

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.3
(862 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
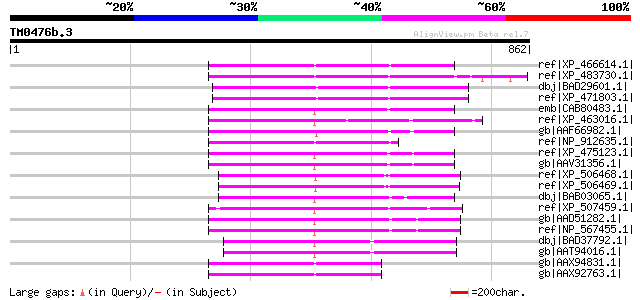
ref|XP_466614.1| putative far-red impaired response protein [Ory... 263 1e-68
ref|XP_483730.1| putative far-red impaired response protein [Ory... 246 2e-63
dbj|BAD29601.1| putative far-red impaired response protein [Oryz... 244 1e-62
ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultiva... 243 2e-62
emb|CAB80483.1| hypothetical protein [Arabidopsis thaliana] gi|4... 238 6e-61
ref|XP_463016.1| putative transposase [Oryza sativa (japonica cu... 237 1e-60
gb|AAF66982.1| transposase [Zea mays] 226 2e-57
ref|NP_912635.1| Putative far-red impaired response protein [Ory... 221 8e-56
ref|XP_475123.1| putative Mutator-like transposase [Oryza sativa... 218 9e-55
gb|AAV31356.1| hypothetical protein [Oryza sativa (japonica cult... 218 9e-55
ref|XP_506468.1| PREDICTED P0616D06.133-1 gene product [Oryza sa... 217 1e-54
ref|XP_506469.1| PREDICTED P0616D06.133-1 gene product [Oryza sa... 217 2e-54
dbj|BAB03065.1| far-red impaired response protein; Mutator-like ... 214 1e-53
ref|XP_507459.1| PREDICTED OJ1135_F06.10-1 gene product [Oryza s... 213 2e-53
gb|AAD51282.1| far-red impaired response protein [Arabidopsis th... 212 5e-53
ref|NP_567455.1| far-red impaired response protein (FAR1) / far-... 212 5e-53
dbj|BAD37792.1| far-red impaired response protein-like [Oryza sa... 212 5e-53
gb|AAT94016.1| hypothetical protein [Oryza sativa (japonica cult... 211 8e-53
gb|AAX94831.1| transposon protein, putative, unclassified [Oryza... 211 1e-52
gb|AAX92763.1| transposon protein, putative, unclassified [Oryza... 211 1e-52
>ref|XP_466614.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|47497275|dbj|BAD19318.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 880
Score = 263 bits (673), Expect = 1e-68
Identities = 138/415 (33%), Positives = 223/415 (53%), Gaps = 9/415 (2%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHV--SDGRSALDYLASLGTKDPSMFVRHTEDADKNLEH 388
G++ T ++ +DL N + R+ H+ D + Y D F + D L+
Sbjct: 207 GFDATGFVSRDLYNYFTRLRKKHILGGDAERVIKYFQWRQKHDMEFFFEYETDEAGRLKR 266
Query: 389 LFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEE 448
LFW D R++Y FGD + FD+TY+ N+Y P + F VNH T + G A++ +EK
Sbjct: 267 LFWSDPQSRIDYDAFGDVVVFDSTYRVNRYNLPFIPFVGVNHHGSTIIFGCAIVADEKVA 326
Query: 449 TYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIP 508
TY W+L+Q L+ M +K P +ITDGD AMR AI V P++ H LC WH+ N +++ P
Sbjct: 327 TYEWILKQFLDCMYQKHPRALITDGDNAMRRAIAAVMPDSEHWLCTWHIEQNMARHLR-P 385
Query: 509 DFLPKFKTCMFGDFQIGDFKRKW-ESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFA 567
D L F+T + + +F+RKW E V+ +N W++ MY R WA AY +G FF
Sbjct: 386 DMLSDFRTLVHAPYDHEEFERKWVEFKVKHKGCEDNQWLVRMYNLRKKWATAYTKGLFFL 445
Query: 568 GFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAP 627
G ++ R E N++L +++ R+ +L ++H+ HCL MR+RE E D ++ P
Sbjct: 446 GMKSNQRSESLNSKLHRYLDRKMSLVLLVEHYEHCLSRMRYREAELDCKASQSIPFTSND 505
Query: 628 PIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNC--EDTLGMSMHTVKRSQESSREW 685
F IE A+RI T +F + + I + D + +C ED L + ++K E +
Sbjct: 506 ASF--IEKDAARIFTPAVFKKLKLVIAKSMDWEVIDCIEEDNLVKYVISMKGDSEMLQIL 563
Query: 686 LVSYYTTTYE-FRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
+Y +T + C+C +M+ L C H++A++ L + IP++ ++ RW+ AK
Sbjct: 564 NCTYVESTMKSINCTCRKMDRECLPCEHMVAIMHHLKLDAIPEACIVRRWTIKAK 618
>ref|XP_483730.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|42407926|dbj|BAD09065.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)] gi|42407459|dbj|BAD10392.1|
putative far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 690
Score = 246 bits (628), Expect = 2e-63
Identities = 154/547 (28%), Positives = 261/547 (47%), Gaps = 24/547 (4%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVSDGR--SALDYLASLGTKDPSMFVRHTEDADKNLEH 388
G+ T ++ KD+ N +Q++ + G + Y+ + D F + D L+
Sbjct: 141 GFETTGFVSKDMYNFFVRQKKKQIVGGDVDRVIKYMQARQKDDMEFFYEYETDEAGRLKR 200
Query: 389 LFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEE 448
LFW D R++Y FGD + FD+TY+ NKY P + F VNH T + A++ +EK
Sbjct: 201 LFWADPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTIIFACAIVADEKIA 260
Query: 449 TYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIP 508
TY W+L++ L+ M +K P +ITD D +MR AI V PN+ HRLC WH+ N +++ P
Sbjct: 261 TYEWVLKRFLDCMCQKHPKGLITDSDNSMRRAIATVMPNSEHRLCTWHIEQNMARHLR-P 319
Query: 509 DFLPKFKTCMFGDFQIGDFKRKW-ESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFA 567
+ F+ + + +F+ KW E ++ +N W+ MY R WAAAY +G +F
Sbjct: 320 KMISDFRVLVHATYSAEEFEEKWVEFKLKRKVAEDNQWLGRMYNLRKKWAAAYTKGMYFL 379
Query: 568 GFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAP 627
G ++ R E N++L + + R+ +L ++H+ HCL MRHRE E D ++ P
Sbjct: 380 GMKSNQRSESLNSKLHRHLDRKMSLVLLVEHYEHCLSRMRHREAELDSKASQSVPFTAND 439
Query: 628 PIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLV 687
IE A+RI T +F + + I+ + D + +C D + + + + + ++
Sbjct: 440 A--SLIEKDAARIFTPSVFKKLKMDIIKSKDCEVIDCIDEENIIKYIICMKGKIDKITIL 497
Query: 688 --SYYTTTYE-FRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLCE 744
SY ++ + C+C +ME L C H+ ++ LN+ IP V+ RW+ AK +
Sbjct: 498 TCSYVESSLKNMSCTCKKMECESLPCQHMFTIMDYLNLQAIPDVCVVPRWTVKAK--IAF 555
Query: 745 TPGSSPSLFRDSAVLVRYVCMLEACRKMCTLACPLIDDF----RQTWEIVGGHTQHLENK 800
++ S + RY M A L C + +++ + +E + T +E
Sbjct: 556 PSDRYGEVYTWSDQMERYRRM-RAIGSEVLLKCSMSEEYTLKVMEMFEKLDLETNAIEGD 614
Query: 801 NNPSVPEGVTGTPSANAGLTTHRRRGRPC-----GPSTSRLRA---KRPLKCSTCKVIGH 852
G S + + + ++ P G T RLR K +C C+V GH
Sbjct: 615 EFDVTLCGHVVAQSLRSDIASVQKVHDPMEVVPKGAPTKRLRGFLEKGNRRCGYCRVRGH 674
Query: 853 NRLTCPL 859
+C +
Sbjct: 675 TVRSCAI 681
>dbj|BAD29601.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|50251625|dbj|BAD29488.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 688
Score = 244 bits (622), Expect = 1e-62
Identities = 138/430 (32%), Positives = 216/430 (50%), Gaps = 8/430 (1%)
Query: 337 YILKDLQNQVGKQRRAHVS--DGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDG 394
+++KDL N + ++ D L YL +D F ++T D + L ++FW D
Sbjct: 170 FVMKDLYNFFTRYEMKNIKGHDAEDVLKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADA 229
Query: 395 IGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLL 454
R++Y+ FG + FD+TY+ NKY P + F VNH TT+ G ++ NE +Y WLL
Sbjct: 230 ESRLDYAAFGGVVIFDSTYRVNKYNLPFIPFIGVNHHRSTTIFGCGILSNESVNSYCWLL 289
Query: 455 EQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIPDFLPKF 514
E LEAM + P ++ITDGD AM AI +V P A HRLC WH+ N + +++ P L +
Sbjct: 290 ETFLEAMRQVHPKSLITDGDLAMAKAISKVMPGAYHRLCTWHIEENMSRHLRKPK-LDEL 348
Query: 515 KTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSR 574
+ ++ +F+R+W E WI MY R WAAAY GK+ G R+ R
Sbjct: 349 RKLIYESMDEEEFERRWADFKENGGTGNEQWIALMYRLREKWAAAYTDGKYLLGMRSNQR 408
Query: 575 CEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIE 634
E N++L +KR +L ++H C++ +R +E + D +S P + +E
Sbjct: 409 SESLNSKLHTLLKRNMSLMCLVKHVKLCIQGLRKKEAQLDAKSTNSVPFCRIDA--DPLE 466
Query: 635 LSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTV--KRSQESSREWLVSYYTT 692
A+RI T +F + R I A + + + + G S++ V K E E VS+
Sbjct: 467 KDAARIYTAVVFKKVRAQIRLIAGLEVISGTNQDGSSLYVVGLKDDNEVWDEVRVSFKGQ 526
Query: 693 TYE-FRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLCETPGSSPS 751
E C C +ME + C+HI V+ L IP+ V++RW+ GAK + P+
Sbjct: 527 ALEGVECHCRKMECEDIPCSHIFVVVKFLGFDTIPRCCVVDRWTMGAKAAFRSDRNADPN 586
Query: 752 LFRDSAVLVR 761
++ + V R
Sbjct: 587 VWSEHMVRYR 596
>ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultivar-group)]
gi|38345091|emb|CAD40514.2| OSJNBa0050F15.2 [Oryza
sativa (japonica cultivar-group)]
Length = 688
Score = 243 bits (620), Expect = 2e-62
Identities = 138/430 (32%), Positives = 216/430 (50%), Gaps = 8/430 (1%)
Query: 337 YILKDLQNQVGKQRRAHVS--DGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDG 394
+++KDL N + ++ D L YL +D F ++T D + L ++FW D
Sbjct: 170 FVMKDLYNFFTRYDMKNIKGHDAEDVLKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADA 229
Query: 395 IGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLL 454
R++Y+ FG + FD+TY+ NKY P + F VNH TT+ G ++ NE +Y WLL
Sbjct: 230 ESRLDYAAFGGVVIFDSTYRVNKYNLPFIPFIGVNHHRSTTIFGCGILSNESVNSYCWLL 289
Query: 455 EQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIPDFLPKF 514
E LEAM + P ++ITDGD AM AI +V P A HRLC WH+ N + +++ P L +
Sbjct: 290 ETFLEAMRQVHPKSLITDGDLAMAKAISKVMPGAYHRLCTWHIEENMSRHLRKPK-LDEL 348
Query: 515 KTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSR 574
+ ++ +F+R+W E WI MY R WAAAY GK+ G R+ R
Sbjct: 349 RKLIYESMDEEEFERRWADFKENGGTGNGQWIALMYRLREKWAAAYTDGKYLLGMRSNQR 408
Query: 575 CEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIE 634
E N++L +KR +L ++H C++ +R +E + D +S P + +E
Sbjct: 409 SESLNSKLHTLLKRNMSLMCLVKHVKLCIQGLRKKEAQLDAKSTNSVPFCRIDA--DPLE 466
Query: 635 LSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTV--KRSQESSREWLVSYYTT 692
A+RI T +F + R I A + + + + G S++ V K E E VS+
Sbjct: 467 KDAARIYTAVVFKKVRAQIRLIAGLEVISGTNQDGSSLYVVGLKDDNEVWDEVRVSFKGQ 526
Query: 693 TYE-FRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLCETPGSSPS 751
E C C +ME + C+HI V+ L IP+ V++RW+ GAK + P+
Sbjct: 527 ALEGVECHCRKMECEDIPCSHIFVVVKFLGFDTIPRCCVVDRWTMGAKAAFRSDRNADPN 586
Query: 752 LFRDSAVLVR 761
++ + V R
Sbjct: 587 VWSEHMVRYR 596
>emb|CAB80483.1| hypothetical protein [Arabidopsis thaliana]
gi|4467124|emb|CAB37558.1| hypothetical protein
[Arabidopsis thaliana] gi|22531237|gb|AAM97122.1|
unknown protein [Arabidopsis thaliana]
gi|15233732|ref|NP_195531.1| far-red impaired responsive
protein, putative [Arabidopsis thaliana]
gi|7485859|pir||T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana
Length = 788
Score = 238 bits (607), Expect = 6e-61
Identities = 127/415 (30%), Positives = 205/415 (48%), Gaps = 9/415 (2%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVS-DGRSALDYLASLGTKDPSMFVRHTEDADKNLEHL 389
G + + D +N + R+ + + + LDYL + +P+ F D+++ ++
Sbjct: 223 GISKVGFTEVDCRNYMRNNRQKSIEGEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNV 282
Query: 390 FWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEET 449
FW D M+++ FGD + FD TY+ N+YR P F+ VNH Q + G A I NE E +
Sbjct: 283 FWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEAS 342
Query: 450 YVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDN----- 504
+VWL L AM P ++ TD D +R AI VFP A HR C WH++ +
Sbjct: 343 FVWLFNTWLAAMSAHPPVSITTDHDAVIRAAIMHVFPGARHRFCKWHILKKCQEKLSHVF 402
Query: 505 IKIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGK 564
+K P F F C+ + DF+R W SL++++ L ++ W+ +Y+ R W Y R
Sbjct: 403 LKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYELRDHEWLQAIYSDRRQWVPVYLRDT 462
Query: 565 FFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGL 624
FFA T R + N+ ++ NL +F + + LE +EV+AD+ + P L
Sbjct: 463 FFADMSLTHRSDSINSYFDGYINASTNLSQFFKLYEKALESRLEKEVKADYDTMNSPPVL 522
Query: 625 KAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSRE 684
K P +E AS + TR+LF RF+ +V + + +D + + V + E+ +
Sbjct: 523 KTP---SPMEKQASELYTRKLFMRFQEELVGTLTFMASKADDDGDLVTYQVAKYGEAHKA 579
Query: 685 WLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
V + CSC E G+ C HI+AV N+ +P +L+RW++ AK
Sbjct: 580 HFVKFNVLEMRANCSCQMFEFSGIICRHILAVFRVTNLLTLPPYYILKRWTRNAK 634
>ref|XP_463016.1| putative transposase [Oryza sativa (japonica cultivar-group)]
gi|38093745|gb|AAR10861.1| putative transposase [Oryza
sativa (japonica cultivar-group)]
Length = 813
Score = 237 bits (605), Expect = 1e-60
Identities = 145/468 (30%), Positives = 228/468 (47%), Gaps = 22/468 (4%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHV--SDGRSALDYLASLGTKDPSMFVR-HTEDADKNLE 387
G + + D N++G++R+ ++ +D ++ L+YL + +DP+ F + D +
Sbjct: 224 GADKVPFSKMDCNNEIGRERKKYLESNDTQTLLEYLRNKQLEDPTFFYAIQVDKEDGRIA 283
Query: 388 HLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKE 447
+ FW DG M+Y+ FGD ++FD T+ NK P NH QT + G A++ N+
Sbjct: 284 NFFWADGQSIMDYACFGDFVSFDTTFDTNKCEMPFAPLLGTNHHKQTIIFGAALLFNQTI 343
Query: 448 ETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI-- 505
E++VWL E L AM K P+T+ TD D AM AI VF N SHRLC WH+ N N+
Sbjct: 344 ESFVWLFETFLTAMSGKHPSTIFTDQDAAMAAAIAFVFRNTSHRLCLWHIYLNGGKNLSH 403
Query: 506 ---KIPD-FLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYF 561
K P+ FL FK C++ D F + W L+ E+NL +N WI ++Y R WA F
Sbjct: 404 VIHKHPNKFLADFKRCVYEDRSEYYFNKMWHELLSEYNLEDNKWISNLYKLREKWAIV-F 462
Query: 562 RGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGD 621
R F A +T R EG N K +R+ L E L +R E++ADF+S + +
Sbjct: 463 RNSFTADITSTQRSEGMNNVYKKRFRRKLGLSELLVECDKVSATLRENELDADFKSRHSN 522
Query: 622 PGLKAP--PIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQ 679
P P P+ + S +R + +L F+ + ++ T G ++ V
Sbjct: 523 PVTYIPNLPMLKIAAESYTRSMYSELEDEFKQQFTLSCKLLKTE-----GATLTFVVMPM 577
Query: 680 ESSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
E E V + T CSC + E IGL C H + V + +P +L RW++ AK
Sbjct: 578 EYDHEATVVFNPTEMTITCSCRKYECIGLLCKHALRVFNMNKVFTLPSHYILNRWTKYAK 637
Query: 740 D--HLCETPGSSPSLFRDSAVLVRYVCMLEACRKMCTLACPLIDDFRQ 785
++ + +L +A + R+ +E C+++ L+DD Q
Sbjct: 638 SGFYIQKQGSEKETLKAHAARISRHATSVEL---KCSVSKELLDDLEQ 682
>gb|AAF66982.1| transposase [Zea mays]
Length = 709
Score = 226 bits (577), Expect = 2e-57
Identities = 132/419 (31%), Positives = 212/419 (50%), Gaps = 17/419 (4%)
Query: 331 GYNNTQYILKDLQNQV-GKQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHL 389
G++N + +DLQN G + + +D + + + + + F D L ++
Sbjct: 168 GFDNIGCMKRDLQNYYRGLREKIKNADAQLFVAQMERKKEANSAFFYDFAVDEHGKLVYI 227
Query: 390 FWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEET 449
W D R +Y+ FGD ++ DATY N+Y F+ VNH Q G A + NEK E+
Sbjct: 228 CWADATCRKSYTHFGDLVSVDATYSTNQYNMRFAPFTGVNHHMQRVFFGAAFLANEKIES 287
Query: 450 YVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIP- 508
Y WL L AM K+P +ITD D ++++AIR P+ HRLC WH+M ++ + P
Sbjct: 288 YEWLFRTFLVAMGGKAPRLIITDEDASIKSAIRTTLPDTIHRLCMWHIMEKVSEKVGHPT 347
Query: 509 ----DFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGK 564
+F TC++G +F+ +W +L++ + L N W+ + Y R +W A+F
Sbjct: 348 SHDKEFWDALNTCVWGSETPEEFEMRWNALMDAYGLESNEWLANRYKIRESWIPAFFMDT 407
Query: 565 FFAG-FRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPG 623
AG RTTSR E N+ +F+ R+ EF F LE RH E++AD S + P
Sbjct: 408 PLAGVLRTTSRSESANSFFNRFIHRKLCFVEFWLRFDTALERQRHEELKADHISIHSTPV 467
Query: 624 LKAPPIFQSIELSASRILTRQLFFRFRPTIVSAAD---MIITNCEDTLGMSMHTVKRSQE 680
L+ P + +E AS + T ++F F+ +++A D ++ T +D ++ V S
Sbjct: 468 LRTPWV---VEKQASILYTHKVFKIFQEEVIAARDHCSVLGTTQQD----AVKFVVVSDG 520
Query: 681 SSREWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
S R+ +V + T+ RCSC E IG+ C HII + ++ +P S +L+RW K
Sbjct: 521 SMRDRVVQWCTSNIFGRCSCKLFEKIGIPCCHIILAMRGEKLYELPSSYILKRWETRCK 579
>ref|NP_912635.1| Putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|20219041|gb|AAM15785.1| Putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 611
Score = 221 bits (563), Expect = 8e-56
Identities = 114/319 (35%), Positives = 172/319 (53%), Gaps = 6/319 (1%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVSDGRS--ALDYLASLGTKDPSMFVRHTEDADKNLEH 388
G+ ++ +DL N K ++ + G + + Y+ + D + + D L+
Sbjct: 166 GFEGAGFVTRDLYNFFVKMKKKRIDGGDADRVIKYMQARQKDDMDFYYEYETDEAGCLKR 225
Query: 389 LFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEE 448
LFW D R++Y FGD + FD+TY+ NKY P + F VNH T + G AV+ +E+
Sbjct: 226 LFWADPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTVIFGCAVVSDERVG 285
Query: 449 TYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIP 508
TY W+L+Q L M +K P +VITDGD AMR AI VFPN+ HRLC WH+ N N+ P
Sbjct: 286 TYEWVLKQFLSCMCQKHPKSVITDGDNAMRRAILLVFPNSDHRLCTWHIEQNMARNLS-P 344
Query: 509 DFLPKFKTCMFGDFQIGDFKRKW-ESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFA 567
L F+ + + +F+RKW E V+ EN W+ MY R WAAAY +G+ F
Sbjct: 345 TMLSDFRVLVHAPLEEDEFERKWVEFKVKHKVSDENRWLNRMYNLRKKWAAAYTKGRVFL 404
Query: 568 GFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAP 627
G ++ R E N++L + + R+ +L ++H+ HCL MRH+E E D ++ P
Sbjct: 405 GMKSNQRSESLNSKLHRLLDRKMSLVILIEHYEHCLSRMRHQEAELDAKASQSVPFTSND 464
Query: 628 PIFQSIELSASRILTRQLF 646
IE A+R+ T ++F
Sbjct: 465 STL--IEKDAARVFTPKIF 481
>ref|XP_475123.1| putative Mutator-like transposase [Oryza sativa (japonica
cultivar-group)] gi|46063440|gb|AAS79743.1| putative
Mutator-like transposase [Oryza sativa (japonica
cultivar-group)]
Length = 1510
Score = 218 bits (554), Expect = 9e-55
Identities = 133/416 (31%), Positives = 202/416 (47%), Gaps = 11/416 (2%)
Query: 331 GYNNTQYILKDLQNQVGKQR-RAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHL 389
G+ N L++LQN R + +D + + L +P+ F D L +
Sbjct: 860 GFQNVGCTLRNLQNYYHDLRCKIKDADAQMFVGQLERKKEVNPAFFYEFMVDKQGRLVRV 919
Query: 390 FWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEET 449
FW D I R NYSVFGD L+ D+TY N+Y V F+ VNH Q+ +G + + +EK E+
Sbjct: 920 FWADAICRKNYSVFGDVLSVDSTYSTNQYNMKFVPFTGVNHHLQSVFLGASFLADEKIES 979
Query: 450 YVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI---- 505
+VWL + L+A +P +ITD D +M+ AI ++ PN HRLC WH+M + +
Sbjct: 980 FVWLFQTFLKATGGVAPRLIITDEDASMKAAIAQILPNTVHRLCMWHIMEKVPEKVGPSI 1039
Query: 506 -KIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGK 564
+ +F + C++G DF+ +W S++ ++ L N W + R +W AYF
Sbjct: 1040 REDGEFWDRLHKCVWGSEDSDDFESEWNSIMAKYGLIGNEWFSTKFDIRQSWILAYFMDI 1099
Query: 565 FFAG-FRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPG 623
AG TTSR E N+ +F+ R+ EF F LE R E++AD S + +P
Sbjct: 1100 PLAGILSTTSRSESANSFFNRFIHRKLTFVEFWLRFDTALECQRQEELKADNISLHTNPK 1159
Query: 624 LKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSR 683
L P +E S I T ++F +F+ ++ A D I G M V S +
Sbjct: 1160 LMTP---WDMEKQCSGIYTHEVFSKFQEQLIVARDHCIIQGISKSG-DMKIVTISSLFEK 1215
Query: 684 EWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
E +V + CSC ES G+ C HII VL + IP +++RW + K
Sbjct: 1216 ERVVQMNKSNMFGTCSCKLYESYGIPCRHIIQVLRGEKQNEIPSIYIMKRWEKICK 1271
>gb|AAV31356.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 896
Score = 218 bits (554), Expect = 9e-55
Identities = 133/416 (31%), Positives = 202/416 (47%), Gaps = 11/416 (2%)
Query: 331 GYNNTQYILKDLQNQVGKQR-RAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHL 389
G+ N L++LQN R + +D + + L +P+ F D L +
Sbjct: 251 GFQNVGCTLRNLQNYYHDLRCKIKDADAQMFVGQLERKKEVNPAFFYEFMVDKQGRLVRV 310
Query: 390 FWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEET 449
FW D I R NYSVFGD L+ D+TY N+Y V F+ VNH Q+ +G + + +EK E+
Sbjct: 311 FWADAICRKNYSVFGDVLSVDSTYSTNQYNMKFVPFTGVNHHLQSVFLGASFLADEKIES 370
Query: 450 YVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI---- 505
+VWL + L+A +P +ITD D +M+ AI ++ PN HRLC WH+M + +
Sbjct: 371 FVWLFQTFLKATGGVAPRLIITDEDASMKAAIAQILPNTVHRLCMWHIMEKVPEKVGPSI 430
Query: 506 -KIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGK 564
+ +F + C++G DF+ +W S++ ++ L N W + R +W AYF
Sbjct: 431 REDGEFWDRLHKCVWGSEDSDDFESEWNSIMAKYGLIGNEWFSTKFDIRQSWILAYFMDI 490
Query: 565 FFAG-FRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPG 623
AG TTSR E N+ +F+ R+ EF F LE R E++AD S + +P
Sbjct: 491 PLAGILSTTSRSESANSFFNRFIHRKLTFVEFWLRFDTALECQRQEELKADNISLHTNPK 550
Query: 624 LKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSR 683
L P +E S I T ++F +F+ ++ A D I G M V S +
Sbjct: 551 LMTP---WDMEKQCSGIYTHEVFSKFQEQLIVARDHCIIQGISKSG-DMKIVTISSLFEK 606
Query: 684 EWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
E +V + CSC ES G+ C HII VL + IP +++RW + K
Sbjct: 607 ERVVQMNKSNMFGTCSCKLYESYGIPCRHIIQVLRGEKQNEIPSIYIMKRWEKICK 662
>ref|XP_506468.1| PREDICTED P0616D06.133-1 gene product [Oryza sativa (japonica
cultivar-group)]
Length = 742
Score = 217 bits (553), Expect = 1e-54
Identities = 123/406 (30%), Positives = 203/406 (49%), Gaps = 10/406 (2%)
Query: 348 KQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDAL 407
+ + + DG + L YL ++ ++P+ F D D L + FW D R +++ FGD L
Sbjct: 287 RMKAMQLGDGGAILKYLQTMQMENPAFFYTMQIDEDDKLTNFFWADPKSREDFNYFGDVL 346
Query: 408 AFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPT 467
D TYK N Y RPL +F VNH QT V G A++ +E E+Y WL E AM K P
Sbjct: 347 CLDTTYKINGYGRPLSLFLGVNHHKQTIVFGAAMLYDESFESYRWLFESFKIAMHGKQPA 406
Query: 468 TVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIK-----IPDFLPKFKTCMFGDF 522
+ D + +A+ +PN + R CAWH+ N+ ++ F F C+FG
Sbjct: 407 VALVDQSIPLASAMAAAWPNTTQRTCAWHVYQNSLKHLNHVFQGSKTFAKDFSRCVFGYE 466
Query: 523 QIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQL 582
+ +F W S++E+++L N W+ ++ R WA AY R F A + + E F++ L
Sbjct: 467 EEEEFLFAWRSMLEKYDLRHNEWLSKLFDERERWALAYERHIFCADIISALQAESFSSVL 526
Query: 583 GKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILT 642
KF+ + +L F +H+ ++ R+ E++ADF++ P + PP + A+ T
Sbjct: 527 KKFLGPQLDLLSFFKHYERAVDEHRYAELQADFQASQSYP--RIPP--AKMLKQAAHTYT 582
Query: 643 RQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMR 702
+F FR D ++ +C + S + V S E +E V + ++ C+C +
Sbjct: 583 PVVFEIFRKEFELFMDSVLFSCGEAGATSEYKVAPS-EKPKEHFVRFDSSDCSCICTCRK 641
Query: 703 MESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLCETPGS 748
E +G+ C H++ VL NI +P+ +L+RW + AK E G+
Sbjct: 642 FEFMGIPCCHMLKVLDYRNIKELPQRYLLKRWRRTAKSANEENQGT 687
>ref|XP_506469.1| PREDICTED P0616D06.133-1 gene product [Oryza sativa (japonica
cultivar-group)]
Length = 732
Score = 217 bits (552), Expect = 2e-54
Identities = 123/405 (30%), Positives = 202/405 (49%), Gaps = 10/405 (2%)
Query: 348 KQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDAL 407
+ + + DG + L YL ++ ++P+ F D D L + FW D R +++ FGD L
Sbjct: 287 RMKAMQLGDGGAILKYLQTMQMENPAFFYTMQIDEDDKLTNFFWADPKSREDFNYFGDVL 346
Query: 408 AFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPT 467
D TYK N Y RPL +F VNH QT V G A++ +E E+Y WL E AM K P
Sbjct: 347 CLDTTYKINGYGRPLSLFLGVNHHKQTIVFGAAMLYDESFESYRWLFESFKIAMHGKQPA 406
Query: 468 TVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIK-----IPDFLPKFKTCMFGDF 522
+ D + +A+ +PN + R CAWH+ N+ ++ F F C+FG
Sbjct: 407 VALVDQSIPLASAMAAAWPNTTQRTCAWHVYQNSLKHLNHVFQGSKTFAKDFSRCVFGYE 466
Query: 523 QIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQL 582
+ +F W S++E+++L N W+ ++ R WA AY R F A + + E F++ L
Sbjct: 467 EEEEFLFAWRSMLEKYDLRHNEWLSKLFDERERWALAYERHIFCADIISALQAESFSSVL 526
Query: 583 GKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILT 642
KF+ + +L F +H+ ++ R+ E++ADF++ P + PP + A+ T
Sbjct: 527 KKFLGPQLDLLSFFKHYERAVDEHRYAELQADFQASQSYP--RIPP--AKMLKQAAHTYT 582
Query: 643 RQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMR 702
+F FR D ++ +C + S + V S E +E V + ++ C+C +
Sbjct: 583 PVVFEIFRKEFELFMDSVLFSCGEAGATSEYKVAPS-EKPKEHFVRFDSSDCSCICTCRK 641
Query: 703 MESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLCETPG 747
E +G+ C H++ VL NI +P+ +L+RW + AK E G
Sbjct: 642 FEFMGIPCCHMLKVLDYRNIKELPQRYLLKRWRRTAKSANEENQG 686
>dbj|BAB03065.1| far-red impaired response protein; Mutator-like transposase-like
protein; phytochrome A signaling protein-like
[Arabidopsis thaliana] gi|22331250|ref|NP_188856.2|
far-red impaired responsive protein, putative
[Arabidopsis thaliana]
Length = 839
Score = 214 bits (545), Expect = 1e-53
Identities = 117/399 (29%), Positives = 207/399 (51%), Gaps = 11/399 (2%)
Query: 347 GKQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDA 406
G+ D + LD+L+ + + + + F D+ ++++FW D R NY F D
Sbjct: 225 GRTLSVETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDV 284
Query: 407 LAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSP 466
++ D TY +NKY+ PL IF VN Q V+G A+I +E TY WL+E L A+ ++P
Sbjct: 285 VSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQAP 344
Query: 467 TTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI-----KIPDFLPKFKTCMFGD 521
+IT+ D M + + +FPN H L WH++ ++N+ + +F+PKF+ C++
Sbjct: 345 KVLITELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKS 404
Query: 522 FQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQ 581
+ DF RKW + F L ++ W+I +Y R WA Y AG T+ R + NA
Sbjct: 405 GKDEDFARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAGMSTSQRADSINAF 464
Query: 582 LGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRIL 641
K++ ++ +++EF++ + L+ E +AD P +K+P F E S S +
Sbjct: 465 FDKYMHKKTSVQEFVKVYDTVLQDRCEEEAKADSEMWNKQPAMKSPSPF---EKSVSEVY 521
Query: 642 TRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVK-RSQESSREWLVSYYTTTYEFRCSC 700
T +F +F+ ++ A + + E+ + T + + E++++++V++ T E C C
Sbjct: 522 TPAVFKKFQIEVLGA--IACSPREENRDATCSTFRVQDFENNQDFMVTWNQTKAEVSCIC 579
Query: 701 MRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
E G C H + VL ++ +IP +L+RW++ AK
Sbjct: 580 RLFEYKGYLCRHTLNVLQCCHLSSIPSQYILKRWTKDAK 618
>ref|XP_507459.1| PREDICTED OJ1135_F06.10-1 gene product [Oryza sativa (japonica
cultivar-group)] gi|51963940|ref|XP_506753.1| PREDICTED
OJ1135_F06.10-1 gene product [Oryza sativa (japonica
cultivar-group)]
Length = 721
Score = 213 bits (542), Expect = 2e-53
Identities = 133/428 (31%), Positives = 204/428 (47%), Gaps = 18/428 (4%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLF 390
G N + KD Q ++ S + L+YL + T++PS F + D + + F
Sbjct: 219 GAENVHLLKKDTQ-----RKYLQPSYAQKLLEYLKNKQTENPSFFYAVQLNDDGRIANFF 273
Query: 391 WCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETY 450
W D ++Y+ FGD ++FD T++ NK+ P F NH Q + G ++I +E E++
Sbjct: 274 WTDCQAIVDYACFGDVVSFDTTFETNKFEMPFAPFVGTNHHKQPILFGASLIYDESSESF 333
Query: 451 VWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI----- 505
WL + L AM K P T+ TD + ++R VFPN+SHRLC H+ NA ++
Sbjct: 334 NWLFQTFLTAMSGKQPATIFTDPSAEIIKSVRLVFPNSSHRLCLRHICHNAVKHLNHVIC 393
Query: 506 KIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKF 565
P+FL FK ++ + + F KW+ LV +NL N W+ ++YA R WAA Y R F
Sbjct: 394 NHPEFLSDFKRRIYEERSVTFFDLKWKELVNAYNLDGNDWMNNLYAMREKWAAVYSRDSF 453
Query: 566 FAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLK 625
+A T EG + L F +R+ L EFL+ + C+ R E+EAD+ S P
Sbjct: 454 YADMMTIQNAEGTSDALKNF-RRKLCLPEFLEEYEKCITSFRQNELEADYNSRQTSPVPY 512
Query: 626 APPIFQSIELSASRILTRQLFFRFRPTI--VSAADMIITNCEDTLGMSMHTVKRSQESSR 683
P + + +A+ TR L+ F + + + + T+ T S+E +
Sbjct: 513 VPDL--PMLKTAAESYTRNLYSHFEEEFKKLFTLSCSLLSQDRTISTYKLTPLNSEEEAY 570
Query: 684 EWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHLC 743
S TT CSC E G+ C H + VL NI P V +RW++ AK L
Sbjct: 571 VVFNSEDTTV---SCSCRMYECTGMLCKHALRVLNYSNIFTFPSHYVYKRWTKYAKAGLF 627
Query: 744 ETPGSSPS 751
+S S
Sbjct: 628 GCRNNSQS 635
>gb|AAD51282.1| far-red impaired response protein [Arabidopsis thaliana]
Length = 827
Score = 212 bits (539), Expect = 5e-53
Identities = 121/426 (28%), Positives = 209/426 (48%), Gaps = 12/426 (2%)
Query: 331 GYNNTQYILK-DLQNQVGKQRRAHVSDGRSA--LDYLASLGTKDPSMFVRHTEDADKNLE 387
GY N +L+ D+ +QV K R + +G S L+Y + ++P F + D+ L
Sbjct: 197 GYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLR 256
Query: 388 HLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKE 447
+LFW D R +Y F D ++FD TY K + PL +F VNH +Q ++G A++ +E
Sbjct: 257 NLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESM 316
Query: 448 ETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI-- 505
ET+VWL++ L AM ++P ++TD DK + +A+ + PN H WH++ +
Sbjct: 317 ETFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSH 376
Query: 506 ---KIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFR 562
+ +FL KF C+F + +F +W +V +F L + W++ ++ R W +
Sbjct: 377 VMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFMS 436
Query: 563 GKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDP 622
F AG T+ R E N+ K++ ++ LKEFL+ + L+ E ADF + + P
Sbjct: 437 DVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEEESVADFDTCHKQP 496
Query: 623 GLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESS 682
LK+P + E + T +F +F+ ++ ++ M+ V + E
Sbjct: 497 ALKSPSPW---EKQMATTYTHTIFKKFQVEVLGVVACHPRKEKEDENMATFRV-QDCEKD 552
Query: 683 REWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHL 742
++LV++ T E C C E G C H + +L +IP +L+RW++ AK +
Sbjct: 553 DDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMCGFASIPPQYILKRWTKDAKSGV 612
Query: 743 CETPGS 748
G+
Sbjct: 613 LAGEGA 618
>ref|NP_567455.1| far-red impaired response protein (FAR1) / far-red impaired
responsive protein (FAR1) [Arabidopsis thaliana]
Length = 768
Score = 212 bits (539), Expect = 5e-53
Identities = 121/426 (28%), Positives = 209/426 (48%), Gaps = 12/426 (2%)
Query: 331 GYNNTQYILK-DLQNQVGKQRRAHVSDGRSA--LDYLASLGTKDPSMFVRHTEDADKNLE 387
GY N +L+ D+ +QV K R + +G S L+Y + ++P F + D+ L
Sbjct: 138 GYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLR 197
Query: 388 HLFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKE 447
+LFW D R +Y F D ++FD TY K + PL +F VNH +Q ++G A++ +E
Sbjct: 198 NLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESM 257
Query: 448 ETYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNI-- 505
ET+VWL++ L AM ++P ++TD DK + +A+ + PN H WH++ +
Sbjct: 258 ETFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSH 317
Query: 506 ---KIPDFLPKFKTCMFGDFQIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFR 562
+ +FL KF C+F + +F +W +V +F L + W++ ++ R W +
Sbjct: 318 VMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFMS 377
Query: 563 GKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRSGYGDP 622
F AG T+ R E N+ K++ ++ LKEFL+ + L+ E ADF + + P
Sbjct: 378 DVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEEESVADFDTCHKQP 437
Query: 623 GLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSMHTVKRSQESS 682
LK+P + E + T +F +F+ ++ ++ M+ V + E
Sbjct: 438 ALKSPSPW---EKQMATTYTHTIFKKFQVEVLGVVACHPRKEKEDENMATFRV-QDCEKD 493
Query: 683 REWLVSYYTTTYEFRCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHL 742
++LV++ T E C C E G C H + +L +IP +L+RW++ AK +
Sbjct: 494 DDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMCGFASIPPQYILKRWTKDAKSGV 553
Query: 743 CETPGS 748
G+
Sbjct: 554 LAGEGA 559
>dbj|BAD37792.1| far-red impaired response protein-like [Oryza sativa (japonica
cultivar-group)] gi|51535335|dbj|BAD38595.1| far-red
impaired response protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 823
Score = 212 bits (539), Expect = 5e-53
Identities = 121/397 (30%), Positives = 200/397 (49%), Gaps = 13/397 (3%)
Query: 355 SDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYK 414
+D + L++ + + + F D ++++FWC + R+++ FGDA+ FD TY+
Sbjct: 215 NDVQKTLNFFREMQCRSKNFFYTIQIDGASRIKNIFWCHSLSRLSFDHFGDAITFDTTYQ 274
Query: 415 KNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGD 474
N+Y P IF VN+ QT + G A++ E E + WL + +AM K P ++TD
Sbjct: 275 TNRYNMPFGIFVGVNNHFQTAIFGCALLREETIEAFKWLFQTFTDAMHGKRPVAILTDNC 334
Query: 475 KAMRNAIRRVFPNASHRLCAWHLMCNATDNI-----KIPDFLPKFKTCMFGDFQIGDFKR 529
M AI+ V+P HR+C WH++ NA +N+ K F +F + +F++
Sbjct: 335 HQMEVAIKAVWPETIHRVCKWHVLKNAKENLGNIYSKRSSFKQEFHRVLNEPQTEAEFEK 394
Query: 530 KWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRR 589
W L+E++NL + ++ M+ + WA AYFR FFA TT R E N L K+VK
Sbjct: 395 AWSDLMEQYNLESSVYLRRMWDMKKKWAPAYFREFFFARMSTTQRSESMNHVLKKYVKPS 454
Query: 590 YNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQS-IELSASRILTRQLFFR 648
+L F + + E + +EA+ + K + S IE ASR+ TR F R
Sbjct: 455 SSLHGFAKRY----ENFYNDRIEAEDAEEHDTYNEKVSTLTSSPIEKHASRVYTRGAFSR 510
Query: 649 FRPTI-VSAADMIITNCEDTLGMSMHTVKRSQES--SREWLVSYYTTTYEFRCSCMRMES 705
F+ +S + M+ + + +H + +S S+E+ V T + C C E
Sbjct: 511 FKEQFKLSFSFMVYHTSDQHVLQLVHIGDDTLQSWGSKEFKVQVDLTEQDLSCGCKLFEH 570
Query: 706 IGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHL 742
+G+ C+HII V+V IPK +L+RW++ A+D +
Sbjct: 571 LGIICSHIIRVMVQYGFTEIPKKYILKRWTKDARDSI 607
>gb|AAT94016.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|51038153|gb|AAT93956.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 909
Score = 211 bits (537), Expect = 8e-53
Identities = 121/397 (30%), Positives = 200/397 (49%), Gaps = 13/397 (3%)
Query: 355 SDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYK 414
+D + L++ + + + F D ++++FWC + R+++ FGDA+ FD TY+
Sbjct: 301 NDVQKTLNFFREMQCRSKNFFYTIQIDGASRIKNIFWCHSLSRLSFDHFGDAITFDTTYQ 360
Query: 415 KNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGD 474
N+Y P IF VN+ QT + G A++ E E + WL + +AM K P ++TD
Sbjct: 361 TNRYNMPFGIFVGVNNHFQTAIFGCALLREETIEAFKWLFQTFTDAMHGKRPAAILTDNC 420
Query: 475 KAMRNAIRRVFPNASHRLCAWHLMCNATDNI-----KIPDFLPKFKTCMFGDFQIGDFKR 529
M AI+ V+P HR+C WH++ NA +N+ K F +F + +F++
Sbjct: 421 HQMEVAIKAVWPETIHRVCKWHVLKNAKENLGNIYSKRSSFKQEFHRVLNEPQTEAEFEK 480
Query: 530 KWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRR 589
W L+E++NL + ++ M+ + WA AYFR FFA TT R E N L K+VK
Sbjct: 481 AWSDLMEQYNLESSVYLRRMWDMKKKWAPAYFREFFFARMSTTQRSESMNHVLKKYVKPS 540
Query: 590 YNLKEFLQHFHHCLEYMRHREVEADFRSGYGDPGLKAPPIFQS-IELSASRILTRQLFFR 648
+L F + + E + +EA+ + K + S IE ASR+ TR F R
Sbjct: 541 SSLHGFAKRY----ENFYNDRIEAEDAEEHDTYNEKVSTLTSSPIEKHASRVYTRGEFSR 596
Query: 649 FRPTI-VSAADMIITNCEDTLGMSMHTVKRSQES--SREWLVSYYTTTYEFRCSCMRMES 705
F+ +S + M+ + + +H + +S S+E+ V T + C C E
Sbjct: 597 FKEQFKLSFSFMVYHTSDQHVLQLVHIGDDTLQSWGSKEFKVQVDLTEQDLSCGCKLFEH 656
Query: 706 IGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKDHL 742
+G+ C+HII V+V IPK +L+RW++ A+D +
Sbjct: 657 LGIICSHIIRVMVQYGFTEIPKKYILKRWTKDARDSI 693
>gb|AAX94831.1| transposon protein, putative, unclassified [Oryza sativa (japonica
cultivar-group)]
Length = 1023
Score = 211 bits (536), Expect = 1e-52
Identities = 105/290 (36%), Positives = 159/290 (54%), Gaps = 4/290 (1%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVSDGRS--ALDYLASLGTKDPSMFVRHTEDADKNLEH 388
GY T + +D+ N K ++ + G + L Y+ + D + + D L+
Sbjct: 727 GYEGTGFTSRDMYNFFVKLKKKRIKGGDADHVLKYMQARQKDDMEFYYDYEIDQAGCLKR 786
Query: 389 LFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEE 448
LFW D R++Y FGD + FD+TY+ NKY P + F VNH T + AV+ +E+ E
Sbjct: 787 LFWSDPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTVIFACAVVSDERVE 846
Query: 449 TYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIP 508
TY W+L Q L M +K P +VITDGD AMR AI VFPN+ HRLC WH+ N N+ P
Sbjct: 847 TYEWVLRQFLTCMCQKHPKSVITDGDNAMRRAILHVFPNSDHRLCTWHIEQNMARNLS-P 905
Query: 509 DFLPKFKTCMFGDFQIGDFKRKW-ESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFA 567
L F+T + +F +F+RKW E ++ +N W+ MY R WAA Y +G+ F
Sbjct: 906 AMLSDFRTLVHSEFDEDEFERKWVEFKIKHKVSDDNKWLKRMYNLRKKWAATYTKGRVFL 965
Query: 568 GFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRS 617
++ R E N++L + + R+ +L ++H+ HCL + H+E E D ++
Sbjct: 966 SMKSNQRSESLNSKLHRLLDRKMSLVILVEHYEHCLSRIHHQEAELDAKA 1015
>gb|AAX92763.1| transposon protein, putative, unclassified [Oryza sativa (japonica
cultivar-group)]
Length = 1185
Score = 211 bits (536), Expect = 1e-52
Identities = 105/290 (36%), Positives = 159/290 (54%), Gaps = 4/290 (1%)
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVSDGRS--ALDYLASLGTKDPSMFVRHTEDADKNLEH 388
GY T + +D+ N K ++ + G + L Y+ + D + + D L+
Sbjct: 727 GYEGTGFTSRDMYNFFVKLKKKRIKGGDADHVLKYMQARQKDDMEFYYDYEIDQAGCLKR 786
Query: 389 LFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEE 448
LFW D R++Y FGD + FD+TY+ NKY P + F VNH T + AV+ +E+ E
Sbjct: 787 LFWSDPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTVIFACAVVSDERVE 846
Query: 449 TYVWLLEQLLEAMDKKSPTTVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIP 508
TY W+L Q L M +K P +VITDGD AMR AI VFPN+ HRLC WH+ N N+ P
Sbjct: 847 TYEWVLRQFLTCMCQKHPKSVITDGDNAMRRAILHVFPNSDHRLCTWHIEQNMARNLS-P 905
Query: 509 DFLPKFKTCMFGDFQIGDFKRKW-ESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFFA 567
L F+T + +F +F+RKW E ++ +N W+ MY R WAA Y +G+ F
Sbjct: 906 AMLSDFRTLVHSEFDEDEFERKWVEFKIKHKVSDDNKWLKRMYNLRKKWAATYTKGRVFL 965
Query: 568 GFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHHCLEYMRHREVEADFRS 617
++ R E N++L + + R+ +L ++H+ HCL + H+E E D ++
Sbjct: 966 SMKSNQRSESLNSKLHRLLDRKMSLVILVEHYEHCLSRIHHQEAELDAKA 1015
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.351 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,378,411,147
Number of Sequences: 2540612
Number of extensions: 57061483
Number of successful extensions: 219866
Number of sequences better than 10.0: 395
Number of HSP's better than 10.0 without gapping: 243
Number of HSP's successfully gapped in prelim test: 152
Number of HSP's that attempted gapping in prelim test: 218956
Number of HSP's gapped (non-prelim): 748
length of query: 862
length of database: 863,360,394
effective HSP length: 137
effective length of query: 725
effective length of database: 515,296,550
effective search space: 373589998750
effective search space used: 373589998750
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0476b.3


