

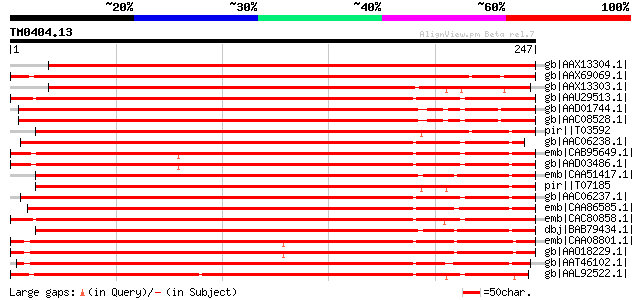
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.13
(247 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAX13304.1| MADS box protein AGb [Lotus corniculatus var. jap... 449 e-125
gb|AAX69069.1| MADS box protein M7 [Pisum sativum] 392 e-108
gb|AAX13303.1| MADS box protein AGa [Lotus corniculatus var. jap... 359 4e-98
gb|AAU29513.1| MADS4; PpMADS4 [Prunus persica] 348 6e-95
gb|AAD01744.1| agamous-like putative transcription factor [Cucum... 345 5e-94
gb|AAC08528.1| CUM1 [Cucumis sativus] gi|7446520|pir||T08039 MAD... 345 5e-94
pir||T03592 floral homeotic protein NAG1 - common tobacco gi|391... 340 3e-92
gb|AAC06238.1| AGAMOUS homolog [Populus balsamifera subsp. trich... 338 8e-92
emb|CAB95649.1| MADS box protein [Betula pendula] 338 1e-91
gb|AAD03486.1| MADS1 [Corylus avellana] 337 1e-91
emb|CAA51417.1| pMADS3 [Petunia x hybrida] gi|478387|pir||JQ2212... 336 3e-91
pir||T07185 floral homeotic protein TAG1 - tomato gi|3913004|sp|... 336 4e-91
gb|AAC06237.1| AGAMOUS homolog [Populus balsamifera subsp. trich... 335 5e-91
emb|CAA86585.1| agamous [Panax ginseng] gi|3913005|sp|Q40872|AG_... 335 5e-91
emb|CAC80858.1| C-type MADS box protein [Malus x domestica] 335 7e-91
dbj|BAB79434.1| PMADS3 [Petunia x hybrida] 334 1e-90
emb|CAA08801.1| MADS-box protein, GAGA2 [Gerbera hybrid cv. 'Ter... 333 3e-90
gb|AAO18229.1| MADS-box transcriptional factor HAM59 [Helianthus... 332 6e-90
gb|AAT46102.1| AGAMOUS-like protein [Akebia trifoliata] 331 1e-89
gb|AAL92522.1| AG-like protein [Gossypium hirsutum] 328 6e-89
>gb|AAX13304.1| MADS box protein AGb [Lotus corniculatus var. japonicus]
Length = 229
Score = 449 bits (1154), Expect = e-125
Identities = 227/229 (99%), Positives = 228/229 (99%)
Query: 19 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNS 78
GRGK+EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNS
Sbjct: 1 GRGKMEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNS 60
Query: 79 VKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNMNA 138
VKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNMNA
Sbjct: 61 VKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNMNA 120
Query: 139 RDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHN 198
RDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHN
Sbjct: 121 RDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHN 180
Query: 199 FNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
FNMLPGTTNFESLQQSQQPFDSRG FQVTGLQPNNNQCARQDQISLQFV
Sbjct: 181 FNMLPGTTNFESLQQSQQPFDSRGSFQVTGLQPNNNQCARQDQISLQFV 229
>gb|AAX69069.1| MADS box protein M7 [Pisum sativum]
Length = 243
Score = 392 bits (1007), Expect = e-108
Identities = 204/247 (82%), Positives = 222/247 (89%), Gaps = 4/247 (1%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
MS PN+SM +SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 1 MSFPNESMP--DSPQRKIGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 58
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
ALIVFS RGRLYEYANNSVKASIERYKKACSD+SG S S NAQ+YQQEAAKLRVQISN
Sbjct: 59 ALIVFSTRGRLYEYANNSVKASIERYKKACSDTSGAKSASETNAQYYQQEAAKLRVQISN 118
Query: 121 LQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNS 180
LQNHNRQM+GEALSNMN ++L+NLE+KLEKGISRIRSKKNEMLFAEIEYMQKREI+LHNS
Sbjct: 119 LQNHNRQMMGEALSNMNGKELRNLESKLEKGISRIRSKKNEMLFAEIEYMQKREIELHNS 178
Query: 181 NQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQD 240
NQ LRAKI+E+D+R NHN N+L G TNFE +Q QQ FDSR +FQV LQP NNQ ARQD
Sbjct: 179 NQALRAKISENDQRNNHNVNVLHGGTNFECIQPQQQ-FDSRSYFQVNELQP-NNQYARQD 236
Query: 241 QISLQFV 247
Q+SLQFV
Sbjct: 237 QMSLQFV 243
>gb|AAX13303.1| MADS box protein AGa [Lotus corniculatus var. japonicus]
Length = 248
Score = 359 bits (921), Expect = 4e-98
Identities = 190/233 (81%), Positives = 211/233 (90%), Gaps = 7/233 (3%)
Query: 19 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNS 78
GRGK+EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLYEYANNS
Sbjct: 1 GRGKMEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANNS 60
Query: 79 VKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNMNA 138
VKA+I+RYKKACSDSSG GS S ANAQFYQQEA KLRVQISNLQN+NRQM+ E+L +MNA
Sbjct: 61 VKATIDRYKKACSDSSGAGSASEANAQFYQQEADKLRVQISNLQNNNRQMMSESLGSMNA 120
Query: 139 RDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHN 198
++LKNLETKLEKGISRIRSKKNE+LFAEIEYMQKREIDLHN+NQLLRAKIAES ER + N
Sbjct: 121 KELKNLETKLEKGISRIRSKKNELLFAEIEYMQKREIDLHNNNQLLRAKIAES-ERNHPN 179
Query: 199 FNMLPG-TTNFESL----QQSQQPFDSRGFFQVTGLQP-NNNQCARQDQISLQ 245
++L G T+N+ES+ QQ QQ FDSRG+FQVTGLQP + Q +RQDQISLQ
Sbjct: 180 LSILAGSTSNYESMQSQQQQQQQQFDSRGYFQVTGLQPTTHTQYSRQDQISLQ 232
>gb|AAU29513.1| MADS4; PpMADS4 [Prunus persica]
Length = 243
Score = 348 bits (894), Expect = 6e-95
Identities = 180/247 (72%), Positives = 211/247 (84%), Gaps = 4/247 (1%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M+ N+SMS + SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 1 MAYENKSMSLD-SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 59
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
ALIVFSNRGRLYEYANNSVK +IERYKKAC++S+ GS S A+ Q+YQQEAAKLR Q N
Sbjct: 60 ALIVFSNRGRLYEYANNSVKETIERYKKACAESTNTGSVSEASTQYYQQEAAKLRAQTGN 119
Query: 121 LQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNS 180
LQN +R M+GE+LS+MN +DLKNLE+KLEKGI+RIRSKKNE+LFAEIEYMQKREIDLHN+
Sbjct: 120 LQNSSRHMMGESLSSMNMKDLKNLESKLEKGINRIRSKKNELLFAEIEYMQKREIDLHNN 179
Query: 181 NQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQD 240
NQLLRAKIAE +ER N N++ G ++E +Q QP+DSR +FQV LQPN+ +RQD
Sbjct: 180 NQLLRAKIAE-NERSQQNINVMAGGGSYEIMQ--SQPYDSRNYFQVNALQPNHQYNSRQD 236
Query: 241 QISLQFV 247
++LQ V
Sbjct: 237 PMALQLV 243
>gb|AAD01744.1| agamous-like putative transcription factor [Cucumis sativus]
Length = 237
Score = 345 bits (886), Expect = 5e-94
Identities = 185/243 (76%), Positives = 206/243 (84%), Gaps = 9/243 (3%)
Query: 5 NQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIV 64
NQ ++SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIV
Sbjct: 4 NQEEKMSDSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIV 63
Query: 65 FSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNH 124
FS+RGRLYEYANNSVKA+I+RYKKA SDSS GSTS AN QFYQQEAAKLRVQI NLQN
Sbjct: 64 FSSRGRLYEYANNSVKATIDRYKKASSDSSNTGSTSEANTQFYQQEAAKLRVQIGNLQNS 123
Query: 125 NRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLL 184
NR MLGE+LS++ A+DLK LETKLEKGISRIRSKKNE+LFAEIEYM+KREIDLHN+NQ+L
Sbjct: 124 NRNMLGESLSSLTAKDLKGLETKLEKGISRIRSKKNELLFAEIEYMRKREIDLHNNNQML 183
Query: 185 RAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISL 244
RAKIAES+ N NM+ G FE +Q P+D R FFQV GLQ +N+Q RQD ++L
Sbjct: 184 RAKIAESE----RNVNMMGG--EFELMQ--SHPYDPRDFFQVNGLQ-HNHQYPRQDNMAL 234
Query: 245 QFV 247
Q V
Sbjct: 235 QLV 237
>gb|AAC08528.1| CUM1 [Cucumis sativus] gi|7446520|pir||T08039 MADS-box protein -
cucumber
Length = 262
Score = 345 bits (886), Expect = 5e-94
Identities = 185/243 (76%), Positives = 206/243 (84%), Gaps = 9/243 (3%)
Query: 5 NQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIV 64
NQ ++SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIV
Sbjct: 29 NQEEKMSDSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIV 88
Query: 65 FSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNH 124
FS+RGRLYEYANNSVKA+I+RYKKA SDSS GSTS AN QFYQQEAAKLRVQI NLQN
Sbjct: 89 FSSRGRLYEYANNSVKATIDRYKKASSDSSNTGSTSEANTQFYQQEAAKLRVQIGNLQNS 148
Query: 125 NRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLL 184
NR MLGE+LS++ A+DLK LETKLEKGISRIRSKKNE+LFAEIEYM+KREIDLHN+NQ+L
Sbjct: 149 NRNMLGESLSSLTAKDLKGLETKLEKGISRIRSKKNELLFAEIEYMRKREIDLHNNNQML 208
Query: 185 RAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISL 244
RAKIAES+ N NM+ G FE +Q P+D R FFQV GLQ +N+Q RQD ++L
Sbjct: 209 RAKIAESE----RNVNMMGG--EFELMQ--SHPYDPRDFFQVNGLQ-HNHQYPRQDNMAL 259
Query: 245 QFV 247
Q V
Sbjct: 260 QLV 262
>pir||T03592 floral homeotic protein NAG1 - common tobacco
gi|3913007|sp|Q43585|AG_TOBAC Floral homeotic protein
AGAMOUS (NAG1) gi|431736|gb|AAA17033.1| NAG1
Length = 248
Score = 340 bits (871), Expect = 3e-92
Identities = 176/239 (73%), Positives = 204/239 (84%), Gaps = 6/239 (2%)
Query: 13 SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLY 72
SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLY
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLY 71
Query: 73 EYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEA 132
EYANNSVKA+IERYKKACSDSS GS S ANAQ+YQQEA+KLR QI NLQN NR MLGE+
Sbjct: 72 EYANNSVKATIERYKKACSDSSNTGSISEANAQYYQQEASKLRAQIGNLQNQNRNMLGES 131
Query: 133 LSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESD 192
L+ ++ RDLKNLE K+EKGIS+IRSKKNE+LFAEIEYMQKREIDLHN+NQ LRAKIAE++
Sbjct: 132 LAALSLRDLKNLEQKIEKGISKIRSKKNELLFAEIEYMQKREIDLHNNNQYLRAKIAETE 191
Query: 193 ----ERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
+++ N++PG++++E + Q FD+R + QV GLQ NN+ RQDQ SLQ V
Sbjct: 192 RAQQQQQQQQMNLMPGSSSYELVPPPHQ-FDTRNYLQVNGLQTNNHY-TRQDQPSLQLV 248
>gb|AAC06238.1| AGAMOUS homolog [Populus balsamifera subsp. trichocarpa]
Length = 238
Score = 338 bits (867), Expect = 8e-92
Identities = 175/237 (73%), Positives = 201/237 (83%), Gaps = 4/237 (1%)
Query: 6 QSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVF 65
Q+ +SP RK+GRGK+EIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVF
Sbjct: 4 QNEPQESSPLRKLGRGKVEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVF 63
Query: 66 SNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHN 125
S+RGRLYEY+NNSVK++IERYKKAC+DSS GS S ANAQFYQQEAAKLR QI NLQN N
Sbjct: 64 SSRGRLYEYSNNSVKSTIERYKKACADSSNNGSVSEANAQFYQQEAAKLRSQIGNLQNSN 123
Query: 126 RQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLR 185
R MLGE+LS ++ ++LK+LE KLEKGI RIRSKKNE+LFAEIEYMQKREIDLHN+NQLLR
Sbjct: 124 RNMLGESLSALSVKELKSLEIKLEKGIGRIRSKKNELLFAEIEYMQKREIDLHNNNQLLR 183
Query: 186 AKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQI 242
AKIAE +ERK + N++PG NFE +Q QPFDSR + QV GL P N+ +DQ+
Sbjct: 184 AKIAE-NERKRQHMNLMPGGVNFEIMQ--SQPFDSRNYSQVNGLPPANHY-PHEDQL 236
>emb|CAB95649.1| MADS box protein [Betula pendula]
Length = 242
Score = 338 bits (866), Expect = 1e-91
Identities = 180/248 (72%), Positives = 207/248 (82%), Gaps = 7/248 (2%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M NQSMS SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAE+
Sbjct: 1 MEFQNQSMSV--SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEI 58
Query: 61 ALIVFSNRGRLYEYANNS-VKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQIS 119
ALIVFS+RGRLYEYANNS VK +IERYKKAC++SS GS S AN QFYQQEAAKLR QI
Sbjct: 59 ALIVFSSRGRLYEYANNSSVKTTIERYKKACAESSNSGSVSEANTQFYQQEAAKLRGQIR 118
Query: 120 NLQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHN 179
++QN NR +LGEALS +N ++LKNLE KLEKGI++IRSKKNE+LFAEIEYMQKRE +LHN
Sbjct: 119 SVQNSNRHLLGEALSELNFKELKNLEIKLEKGINKIRSKKNELLFAEIEYMQKREAELHN 178
Query: 180 SNQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQ 239
+NQ+LRAKIAE +ER N N++PG N+E +Q Q +DSR +FQV LQPN++ RQ
Sbjct: 179 NNQILRAKIAE-NERNQQNLNVMPGGGNYELMQ--SQSYDSRTYFQVDALQPNHHY-PRQ 234
Query: 240 DQISLQFV 247
DQI LQ V
Sbjct: 235 DQIPLQLV 242
>gb|AAD03486.1| MADS1 [Corylus avellana]
Length = 242
Score = 337 bits (865), Expect = 1e-91
Identities = 180/248 (72%), Positives = 205/248 (82%), Gaps = 7/248 (2%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M NQSMS SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAE+
Sbjct: 1 MEFQNQSMSV--SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEI 58
Query: 61 ALIVFSNRGRLYEYANNS-VKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQIS 119
ALIVFS+RGRLYEYANNS VK +IERYKKAC+DSS GS S AN QFYQQEAAKLR QI
Sbjct: 59 ALIVFSSRGRLYEYANNSSVKTTIERYKKACADSSNSGSVSEANTQFYQQEAAKLRGQIR 118
Query: 120 NLQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHN 179
++Q+ NR MLGEALS +N ++LKNLE LEKGI+RIRSKKNE+L AEIEYM KRE+DLHN
Sbjct: 119 SVQDSNRHMLGEALSELNFKELKNLEKNLEKGINRIRSKKNELLLAEIEYMHKREVDLHN 178
Query: 180 SNQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQ 239
+NQ LRAKIAE +ER N N++PG N+E +Q Q FDSR +FQV LQPN++ RQ
Sbjct: 179 NNQFLRAKIAE-NERNQQNLNVMPGGGNYELMQ--SQSFDSRNYFQVDALQPNHHY-PRQ 234
Query: 240 DQISLQFV 247
DQ++LQ V
Sbjct: 235 DQMALQLV 242
>emb|CAA51417.1| pMADS3 [Petunia x hybrida] gi|478387|pir||JQ2212 pMADS3 protein -
garden petunia gi|3913006|sp|Q40885|AG_PETHY Floral
homeotic protein AGAMOUS (pMADS3)
Length = 242
Score = 336 bits (862), Expect = 3e-91
Identities = 173/235 (73%), Positives = 202/235 (85%), Gaps = 4/235 (1%)
Query: 13 SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLY 72
SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLY
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLY 71
Query: 73 EYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEA 132
EYANNSVKA+IERYKKACSDSS GS + ANAQ+YQQEA+KLR QI NLQN NR LGE+
Sbjct: 72 EYANNSVKATIERYKKACSDSSNTGSIAEANAQYYQQEASKLRAQIGNLQNQNRNFLGES 131
Query: 133 LSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESD 192
L+ +N RDL+NLE K+EKGIS+IR+KKNE+LFAEIEYMQKREIDLHN+NQ LRAKIAE++
Sbjct: 132 LAALNLRDLRNLEQKIEKGISKIRAKKNELLFAEIEYMQKREIDLHNNNQYLRAKIAETE 191
Query: 193 ERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
++ N++PG+++++ L QQ FD+R + QV GLQ NN+ RQDQ LQ V
Sbjct: 192 --RSQQMNLMPGSSSYD-LVPPQQSFDARNYLQVNGLQTNNHY-PRQDQPPLQLV 242
>pir||T07185 floral homeotic protein TAG1 - tomato gi|3913004|sp|Q40168|AG_LYCES
Floral homeotic protein AGAMOUS (TAG1)
gi|457382|gb|AAA34197.1| TAG1
Length = 248
Score = 336 bits (861), Expect = 4e-91
Identities = 171/238 (71%), Positives = 200/238 (83%), Gaps = 4/238 (1%)
Query: 13 SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLY 72
SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVAL+VFSNRGRLY
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSNRGRLY 71
Query: 73 EYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEA 132
EYANNSVKA+IERYKKACSDSS GS S ANAQ+YQQEA+KLR QI NL N NR M+GEA
Sbjct: 72 EYANNSVKATIERYKKACSDSSNTGSVSEANAQYYQQEASKLRAQIGNLMNQNRNMMGEA 131
Query: 133 LSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESD 192
L+ M ++LKNLE ++EKGIS+IRSKKNE+LFAEIEYMQKRE+DLHN+NQ LRAKIAE++
Sbjct: 132 LAGMKLKELKNLEQRIEKGISKIRSKKNELLFAEIEYMQKREVDLHNNNQYLRAKIAETE 191
Query: 193 --ERKNHNFNMLPG-TTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
+ ++ N++PG ++N+ L Q FD+R + QV GLQ NN+ RQDQ +Q V
Sbjct: 192 RAQHQHQQMNLMPGSSSNYHELVPPPQQFDTRNYLQVNGLQTNNHY-PRQDQPPIQLV 248
>gb|AAC06237.1| AGAMOUS homolog [Populus balsamifera subsp. trichocarpa]
Length = 241
Score = 335 bits (860), Expect = 5e-91
Identities = 173/242 (71%), Positives = 209/242 (85%), Gaps = 4/242 (1%)
Query: 6 QSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVF 65
Q+ S +SP RK+GRGK+EIKRIENTTNRQVTFCKRR+GLLKKAYELSVLCDAEVALIVF
Sbjct: 4 QNESLESSPLRKLGRGKVEIKRIENTTNRQVTFCKRRSGLLKKAYELSVLCDAEVALIVF 63
Query: 66 SNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHN 125
S+RGRLYEY+N+SVK++IERYKKA +DSS GS S ANAQ+YQQEAAKLR QI NLQN N
Sbjct: 64 SSRGRLYEYSNDSVKSTIERYKKASADSSNTGSVSEANAQYYQQEAAKLRSQIGNLQNSN 123
Query: 126 RQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLR 185
R MLGEALS+++ ++LK+LE +LEKGISRIRSKKNE+LFAEIEYMQKRE+DLHN+NQLLR
Sbjct: 124 RHMLGEALSSLSVKELKSLEIRLEKGISRIRSKKNELLFAEIEYMQKREVDLHNNNQLLR 183
Query: 186 AKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQ 245
AKI+E +ERK + N++PG +FE +Q QP+DSR + QV GLQP ++ + QDQ++LQ
Sbjct: 184 AKISE-NERKRQSMNLMPGGADFEIVQ--SQPYDSRNYSQVNGLQPASHY-SHQDQMALQ 239
Query: 246 FV 247
V
Sbjct: 240 LV 241
>emb|CAA86585.1| agamous [Panax ginseng] gi|3913005|sp|Q40872|AG_PANGI Floral
homeotic protein AGAMOUS (GAG2)
Length = 242
Score = 335 bits (860), Expect = 5e-91
Identities = 173/239 (72%), Positives = 201/239 (83%), Gaps = 4/239 (1%)
Query: 9 SANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNR 68
S N SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS R
Sbjct: 8 SGNLSPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTR 67
Query: 69 GRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQM 128
GRLYEYANNSVK +IERYKKAC+DS S S ANAQFYQQEA+KLR +IS++Q +NR M
Sbjct: 68 GRLYEYANNSVKGTIERYKKACTDSPNTSSVSEANAQFYQQEASKLRQEISSIQKNNRNM 127
Query: 129 LGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKI 188
+GE+L ++ RDLK LETKLEKGISRIRSKKNE+LFAEIEYMQK+EIDLHN+NQ LRAKI
Sbjct: 128 MGESLGSLTVRDLKGLETKLEKGISRIRSKKNELLFAEIEYMQKKEIDLHNNNQYLRAKI 187
Query: 189 AESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
AE +ER + N++PG++++E Q FD R + Q+ GLQPNN+ +RQDQ +LQ V
Sbjct: 188 AE-NERAQQHMNLMPGSSDYE--LAPPQSFDGRNYIQLNGLQPNNHY-SRQDQTALQLV 242
>emb|CAC80858.1| C-type MADS box protein [Malus x domestica]
Length = 245
Score = 335 bits (859), Expect = 7e-91
Identities = 177/249 (71%), Positives = 209/249 (83%), Gaps = 6/249 (2%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M+ ++S+S + SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 1 MAYESKSLSLD-SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 59
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
ALIVFSNRGRLYEYANNSVK +IERYKKA +DSS GS S A+ Q+YQQEAAKLR +I
Sbjct: 60 ALIVFSNRGRLYEYANNSVKGTIERYKKASADSSNTGSVSEASTQYYQQEAAKLRARIVK 119
Query: 121 LQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNS 180
LQN NR M+G+AL++M+ +DLK+LE KLEK ISRIRSKKNE+LFAEIEYMQKRE+DLHN+
Sbjct: 120 LQNDNRNMMGDALNSMSVKDLKSLENKLEKAISRIRSKKNELLFAEIEYMQKRELDLHNN 179
Query: 181 NQLLRAKIAESDERKNHNFNMLP--GTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCAR 238
NQLLRAKIAE +ER + N++ GT++++ LQ QP+DSR +FQV LQPN+ R
Sbjct: 180 NQLLRAKIAE-NERASRTLNVMAGGGTSSYDILQ--SQPYDSRNYFQVNALQPNHQYNPR 236
Query: 239 QDQISLQFV 247
DQISLQ V
Sbjct: 237 HDQISLQLV 245
>dbj|BAB79434.1| PMADS3 [Petunia x hybrida]
Length = 242
Score = 334 bits (857), Expect = 1e-90
Identities = 172/235 (73%), Positives = 201/235 (85%), Gaps = 4/235 (1%)
Query: 13 SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLY 72
SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLY
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLY 71
Query: 73 EYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEA 132
EYANNSVKA+IERYKKACSDSS GS + ANAQ+YQQEA+KLR QI N QN NR LGE+
Sbjct: 72 EYANNSVKATIERYKKACSDSSNTGSIAEANAQYYQQEASKLRAQIGNXQNQNRNFLGES 131
Query: 133 LSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESD 192
L+ +N RDL+NLE K+EKGIS+IR+KKNE+LFAEIEYMQKREIDLHN+NQ LRAKIAE++
Sbjct: 132 LAALNLRDLRNLEQKIEKGISKIRAKKNELLFAEIEYMQKREIDLHNNNQYLRAKIAETE 191
Query: 193 ERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
++ N++PG+++++ L QQ FD+R + QV GLQ NN+ RQDQ LQ V
Sbjct: 192 --RSQQMNLMPGSSSYD-LVPPQQSFDARNYLQVNGLQTNNHY-PRQDQPPLQLV 242
>emb|CAA08801.1| MADS-box protein, GAGA2 [Gerbera hybrid cv. 'Terra Regina']
Length = 246
Score = 333 bits (853), Expect = 3e-90
Identities = 178/251 (70%), Positives = 203/251 (79%), Gaps = 9/251 (3%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
MS PN S SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 1 MSFPNDS--GEMSPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 58
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
ALIVFS+RGRLYEYANNSVK +I+RYKKAC D GS + ANAQFYQQEAAKLR QI+N
Sbjct: 59 ALIVFSSRGRLYEYANNSVKGTIDRYKKACLDPPSSGSVAEANAQFYQQEAAKLRQQIAN 118
Query: 121 LQNHNRQ----MLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREID 176
LQN NRQ ++GE+L NM A+DLKNLE+KLEKGI +IRSKKNE+LFAEIEYMQKRE +
Sbjct: 119 LQNQNRQFYRNIMGESLGNMPAKDLKNLESKLEKGIGKIRSKKNEILFAEIEYMQKRENE 178
Query: 177 LHNSNQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQC 236
LHNSNQ LR+KIAE +ER + +++PG++++E L QPFD R + QV LQPNNN
Sbjct: 179 LHNSNQFLRSKIAE-NERAQQHMSLMPGSSDYE-LVAPHQPFDGRNYLQVNDLQPNNNYS 236
Query: 237 ARQDQISLQFV 247
QDQ LQ V
Sbjct: 237 C-QDQTPLQLV 246
>gb|AAO18229.1| MADS-box transcriptional factor HAM59 [Helianthus annuus]
Length = 247
Score = 332 bits (851), Expect = 6e-90
Identities = 175/251 (69%), Positives = 203/251 (80%), Gaps = 8/251 (3%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
MS PN+S SPQRK+GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 1 MSFPNES--GEMSPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 58
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
ALIVFS+RGRLYEYANNSVK +I+RYKKAC D GS + ANAQFYQQEAAKLR QI+N
Sbjct: 59 ALIVFSSRGRLYEYANNSVKGTIDRYKKACLDPPSSGSVAEANAQFYQQEAAKLRQQIAN 118
Query: 121 LQNHNRQ----MLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREID 176
LQN NRQ ++GE+L NM A+DLKNLE KLEKGISRIRSKKNE+LFAEIEYM KRE +
Sbjct: 119 LQNQNRQFYRNIMGESLGNMPAKDLKNLEGKLEKGISRIRSKKNELLFAEIEYMPKRENE 178
Query: 177 LHNSNQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQC 236
LHN+NQ LRAKIAE++ + + +++PG+++++ L QPFD R + QV LQPNN+
Sbjct: 179 LHNNNQFLRAKIAENERSQQQHMSLMPGSSDYD-LVPPHQPFDGRNYLQVNDLQPNNSYS 237
Query: 237 ARQDQISLQFV 247
QDQ LQ V
Sbjct: 238 C-QDQTPLQLV 247
>gb|AAT46102.1| AGAMOUS-like protein [Akebia trifoliata]
Length = 245
Score = 331 bits (848), Expect = 1e-89
Identities = 175/242 (72%), Positives = 198/242 (81%), Gaps = 7/242 (2%)
Query: 4 PNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALI 63
PN+S +NNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVAL+
Sbjct: 9 PNRS--SNNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALV 66
Query: 64 VFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQN 123
VFSNRGRLYEY+NNSVK +IERYKKAC DSS GS S ANAQFYQQE+ KLR QI NLQN
Sbjct: 67 VFSNRGRLYEYSNNSVKTTIERYKKACVDSSNSGSVSEANAQFYQQESLKLRQQIGNLQN 126
Query: 124 HNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQL 183
NR ++GEAL +M+ ++LK LET++EKGISRIRSKKNE+LFAEIEYMQKREIDL N N
Sbjct: 127 LNRHLMGEALGSMSIKELKQLETRIEKGISRIRSKKNELLFAEIEYMQKREIDLQNDNMY 186
Query: 184 LRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQIS 243
LRAKIAE +ER + N++PG E S PFDSR F QV L+PNN+ + DQ +
Sbjct: 187 LRAKIAE-NERAGQHMNLMPGN---EYEVMSSAPFDSRNFLQVNLLEPNNHY-SHTDQTA 241
Query: 244 LQ 245
LQ
Sbjct: 242 LQ 243
>gb|AAL92522.1| AG-like protein [Gossypium hirsutum]
Length = 244
Score = 328 bits (842), Expect = 6e-89
Identities = 178/249 (71%), Positives = 206/249 (82%), Gaps = 11/249 (4%)
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M PN+S+ +SPQ+KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 1 MVYPNESLE--DSPQKKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 58
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
AL+ FS+RGRLYEYANNSVKA+IERYKKA SDSS GS + NAQFYQQEA KLR QI N
Sbjct: 59 ALVAFSSRGRLYEYANNSVKATIERYKKA-SDSSNTGSVAEVNAQFYQQEADKLRNQIRN 117
Query: 121 LQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNS 180
LQN NR MLGE++ + ++LK+LE++LEKGISRIRSKKNE+LFAEIEYMQKREIDLHN+
Sbjct: 118 LQNANRHMLGESIGGLPMKELKSLESRLEKGISRIRSKKNELLFAEIEYMQKREIDLHNN 177
Query: 181 NQLLRAKIAESDERKNHNFNMLPG--TTNFESLQQSQQPFDSRGFFQVTGLQPNNNQC-- 236
NQLLRAKIAE +ERK + N++PG + NFE+L QP+DSR +FQV LQP N
Sbjct: 178 NQLLRAKIAE-NERKQQSMNLMPGGSSANFEALH--SQPYDSRNYFQVDALQPATNYYNP 234
Query: 237 -ARQDQISL 244
+QDQI+L
Sbjct: 235 QLQQDQIAL 243
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 368,705,141
Number of Sequences: 2540612
Number of extensions: 13965999
Number of successful extensions: 42074
Number of sequences better than 10.0: 2157
Number of HSP's better than 10.0 without gapping: 1873
Number of HSP's successfully gapped in prelim test: 284
Number of HSP's that attempted gapping in prelim test: 39046
Number of HSP's gapped (non-prelim): 2274
length of query: 247
length of database: 863,360,394
effective HSP length: 125
effective length of query: 122
effective length of database: 545,783,894
effective search space: 66585635068
effective search space used: 66585635068
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0404.13


