

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.7
(109 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
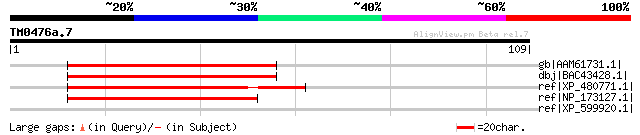
gb|AAM61731.1| unknown [Arabidopsis thaliana] 57 8e-08
dbj|BAC43428.1| unknown protein [Arabidopsis thaliana] gi|289731... 57 8e-08
ref|XP_480771.1| pentatricopeptide (PPR) repeat-containing prote... 56 2e-07
ref|NP_173127.1| pentatricopeptide (PPR) repeat-containing prote... 55 5e-07
ref|XP_599920.1| PREDICTED: similar to hypothetical protein, par... 32 4.9
>gb|AAM61731.1| unknown [Arabidopsis thaliana]
Length = 161
Score = 57.4 bits (137), Expect = 8e-08
Identities = 31/44 (70%), Positives = 33/44 (74%)
Query: 13 IYQDTVKAIVACLANTVDSAESVLRVAATGHDKRLYFKVSPSLY 56
I QDT IVA LANTV +AE VLRVAATGHDKRL+ KV LY
Sbjct: 63 ISQDTASNIVARLANTVGAAEGVLRVAATGHDKRLFVKVVICLY 106
>dbj|BAC43428.1| unknown protein [Arabidopsis thaliana] gi|28973105|gb|AAO63877.1|
unknown protein [Arabidopsis thaliana]
gi|18412164|ref|NP_565194.1| expressed protein
[Arabidopsis thaliana]
Length = 164
Score = 57.4 bits (137), Expect = 8e-08
Identities = 31/44 (70%), Positives = 33/44 (74%)
Query: 13 IYQDTVKAIVACLANTVDSAESVLRVAATGHDKRLYFKVSPSLY 56
I QDT IVA LANTV +AE VLRVAATGHDKRL+ KV LY
Sbjct: 66 ISQDTASNIVARLANTVGAAEGVLRVAATGHDKRLFVKVVICLY 109
>ref|XP_480771.1| pentatricopeptide (PPR) repeat-containing protein-like [Oryza
sativa (japonica cultivar-group)]
gi|38637178|dbj|BAD03430.1| pentatricopeptide (PPR)
repeat-containing protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 135
Score = 55.8 bits (133), Expect = 2e-07
Identities = 29/50 (58%), Positives = 38/50 (76%), Gaps = 2/50 (4%)
Query: 13 IYQDTVKAIVACLANTVDSAESVLRVAATGHDKRLYFKVSPSLY*ILTHF 62
I Q+ +IVA LANT+ +AESVLRVAATGHDK+L+FK P L+ ++ F
Sbjct: 84 ISQEMANSIVASLANTIGAAESVLRVAATGHDKKLFFK--PELFDMIAIF 131
>ref|NP_173127.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana]
Length = 738
Score = 54.7 bits (130), Expect = 5e-07
Identities = 29/40 (72%), Positives = 32/40 (79%)
Query: 13 IYQDTVKAIVACLANTVDSAESVLRVAATGHDKRLYFKVS 52
I QDT IVA LANTV +AESVLRVAATGHDKRL+ K +
Sbjct: 66 ISQDTACNIVARLANTVGAAESVLRVAATGHDKRLFVKAT 105
>ref|XP_599920.1| PREDICTED: similar to hypothetical protein, partial [Bos taurus]
Length = 111
Score = 31.6 bits (70), Expect = 4.9
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Query: 6 CGSSGVAIYQDTVKAI-VACLANTVDSAESVLRVAATGHDKRLYFKVSPSLY 56
CGSSG+ YQ V I AC+ TV+ AE + R+A+ G+ + K+ Y
Sbjct: 42 CGSSGIQEYQKGVPKIPTACI--TVEDAEMMSRMASRGNRIVVQLKMGAKSY 91
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.366 0.161 0.597
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 141,427,646
Number of Sequences: 2540612
Number of extensions: 3783682
Number of successful extensions: 30136
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 30132
Number of HSP's gapped (non-prelim): 5
length of query: 109
length of database: 863,360,394
effective HSP length: 85
effective length of query: 24
effective length of database: 647,408,374
effective search space: 15537800976
effective search space used: 15537800976
T: 11
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0476a.7


