGVLAGQLP 134
Query: 137 ----FYIS--LGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAY 190
F++ +G + L L S GL + F Y R+ +HEA HFL+AY
Sbjct: 135 GDWGFFVPYLIGSISLIVLAIGSISPGLLQAAIGAFSTVFPD-YQERIARHEAAHFLVAY 193
Query: 191 LVGILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAG 250
L+G+ GY SLD K+ +L +D + + + +G++ K +++ + +++AG
Sbjct: 194 LIGLPILGY---SLDIGKEHVNL--------IDDQLQKLIYSGQLDQKEIDRLAVVSMAG 242
Query: 251 VCTEYLIYGFSEGGLDDIRKLDSLLSGL--GFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
+ E L Y G D+ L ++ T+ + + RW+VL + LL+ ++AA
Sbjct: 243 LAAEGLEYDKVVGQSADLFTLQRFINRTKPPLTKDQQQNLTRWAVLFSASLLKNNKAA 300
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.139 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 503,177,564
Number of Sequences: 2540612
Number of extensions: 20788712
Number of successful extensions: 49385
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 49333
Number of HSP's gapped (non-prelim): 37
length of query: 309
length of database: 863,360,394
effective HSP length: 127
effective length of query: 182
effective length of database: 540,702,670
effective search space: 98407885940
effective search space used: 98407885940
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0544.1


