

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
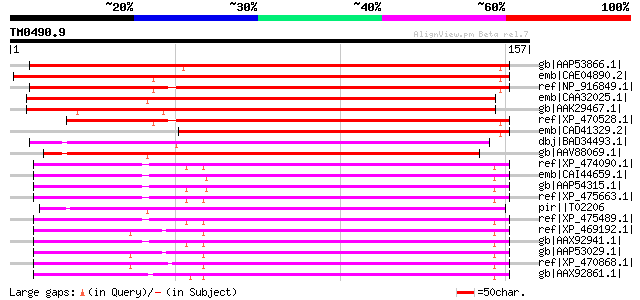
Query= TM0490.9
(157 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP53866.1| putative polyprotein [Oryza sativa (japonica cult... 145 3e-34
emb|CAE04890.2| OSJNBa0042I15.12 [Oryza sativa (japonica cultiva... 143 1e-33
ref|NP_916849.1| retrovirus-related pol polyprotein from transpo... 140 1e-32
emb|CAA32025.1| unnamed protein product [Nicotiana tabacum] gi|1... 134 8e-31
gb|AAK29467.1| polyprotein-like [Lycopersicon chilense] 131 7e-30
ref|XP_470528.1| Putative retrovirus-related pol polyprotein [Or... 124 6e-28
emb|CAD41329.2| OJ991113_30.13 [Oryza sativa (japonica cultivar-... 102 3e-21
dbj|BAD34493.1| Gag-Pol [Ipomoea batatas] 94 9e-19
gb|AAV88069.1| hypothetical retrotransposon [Ipomoea batatas] 87 2e-16
ref|XP_474090.1| OSJNBa0033G05.13 [Oryza sativa (japonica cultiv... 86 4e-16
emb|CAI44659.1| OSJNBa0061C06.9 [Oryza sativa (japonica cultivar... 85 5e-16
gb|AAP54315.1| putative polyprotein [Oryza sativa (japonica cult... 84 9e-16
ref|XP_475663.1| putative polyprotein [Oryza sativa (japonica cu... 84 9e-16
pir||T02206 hypothetical protein - common tobacco retrotransposo... 83 3e-15
ref|XP_475489.1| putative polyprotein [Oryza sativa (japonica cu... 82 6e-15
ref|XP_469192.1| putative polyprotein [Oryza sativa (japonica cu... 81 8e-15
gb|AAX92941.1| retrotransposon protein, putative, Ty1-copia sub-... 81 8e-15
gb|AAP53029.1| putative retrotransposon-related protein [Oryza s... 81 1e-14
ref|XP_470868.1| Putative retroelement pol polyprotein [Oryza sa... 80 2e-14
gb|AAX92861.1| retrotransposon protein, putative, Ty1-copia sub-... 78 9e-14
>gb|AAP53866.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37534554|ref|NP_921579.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 382
Score = 145 bits (367), Expect = 3e-34
Identities = 75/147 (51%), Positives = 105/147 (71%), Gaps = 2/147 (1%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVD-WNEMKKKAAGLIKL 65
+KFEV + +GT NF LWQMR+KDLLAQQG+ KAL++ + D WNEMK +AA I+L
Sbjct: 7 SKFEVVKFDGTGNFVLWQMRLKDLLAQQGISKALQETMPEKIDADKWNEMKAQAAATIRL 66
Query: 66 CVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAF 125
+SD VM +D + ++WD L S +MSK+L KL +KQ+ Y L++QE DL+ HV+ F
Sbjct: 67 SLSDSVMYQVMDEKSPKEIWDKLASLHMSKSLTSKLYLKQQLYGLQVQEESDLRKHVDVF 126
Query: 126 NNILADLTRLGVKVDDENKAII-FCAL 151
N ++ DL++L VK+DDE+KAII C+L
Sbjct: 127 NQLVVDLSKLDVKLDDEDKAIILLCSL 153
>emb|CAE04890.2| OSJNBa0042I15.12 [Oryza sativa (japonica cultivar-group)]
Length = 432
Score = 143 bits (361), Expect = 1e-33
Identities = 74/152 (48%), Positives = 104/152 (67%), Gaps = 2/152 (1%)
Query: 2 LNPGGAKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRD-KSLDIAIVDWNEMKKKAA 60
L P KF++ + +G NFGLWQ RVKDLLAQQ + KAL+ K + D EM+ +AA
Sbjct: 37 LAPTCMKFDLVKFDGFGNFGLWQTRVKDLLAQQEVSKALKGVKPAKMEDDDCEEMQLQAA 96
Query: 61 GLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQD 120
I+ C+SD +M H ++ T+ +W+ LE+Q+MSKTL +K +KQ+ Y LKMQEG DL
Sbjct: 97 ATIRRCLSDQIMYHVMEETSPKIIWEKLEAQFMSKTLTNKRYLKQKLYGLKMQEGADLTA 156
Query: 121 HVNAFNNILADLTRLGVKVDDENKAII-FCAL 151
HVN FN ++ DL ++ V+VDDE+KAI+ C+L
Sbjct: 157 HVNVFNQLVTDLVKMDVEVDDEDKAIVLLCSL 188
>ref|NP_916849.1| retrovirus-related pol polyprotein from transposon TNT 1-94-like
[Oryza sativa (japonica cultivar-group)]
Length = 425
Score = 140 bits (352), Expect = 1e-32
Identities = 74/149 (49%), Positives = 102/149 (67%), Gaps = 6/149 (4%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRD---KSLDIAIVDWNEMKKKAAGLI 63
+KFE+ + +GT NF LWQMR+KDLLAQQG+ KAL + + +D W EMK +AA I
Sbjct: 7 SKFEMVKFDGTGNFVLWQMRLKDLLAQQGISKALEETMPEKMDAG--KWEEMKAQAAATI 64
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
+L +SD VM +D T ++WD L S YMSK+L KL +KQ+ Y L+MQE DL+ HV+
Sbjct: 65 RLSLSDSVMYQVMDEKTPKEIWDKLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRKHVD 124
Query: 124 AFNNILADLTRLGVKVDDENKAII-FCAL 151
FN ++ DL++L V + DE+KAII C+L
Sbjct: 125 VFNQLVVDLSKLDVNLYDEDKAIILLCSL 153
>emb|CAA32025.1| unnamed protein product [Nicotiana tabacum]
gi|130582|sp|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease]
Length = 1328
Score = 134 bits (337), Expect = 8e-31
Identities = 65/145 (44%), Positives = 94/145 (64%), Gaps = 3/145 (2%)
Query: 6 GAKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKAL---RDKSLDIAIVDWNEMKKKAAGL 62
G K+EV + NG F WQ R++DLL QQGLHK L K + DW ++ ++AA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHV 122
I+L +SDDV+N+ +D T +W LES YMSKTL +KL +K++ Y+L M EG + H+
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 NAFNNILADLTRLGVKVDDENKAII 147
N FN ++ L LGVK+++E+KAI+
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAIL 147
>gb|AAK29467.1| polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 131 bits (329), Expect = 7e-30
Identities = 65/146 (44%), Positives = 97/146 (65%), Gaps = 4/146 (2%)
Query: 6 GAKFEVTRLNGTRN-FGLWQMRVKDLLAQQGLHKALRDKSL---DIAIVDWNEMKKKAAG 61
G K+EV + NG + F +WQ R+KDLL QQGLHKAL KS + + DW E+ +KAA
Sbjct: 3 GVKYEVAKFNGDKPVFSMWQRRMKDLLIQQGLHKALGGKSKKPESMKLEDWEELDEKAAS 62
Query: 62 LIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDH 121
I+L ++DDV+N+ +D + +W LE+ YMSKTL +KL +K++ Y+L M EG + H
Sbjct: 63 AIRLHLTDDVVNNIVDEESACGIWTKLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSH 122
Query: 122 VNAFNNILADLTRLGVKVDDENKAII 147
+N N ++ L LGVK+++E+K I+
Sbjct: 123 LNVLNGLITQLANLGVKIEEEDKRIV 148
>ref|XP_470528.1| Putative retrovirus-related pol polyprotein [Oryza sativa (japonica
cultivar-group)] gi|27436748|gb|AAO13467.1| Putative
retrovirus-related pol polyprotein [Oryza sativa
(japonica cultivar-group)]
Length = 556
Score = 124 bits (312), Expect = 6e-28
Identities = 68/138 (49%), Positives = 94/138 (67%), Gaps = 6/138 (4%)
Query: 18 RNFGLWQMRVKDLLAQQGLHKALRD---KSLDIAIVDWNEMKKKAAGLIKLCVSDDVMNH 74
R G QMR+KDLLAQQG+ KAL + + +D+ W EMK +A +I+L +SD VM
Sbjct: 149 RRDGSSQMRLKDLLAQQGISKALEETMPEKMDVG--KWVEMKAQATAIIRLSLSDFVMYQ 206
Query: 75 TLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTR 134
+D T ++WD L S YMSK+L KL +KQ+ Y L+MQE DL+ HV+ FN ++ DL++
Sbjct: 207 VMDEKTPKEIWDKLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRKHVDVFNQLVVDLSK 266
Query: 135 LGVKVDDENKAII-FCAL 151
L VK+DDE+KAII C+L
Sbjct: 267 LDVKLDDEDKAIILLCSL 284
>emb|CAD41329.2| OJ991113_30.13 [Oryza sativa (japonica cultivar-group)]
gi|50925386|ref|XP_472962.1| OJ991113_30.13 [Oryza
sativa (japonica cultivar-group)]
Length = 353
Score = 102 bits (254), Expect = 3e-21
Identities = 51/101 (50%), Positives = 70/101 (68%), Gaps = 1/101 (0%)
Query: 52 WNEMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLK 111
W+EMK +AA I+L + D VM +D T ++WD L S YMSK+L KL +KQ+ Y L+
Sbjct: 6 WDEMKAQAAATIRLSLLDSVMYQVMDEKTPKEIWDKLASLYMSKSLTSKLYLKQQSYGLQ 65
Query: 112 MQEGGDLQDHVNAFNNILADLTRLGVKVDDENKAII-FCAL 151
MQE DL+ HV+ FN ++ DL +L VK+DDE+KAII C+L
Sbjct: 66 MQEESDLRKHVDIFNQLVVDLRKLDVKLDDEDKAIILLCSL 106
>dbj|BAD34493.1| Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 94.4 bits (233), Expect = 9e-19
Identities = 44/140 (31%), Positives = 84/140 (59%), Gaps = 2/140 (1%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAI-VDWNEMKKKAAGLIKL 65
AKFE+ + NG +NF LW+++VK +L + A+ ++ +D W+EM + A + L
Sbjct: 3 AKFEIEKFNG-KNFSLWKLKVKAILRKDNCLAAISERPVDFTDDKKWSEMNEDAMADLYL 61
Query: 66 CVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAF 125
++D V++ + T N++WD L Y +K+L +K+ +K++ Y+L+M E + +H+N
Sbjct: 62 SIADGVLSSIEEKKTANEIWDHLNRLYEAKSLHNKIFLKRKLYTLRMSESTSVTEHLNTL 121
Query: 126 NNILADLTRLGVKVDDENKA 145
N + + LT L K++ + +A
Sbjct: 122 NTLFSQLTSLSCKIEPQERA 141
>gb|AAV88069.1| hypothetical retrotransposon [Ipomoea batatas]
Length = 1415
Score = 86.7 bits (213), Expect = 2e-16
Identities = 45/134 (33%), Positives = 82/134 (60%), Gaps = 3/134 (2%)
Query: 11 VTRLNGTRNFGLWQMRVKDLLAQQGLHKAL--RDKSLDIAIVDWNEMKKKAAGLIKLCVS 68
+ RLNG RN+ +W+ ++KDLL + LH + K +++ +W+ ++ G I+ V
Sbjct: 8 MVRLNG-RNYHIWKAKMKDLLFVKKLHLPVFASAKPENMSDEEWDFEHQQVCGYIRQWVE 66
Query: 69 DDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFNNI 128
D+V+NH ++ T +W+ LE+ Y SKT +KL + ++ +++ +EG + DHVN F +
Sbjct: 67 DNVLNHIINETHARSLWNKLETLYASKTGNNKLFLLKQMMNIRYREGTLINDHVNDFQGV 126
Query: 129 LADLTRLGVKVDDE 142
L L+ +G+K +DE
Sbjct: 127 LDQLSGMGIKFEDE 140
>ref|XP_474090.1| OSJNBa0033G05.13 [Oryza sativa (japonica cultivar-group)]
gi|38344889|emb|CAD41912.2| OSJNBa0033G05.13 [Oryza
sativa (japonica cultivar-group)]
Length = 1181
Score = 85.5 bits (210), Expect = 4e-16
Identities = 53/149 (35%), Positives = 86/149 (57%), Gaps = 7/149 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDW-NEMKK---KAAGLI 63
K+++ L+ F LWQ++++ +LAQQ L AL D DW N+ KK KA I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQELDDAL--SGFDKRTQDWSNDEKKRDRKAMSYI 62
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
L +S++++ L T +W LE M+K L K+ +KQ+ + K+Q+ G + DH++
Sbjct: 63 HLHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLS 122
Query: 124 AFNNILADLTRLGVKVDDENKA-IIFCAL 151
AF I+ADL + VK D+++ A I+ C+L
Sbjct: 123 AFKEIVADLESMEVKYDEKDLALILLCSL 151
>emb|CAI44659.1| OSJNBa0061C06.9 [Oryza sativa (japonica cultivar-group)]
Length = 1007
Score = 85.1 bits (209), Expect = 5e-16
Identities = 51/149 (34%), Positives = 84/149 (56%), Gaps = 7/149 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKK----AAGLI 63
K+++ L+ F LWQ++++ +LAQQ L AL D DW++ +KK A I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDAL--SGFDKRTQDWSKDEKKRDCKAMSYI 62
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
+ +S++++ L T +W LE M+K L K+ +KQ+ + K+Q+ G + DH+
Sbjct: 63 HVHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLF 122
Query: 124 AFNNILADLTRLGVKVDDENKA-IIFCAL 151
AF I+ADL + VK D+EN I+ C+L
Sbjct: 123 AFKEIVADLESMEVKYDEENLGLILLCSL 151
>gb|AAP54315.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37535452|ref|NP_922028.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
gi|22094359|gb|AAM91886.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1280
Score = 84.3 bits (207), Expect = 9e-16
Identities = 52/149 (34%), Positives = 84/149 (55%), Gaps = 7/149 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDW-NEMKKK---AAGLI 63
K+++ L+ F LWQ++++ +LAQQ L AL D DW N+ KKK A I
Sbjct: 40 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDAL--SGFDKRTQDWSNDEKKKDRKAMSYI 97
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
L +S++++ L T +W LE M+K L K+ +KQ+ + K+Q+ G + DH++
Sbjct: 98 HLHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLS 157
Query: 124 AFNNILADLTRLGVKVDDENKA-IIFCAL 151
F I+ADL + VK D+E+ I+ C+L
Sbjct: 158 TFKEIVADLESIEVKYDEEDLGLILLCSL 186
>ref|XP_475663.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|48475188|gb|AAT44257.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1211
Score = 84.3 bits (207), Expect = 9e-16
Identities = 52/149 (34%), Positives = 84/149 (55%), Gaps = 7/149 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDW-NEMKK---KAAGLI 63
K+++ L+ F LWQ++++ +LAQQ L AL D DW N+ KK K I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDAL--SGFDKRTQDWSNDEKKRDRKTMSYI 62
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
L +S++++ L T +W LE M+K L K+ +KQ+ + K+Q+ G + DH++
Sbjct: 63 HLHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLS 122
Query: 124 AFNNILADLTRLGVKVDDENKA-IIFCAL 151
AF I+ADL + VK D+E+ I+ C+L
Sbjct: 123 AFKKIVADLESMEVKYDEEDLCLILLCSL 151
>pir||T02206 hypothetical protein - common tobacco retrotransposon Tto1
gi|1167523|dbj|BAA11674.1| ORF(AA 1-1338) [Nicotiana
tabacum]
Length = 1338
Score = 82.8 bits (203), Expect = 3e-15
Identities = 49/143 (34%), Positives = 74/143 (51%), Gaps = 3/143 (2%)
Query: 10 EVTRLNGTRNFGLWQMRVKDLLAQQGLHKAL--RDKSLDIAIVDWNEMKKKAAGLIKLCV 67
++ LNGT N+ LW+ ++KDLL +H + K D + DW + G I+ V
Sbjct: 7 KMVNLNGT-NYHLWRNKMKDLLFVTKMHLPVFSSQKPEDKSDEDWEFEHNQVCGYIRQFV 65
Query: 68 SDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFNN 127
D+V NH +T +WD LE Y SKT +KL + +K EG + DH+N
Sbjct: 66 EDNVYNHISGVTHARSLWDKLEELYASKTGNNKLFYLTKLMQVKYVEGTTVADHLNEIQG 125
Query: 128 ILADLTRLGVKVDDENKAIIFCA 150
I+ L+ +G+K DDE A++ A
Sbjct: 126 IVDQLSGMGIKFDDEVLALMVLA 148
>ref|XP_475489.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|48475213|gb|AAT44282.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1243
Score = 81.6 bits (200), Expect = 6e-15
Identities = 51/149 (34%), Positives = 84/149 (56%), Gaps = 7/149 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDW-NEMKK---KAAGLI 63
K+++ L+ F LWQ++++ +LAQQ L AL D DW N+ KK KA I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDAL--SGFDKRTQDWSNDEKKRDRKAISYI 62
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
L +S++++ L T +W LE M+K L K+ +KQ+ + K+Q+ + DH++
Sbjct: 63 HLHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLS 122
Query: 124 AFNNILADLTRLGVKVDDENKA-IIFCAL 151
AF I+ADL + VK D+++ I+ C+L
Sbjct: 123 AFKEIVADLESMEVKYDEDDLGLILLCSL 151
>ref|XP_469192.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|53370655|gb|AAU89150.1| integrase core domain
containing protein [Oryza sativa (japonica
cultivar-group)] gi|40538906|gb|AAR87163.1| putative
polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1322
Score = 81.3 bits (199), Expect = 8e-15
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 7/150 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQG-LHKALRDKSLDIAIVDWNEMKK----KAAGL 62
K+++ L+ F LWQ++++ +LAQ L +AL +W +K KA L
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKK-KTTEWTAEEKRKDRKALSL 63
Query: 63 IKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHV 122
I+L +S+D++ L T ++W LES MSK L K+ +K + +S K+ E G + +H+
Sbjct: 64 IQLHLSNDILQEVLQKKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLHESGSVLNHI 123
Query: 123 NAFNNILADLTRLGVKVDDENKA-IIFCAL 151
+ F I+ADL + V+ DDE+ ++ C+L
Sbjct: 124 SVFKEIVADLVSMEVQFDDEDLGLLLLCSL 153
>gb|AAX92941.1| retrotransposon protein, putative, Ty1-copia sub-class [Oryza
sativa (japonica cultivar-group)]
Length = 2340
Score = 81.3 bits (199), Expect = 8e-15
Identities = 52/149 (34%), Positives = 82/149 (54%), Gaps = 7/149 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDW-NEMKK---KAAGLI 63
K+++ L F LWQ++++ +LAQQ L AL D DW N+ KK KA I
Sbjct: 212 KYDLPLLYRDTRFSLWQVKMRAVLAQQDLDDAL--SGFDKRTHDWSNDEKKRDRKAMSYI 269
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
L +S++++ L +W LE M+K L K+ +KQ + K+Q+ G + DH++
Sbjct: 270 HLHLSNNILQEVLKEEIAAGLWLKLEQICMTKDLTSKMHLKQTLFLHKLQDDGSVMDHLS 329
Query: 124 AFNNILADLTRLGVKVDDENKA-IIFCAL 151
AF I+ADL + VK D+E+ I+ C+L
Sbjct: 330 AFKEIIADLESMEVKYDEEDLGLILLCSL 358
>gb|AAP53029.1| putative retrotransposon-related protein [Oryza sativa (japonica
cultivar-group)] gi|37532880|ref|NP_920742.1| putative
retrotransposon-related protein [Oryza sativa (japonica
cultivar-group)] gi|22655747|gb|AAN04164.1| Putative
retrotransposon protein [Oryza sativa (japonica
cultivar-group)] gi|16905223|gb|AAL31093.1| putative
retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 80.9 bits (198), Expect = 1e-14
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 7/150 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQG-LHKALRDKSLDIAIVDWNEMKK----KAAGL 62
K+++ + F LWQ++++ +LAQ L +AL +W +K KA L
Sbjct: 2 KYDLPLQDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKK-KTTEWTAEEKRKDRKALSL 60
Query: 63 IKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHV 122
I+L +S+D++ L T ++W LES MSK L K+ +K + +S K+QE G + +H+
Sbjct: 61 IQLHLSNDILQKVLQEKTAAELWFKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHI 120
Query: 123 NAFNNILADLTRLGVKVDDENKA-IIFCAL 151
+ F I+ADL + V+ DDE+ ++ C+L
Sbjct: 121 SVFKEIIADLVSMEVQFDDEDLGLLLLCSL 150
>ref|XP_470868.1| Putative retroelement pol polyprotein [Oryza sativa]
gi|14029020|gb|AAK52561.1| Putative retroelement pol
polyprotein [Oryza sativa]
Length = 1326
Score = 80.1 bits (196), Expect = 2e-14
Identities = 49/150 (32%), Positives = 85/150 (56%), Gaps = 7/150 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQG-LHKALRDKSLDIAIVDWNEMKK----KAAGL 62
K+++ L+ F LWQ++++ +LAQ L +AL + +W +K KA L
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAILAQTSDLDEALESFGKKKS-TEWTAEEKRKDRKALLL 63
Query: 63 IKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHV 122
I+L +S+D++ L T ++W LES MSK L K+ +K + +S K+QE G + +H+
Sbjct: 64 IQLHLSNDILQEVLQEKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHI 123
Query: 123 NAFNNILADLTRLGVKVDDENKA-IIFCAL 151
+ F I+ DL + V+ DDE+ ++ C+L
Sbjct: 124 SVFKEIVVDLVSIEVQFDDEDLGLLLLCSL 153
>gb|AAX92861.1| retrotransposon protein, putative, Ty1-copia sub-class [Oryza
sativa (japonica cultivar-group)]
Length = 1373
Score = 77.8 bits (190), Expect = 9e-14
Identities = 50/149 (33%), Positives = 83/149 (55%), Gaps = 6/149 (4%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWN--EMKK--KAAGLI 63
KF++ LN F LWQ++++ +LAQ + D + +W E++K KA LI
Sbjct: 2 KFDLPLLNYDTRFSLWQVKMRGILAQTHDYDEALD-NFGKRRAEWTAEEIRKDQKALALI 60
Query: 64 KLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVN 123
+L + +D++ L T ++W LES MSK L K+ +K + ++LKM+E + H+
Sbjct: 61 QLHLHNDILQECLTEKTSAELWLKLESICMSKDLTSKMQMKMKLFTLKMKEEDSVITHMA 120
Query: 124 AFNNILADLTRLGVKVDDENKA-IIFCAL 151
F I+ADL + VK DDE+ ++ C+L
Sbjct: 121 EFKKIVADLVSMEVKYDDEDLGLLLLCSL 149
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 253,403,890
Number of Sequences: 2540612
Number of extensions: 8971794
Number of successful extensions: 23450
Number of sequences better than 10.0: 222
Number of HSP's better than 10.0 without gapping: 87
Number of HSP's successfully gapped in prelim test: 135
Number of HSP's that attempted gapping in prelim test: 23275
Number of HSP's gapped (non-prelim): 246
length of query: 157
length of database: 863,360,394
effective HSP length: 117
effective length of query: 40
effective length of database: 566,108,790
effective search space: 22644351600
effective search space used: 22644351600
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0490.9


