

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.8
(721 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
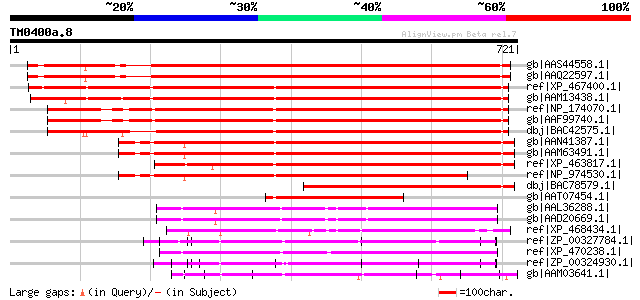
Score E
Sequences producing significant alignments: (bits) Value
gb|AAS44558.1| kinesin light chain-like protein [Arabidopsis tha... 846 0.0
gb|AAQ22597.1| At3g27960 [Arabidopsis thaliana] 843 0.0
ref|XP_467400.1| putative kinesin light chain [Oryza sativa (jap... 829 0.0
gb|AAM13438.1| similar to A. thaliana protein BAB01483 similar ... 801 0.0
ref|NP_174070.1| kinesin light chain-related [Arabidopsis thaliana] 691 0.0
gb|AAF99740.1| F17L21.29 [Arabidopsis thaliana] 691 0.0
dbj|BAC42575.1| unknown protein [Arabidopsis thaliana] gi|290289... 691 0.0
gb|AAN41387.1| unknown protein [Arabidopsis thaliana] gi|7267782... 645 0.0
gb|AAM63491.1| putative kinesin light chain [Arabidopsis thaliana] 645 0.0
ref|XP_463817.1| kinesin light chain-like protein [Oryza sativa ... 629 e-178
ref|NP_974530.1| kinesin light chain-related [Arabidopsis thaliana] 564 e-159
dbj|BAC78579.1| hypothetical protein [Oryza sativa (japonica cul... 405 e-111
gb|AAT07454.1| kinesin-like protein [Mirabilis jalapa] 297 7e-79
gb|AAL36288.1| putative kinesin light chain [Arabidopsis thalian... 170 2e-40
gb|AAD20669.1| putative kinesin light chain [Arabidopsis thalian... 170 2e-40
ref|XP_468434.1| kinesin light chain-like [Oryza sativa (japonic... 152 4e-35
ref|ZP_00327784.1| COG0457: FOG: TPR repeat [Trichodesmium eryth... 128 6e-28
ref|XP_470238.1| Hypothetical protein [Oryza sativa (japonica cu... 127 1e-27
ref|ZP_00324930.1| COG0457: FOG: TPR repeat [Trichodesmium eryth... 121 9e-26
gb|AAM03641.1| hypothetical protein (multi-domain) [Methanosarci... 115 5e-24
>gb|AAS44558.1| kinesin light chain-like protein [Arabidopsis thaliana]
gi|9294312|dbj|BAB01483.1| unnamed protein product
[Arabidopsis thaliana] gi|15232873|ref|NP_189435.1|
kinesin light chain-related [Arabidopsis thaliana]
Length = 663
Score = 846 bits (2186), Expect = 0.0
Identities = 448/695 (64%), Positives = 539/695 (77%), Gaps = 58/695 (8%)
Query: 26 KDNFNQQASPRSTLSPRSIQSSDSIDLAIDGVVDTSIEQLYYNVCEMRSSD-QSPSRASF 84
KD+ QASPRS LS SIDLAIDG ++ SIEQLY+NVCEM SSD QSPSRASF
Sbjct: 11 KDDSALQASPRSPLS--------SIDLAIDGAMNASIEQLYHNVCEMESSDDQSPSRASF 62
Query: 85 YSYGEESRIDSELGHLVGGILE---ITKEVVTENKEESNGNAA--EKDIVSCGKEATKKD 139
SYG ESRID EL HLVG + E KE++ E KEESNG + +K +S GK+ K
Sbjct: 63 ISYGAESRIDLELRHLVGDVGEEGESKKEIILEKKEESNGEGSLSQKKPLSNGKKVAKTS 122
Query: 140 NNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIE 199
N P SRI S+ KS +L
Sbjct: 123 PNNPKMPGSRI------SSRKSPDLG---------------------------------- 142
Query: 200 ERIATGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSL--ELVM 257
+++ ++P+LG LLKQ R+++SSGEN KALDLALRA+K FE C +G+ L LVM
Sbjct: 143 -KVSVDEESPELGVVLLKQARELVSSGENLNKALDLALRAVKVFEKCGEGEKQLGLNLVM 201
Query: 258 CLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIEN 317
LH+LA IY LG+YN+A+P+LERSI+IP++EDG+DHALAKFAGCMQLGD Y +MG +EN
Sbjct: 202 SLHILAAIYAGLGRYNDAVPVLERSIEIPMIEDGEDHALAKFAGCMQLGDMYGLMGQVEN 261
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
SI+ YTAGLEIQ QVLGE+D R GETCRY+AEAHVQA+QF+EA RLCQMAL+IH+ NG
Sbjct: 262 SIMLYTAGLEIQRQVLGESDARVGETCRYLAEAHVQAMQFEEASRLCQMALDIHKENGAA 321
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLA 437
++AS+EE+ADR+LMGLI D+KGDYE ALEHYVLASMAMS+ +DVA+VD SIGDAY++
Sbjct: 322 ATASIEEAADRKLMGLICDAKGDYEVALEHYVLASMAMSSQNHREDVAAVDCSIGDAYMS 381
Query: 438 LARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIF 497
LAR+DEA+F+YQKAL VFK KGE H +VA VYVRLADLYNKIGK +DSKSYCENAL+I+
Sbjct: 382 LARFDEAIFAYQKALAVFKQGKGETHSSVALVYVRLADLYNKIGKTRDSKSYCENALKIY 441
Query: 498 GKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQM 557
K PG P EE+A G I+++AIYQSMN+L++ LKLL++ALKIY +APGQQ+TIAGIEAQM
Sbjct: 442 LKPTPGTPMEEVATGFIEISAIYQSMNELDQALKLLRRALKIYANAPGQQNTIAGIEAQM 501
Query: 558 GVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFE 617
GV+ YM+GNYS+SY+IFKSA++KFR SGEKK+ALFGIALNQMGLACVQRYAINEAADLFE
Sbjct: 502 GVVTYMMGNYSESYDIFKSAISKFRNSGEKKTALFGIALNQMGLACVQRYAINEAADLFE 561
Query: 618 EARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVD 677
EA+TILEKE G YHPDTL VYSNLAGTYDAMGR+DDAIEILE+VVG REEKLGTANP+V+
Sbjct: 562 EAKTILEKECGPYHPDTLAVYSNLAGTYDAMGRLDDAIEILEYVVGTREEKLGTANPEVE 621
Query: 678 DEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRL 712
DEK+RLA LLKEAGRGR++++ R+L TLLD N +
Sbjct: 622 DEKQRLAALLKEAGRGRSKRN-RALLTLLDNNPEI 655
>gb|AAQ22597.1| At3g27960 [Arabidopsis thaliana]
Length = 663
Score = 843 bits (2179), Expect = 0.0
Identities = 447/695 (64%), Positives = 538/695 (77%), Gaps = 58/695 (8%)
Query: 26 KDNFNQQASPRSTLSPRSIQSSDSIDLAIDGVVDTSIEQLYYNVCEMRSSD-QSPSRASF 84
KD+ QASPRS LS SIDLAIDG ++ SIEQLY+NVCEM SSD QSPSRASF
Sbjct: 11 KDDSALQASPRSPLS--------SIDLAIDGAMNASIEQLYHNVCEMESSDDQSPSRASF 62
Query: 85 YSYGEESRIDSELGHLVGGILE---ITKEVVTENKEESNGNAA--EKDIVSCGKEATKKD 139
SYG ESRID EL HLVG + E KE++ E KEESNG + +K +S GK+ K
Sbjct: 63 ISYGAESRIDLELRHLVGDVGEEGESKKEIILEKKEESNGEGSLSQKKPLSNGKKVAKTS 122
Query: 140 NNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIE 199
N P SRI S+ KS +L
Sbjct: 123 PNNPKMPGSRI------SSRKSPDLG---------------------------------- 142
Query: 200 ERIATGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSL--ELVM 257
+++ ++P+LG LLKQ R+++SSGEN KALDLALRA+K FE C +G+ L LVM
Sbjct: 143 -KVSVDEESPELGVVLLKQARELVSSGENLNKALDLALRAVKVFEKCGEGEKQLGLNLVM 201
Query: 258 CLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIEN 317
LH+LA IY LG+YN+A+P+L RSI+IP++EDG+DHALAKFAGCMQLGD Y +MG +EN
Sbjct: 202 SLHILAAIYAGLGRYNDAVPVLGRSIEIPMIEDGEDHALAKFAGCMQLGDMYGLMGQVEN 261
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
SI+ YTAGLEIQ QVLGE+D R GETCRY+AEAHVQA+QF+EA RLCQMAL+IH+ NG
Sbjct: 262 SIMLYTAGLEIQRQVLGESDARVGETCRYLAEAHVQAMQFEEASRLCQMALDIHKENGAA 321
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLA 437
++AS+EE+ADR+LMGLI D+KGDYE ALEHYVLASMAMS+ +DVA+VD SIGDAY++
Sbjct: 322 ATASIEEAADRKLMGLICDAKGDYEVALEHYVLASMAMSSQNHREDVAAVDCSIGDAYMS 381
Query: 438 LARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIF 497
LAR+DEA+F+YQKAL VFK KGE H +VA VYVRLADLYNKIGK +DSKSYCENAL+I+
Sbjct: 382 LARFDEAIFAYQKALAVFKQGKGETHSSVALVYVRLADLYNKIGKTRDSKSYCENALKIY 441
Query: 498 GKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQM 557
K PG P EE+A G I+++AIYQSMN+L++ LKLL++ALKIY +APGQQ+TIAGIEAQM
Sbjct: 442 LKPTPGTPMEEVATGFIEISAIYQSMNELDQALKLLRRALKIYANAPGQQNTIAGIEAQM 501
Query: 558 GVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFE 617
GV+ YM+GNYS+SY+IFKSA++KFR SGEKK+ALFGIALNQMGLACVQRYAINEAADLFE
Sbjct: 502 GVVTYMMGNYSESYDIFKSAISKFRNSGEKKTALFGIALNQMGLACVQRYAINEAADLFE 561
Query: 618 EARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVD 677
EA+TILEKE G YHPDTL VYSNLAGTYDAMGR+DDAIEILE+VVG REEKLGTANP+V+
Sbjct: 562 EAKTILEKECGPYHPDTLAVYSNLAGTYDAMGRLDDAIEILEYVVGTREEKLGTANPEVE 621
Query: 678 DEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRL 712
DEK+RLA LLKEAGRGR++++ R+L TLLD N +
Sbjct: 622 DEKQRLAALLKEAGRGRSKRN-RALLTLLDNNPEI 655
>ref|XP_467400.1| putative kinesin light chain [Oryza sativa (japonica
cultivar-group)] gi|41053168|dbj|BAD08110.1| putative
kinesin light chain [Oryza sativa (japonica
cultivar-group)] gi|19387245|gb|AAL87157.1| putative
kinesin light chain gene [Oryza sativa (japonica
cultivar-group)]
Length = 711
Score = 829 bits (2141), Expect = 0.0
Identities = 440/686 (64%), Positives = 536/686 (77%), Gaps = 13/686 (1%)
Query: 28 NFNQQASPRSTLSPRSIQSSDSIDLAIDG--VVDTSIEQLYYNVCEMRSSDQ--SPSRAS 83
N + ++PRS ++ +S+ S +++++ ++G + SIEQLY NVCEM SS + SPSR S
Sbjct: 23 NKDNSSAPRSPVASKSMHS-EALEMHVEGSGAGEPSIEQLYNNVCEMESSSEGGSPSRES 81
Query: 84 FYSYGEESRIDSELGHLVGGILEITKEVVTENKEESNGNAAEKDIVSCGKEATKKDNNQS 143
F S GEESRIDSEL HLV G +E K V+ E + E +GNAA + K ++ S
Sbjct: 82 FGSDGEESRIDSELRHLVAGEMEAMK-VIEEEEGEGSGNAANAVTAAENGTPVKAMSSNS 140
Query: 144 HSPSSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIA 203
SS+ S + S + ST++ S+ GS R+ S E+
Sbjct: 141 SKKSSKKAAKSQLESESSVGPNGKASTEEGEAEVSKPGSRVGRRRKASPNPHNGTED--- 197
Query: 204 TGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLA 263
GL+NPDLGPFLLK RD+I+S +NP++AL ALRA KSFE CA GKPSL LVM LHV+A
Sbjct: 198 AGLNNPDLGPFLLKHARDLIAS-DNPRRALKYALRATKSFEKCAGGKPSLNLVMSLHVVA 256
Query: 264 TIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYT 323
IYCNLG+Y+EA+P+L+RS++IPV+E+GQ+HALAKF+GCMQLGDTY M+G S+ +Y
Sbjct: 257 AIYCNLGKYDEAVPVLQRSLEIPVIEEGQEHALAKFSGCMQLGDTYGMLGQTALSLQWYA 316
Query: 324 AGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVE 383
AGLEIQ Q LGE DPR GETCRY+AEAHVQALQ DEA+RLCQ AL+IHR NG AS+E
Sbjct: 317 AGLEIQKQTLGEQDPRVGETCRYLAEAHVQALQLDEAQRLCQKALDIHRENGE--PASLE 374
Query: 384 ESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDE 443
E+ADRRLMGLI D+KGD+EAALEH V+ASMAM ANGQE +VASVD SIGD YL+L RYDE
Sbjct: 375 ETADRRLMGLICDTKGDHEAALEHLVMASMAMVANGQETEVASVDCSIGDIYLSLGRYDE 434
Query: 444 AVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPG 503
AVFSYQKALTVFK++KGENH VASV+VRLADLYNK GK ++SKSYCENAL+I+ K PG
Sbjct: 435 AVFSYQKALTVFKTSKGENHATVASVFVRLADLYNKTGKLRESKSYCENALKIYQKPIPG 494
Query: 504 IPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYM 563
EEIA GL DV+AIY++MN+ E+ LKLL+KALK+Y ++ GQQSTIAGIEAQMGV++Y+
Sbjct: 495 TSLEEIATGLTDVSAIYETMNEHEQALKLLQKALKMYNNSAGQQSTIAGIEAQMGVLHYI 554
Query: 564 LGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTIL 623
LGNY ++Y+ FKSA+AK R GEKK+A FG+ALNQMGLACVQRY+INEAA+LFEEAR +L
Sbjct: 555 LGNYGEAYDSFKSAIAKLRTCGEKKTAFFGVALNQMGLACVQRYSINEAAELFEEARAVL 614
Query: 624 EKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRL 683
E+EYG YHPDTLGVYSNLAGTYDAMGR+D+AIEILE VVGMREEKLGTANPDVDDEKRRL
Sbjct: 615 EQEYGPYHPDTLGVYSNLAGTYDAMGRLDEAIEILEHVVGMREEKLGTANPDVDDEKRRL 674
Query: 684 AELLKEAGRGRNRKSKRSLETLLDAN 709
AELLKEAGRGR+RK+K SLE LL+ N
Sbjct: 675 AELLKEAGRGRSRKAK-SLENLLETN 699
>gb|AAM13438.1| similar to A. thaliana protein BAB01483 similar to kinesin light
chain [Hordeum vulgare subsp. vulgare]
Length = 708
Score = 801 bits (2069), Expect = 0.0
Identities = 424/683 (62%), Positives = 519/683 (75%), Gaps = 12/683 (1%)
Query: 30 NQQASPRSTLSPRSIQSSDSIDLAIDGVVDTSIEQLYYNVCEMRSSDQ---SPSRASFYS 86
N++ T++P + + + G + SIEQLY NVCEM SS + SPSR SF S
Sbjct: 23 NKENLTAPTMAPSMQSEALEMHVEDSGAGEPSIEQLYNNVCEMESSSEGGGSPSRESFGS 82
Query: 87 YGEESRIDSELGHLVGGILEITKEVVTENKEESNGNAAEKDIVSCGKEATKKDNNQSHSP 146
GEESRIDSEL HLV G +E K V+ E +E+ G+ + + T S+S
Sbjct: 83 DGEESRIDSELRHLVAGEMEAMK-VIEEEEEKEKGSGSVTNAAPTAGNGTPARPQSSNSS 141
Query: 147 SSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIATGL 206
+ + +S S + S ++ S+ GS R+ + + E+ GL
Sbjct: 142 KKKAAKSQLESDA-SVGPNGKASPEEGESEVSKPGSRVGRRRKANAKSQNGTED---AGL 197
Query: 207 DNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLATIY 266
DNPDLGPFLLK RD+I+S +NP++AL ALRA KSFE CA GKPSL LVM LHV+A I+
Sbjct: 198 DNPDLGPFLLKHARDLIAS-DNPRRALKYALRATKSFEKCAGGKPSLNLVMSLHVVAAIH 256
Query: 267 CNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGL 326
CN+G+Y EA+P+L+RS++IPV E+GQ+HALAKF+GCMQLGDTY M+G I S+ +Y GL
Sbjct: 257 CNMGKYEEAVPVLQRSLEIPVTEEGQEHALAKFSGCMQLGDTYGMLGQIALSLQWYAKGL 316
Query: 327 EIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESA 386
+IQ Q LGE DPR GETCRY+AEAHVQALQ DEA++LCQMAL+IHR NG AS+EE+A
Sbjct: 317 DIQKQTLGEQDPRVGETCRYLAEAHVQALQLDEAQKLCQMALDIHRDNGQ--PASLEETA 374
Query: 387 DRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVF 446
DRRLMGLI D+KGD+EAALEH V+ASMAM ANGQE +VASVD SIGD YL+L RYDEAV
Sbjct: 375 DRRLMGLICDTKGDHEAALEHLVMASMAMVANGQETEVASVDCSIGDIYLSLGRYDEAVC 434
Query: 447 SYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPP 506
+YQKALTVFK++KGENH VASV++RLADLYNK GK ++SKSYCENAL+I+ K PG
Sbjct: 435 AYQKALTVFKTSKGENHATVASVFLRLADLYNKTGKLRESKSYCENALKIYQKPIPGTSL 494
Query: 507 EEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGN 566
EEIA GL DV+AIY++MN+ ++ LKLL+KALK+Y ++ GQQSTIAGIEAQ+GV++Y+ GN
Sbjct: 495 EEIATGLTDVSAIYETMNEHDQALKLLQKALKMYNNSAGQQSTIAGIEAQIGVLHYISGN 554
Query: 567 YSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKE 626
Y D+Y+ FKSA+ K R GEKKSA FGIALNQMGLACVQRY+INEAA+LFEEART+LE+E
Sbjct: 555 YGDAYDSFKSAITKLRTCGEKKSAFFGIALNQMGLACVQRYSINEAAELFEEARTVLEQE 614
Query: 627 YGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAEL 686
YG YH DTLGVYSNLAGTYDAMGR+D+AIEILE+VV MREEKLGTANPDVDDEKRRLAEL
Sbjct: 615 YGPYHADTLGVYSNLAGTYDAMGRLDEAIEILEYVVEMREEKLGTANPDVDDEKRRLAEL 674
Query: 687 LKEAGRGRNRKSKRSLETLLDAN 709
LKEAGRGR+RK+K SLE LL N
Sbjct: 675 LKEAGRGRSRKAK-SLENLLGTN 696
>ref|NP_174070.1| kinesin light chain-related [Arabidopsis thaliana]
Length = 650
Score = 691 bits (1784), Expect = 0.0
Identities = 370/658 (56%), Positives = 485/658 (73%), Gaps = 32/658 (4%)
Query: 55 DGVVDTSIEQLYYNVCEMRSSDQSPSRASFYSYGEESRIDSELGHLVGGILEITKEVVTE 114
D + DT+IE+L N+CE++SS+QSPSR SF SYG+ES+IDS+L HL G + + E
Sbjct: 14 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILEDE 73
Query: 115 NKEESNGNAAEKDIVSCGKEATKKDNNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRS 174
E+ E D+ S + S+ +E + K T K++
Sbjct: 74 GDEDEVAKPEEFDVKS--------------NSSNLDLEVMPRDMEKQ--------TGKKN 111
Query: 175 ERSSRKGSLYPMRKHR--SLALRGIIEERIATGLDNPDLGPFLLKQTRDMISSGENPQKA 232
S G + MRK + L+ EE + +N +L FLL Q R+++SSG++ KA
Sbjct: 112 VTKSNVG-VGGMRKKKVGGTKLQNGNEEPSS---ENVELARFLLNQARNLVSSGDSTHKA 167
Query: 233 LDLALRALKSFEMCAD-GKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDG 291
L+L RA K FE A+ GKP LE +MCLHV A ++C L +YNEAIP+L+RS++IPV+E+G
Sbjct: 168 LELTHRAAKLFEASAENGKPCLEWIMCLHVTAAVHCKLKEYNEAIPVLQRSVEIPVVEEG 227
Query: 292 QDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAH 351
++HALAKFAG MQLGDTYAM+G +E+SI YT GL IQ +VLGE DPR GETCRY+AEA
Sbjct: 228 EEHALAKFAGLMQLGDTYAMVGQLESSISCYTEGLNIQKKVLGENDPRVGETCRYLAEAL 287
Query: 352 VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA 411
VQAL+FDEA+++C+ AL IHR +G S+ E+ADRRLMGLI ++KGD+E ALEH VLA
Sbjct: 288 VQALRFDEAQQVCETALSIHRESG--LPGSIAEAADRRLMGLICETKGDHENALEHLVLA 345
Query: 412 SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYV 471
SMAM+ANGQE +VA VD+SIGD+YL+L+R+DEA+ +YQK+LT K+ KGENHP V SVY+
Sbjct: 346 SMAMAANGQESEVAFVDTSIGDSYLSLSRFDEAICAYQKSLTALKTAKGENHPAVGSVYI 405
Query: 472 RLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLK 531
RLADLYN+ GK +++KSYCENALRI+ I PEEIA+GL D++ I +SMN++E+ +
Sbjct: 406 RLADLYNRTGKVREAKSYCENALRIYESHNLEISPEEIASGLTDISVICESMNEVEQAIT 465
Query: 532 LLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSAL 591
LL+KALKIY +PGQ+ IAGIEAQMGV+YYM+G Y +SYN FKSA++K RA+G+K+S
Sbjct: 466 LLQKALKIYADSPGQKIMIAGIEAQMGVLYYMMGKYMESYNTFKSAISKLRATGKKQSTF 525
Query: 592 FGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRV 651
FGIALNQMGLAC+Q AI EA +LFEEA+ ILE+E G YHP+TLG+YSNLAG YDA+GR+
Sbjct: 526 FGIALNQMGLACIQLDAIEEAVELFEEAKCILEQECGPYHPETLGLYSNLAGAYDAIGRL 585
Query: 652 DDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDAN 709
DDAI++L VVG+REEKLGTANP +DEKRRLA+LLKEAG RK+K SL+TL+D++
Sbjct: 586 DDAIKLLGHVVGVREEKLGTANPVTEDEKRRLAQLLKEAGNVTGRKAK-SLKTLIDSD 642
>gb|AAF99740.1| F17L21.29 [Arabidopsis thaliana]
Length = 684
Score = 691 bits (1784), Expect = 0.0
Identities = 370/658 (56%), Positives = 485/658 (73%), Gaps = 32/658 (4%)
Query: 55 DGVVDTSIEQLYYNVCEMRSSDQSPSRASFYSYGEESRIDSELGHLVGGILEITKEVVTE 114
D + DT+IE+L N+CE++SS+QSPSR SF SYG+ES+IDS+L HL G + + E
Sbjct: 48 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILEDE 107
Query: 115 NKEESNGNAAEKDIVSCGKEATKKDNNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRS 174
E+ E D+ S + S+ +E + K T K++
Sbjct: 108 GDEDEVAKPEEFDVKS--------------NSSNLDLEVMPRDMEKQ--------TGKKN 145
Query: 175 ERSSRKGSLYPMRKHR--SLALRGIIEERIATGLDNPDLGPFLLKQTRDMISSGENPQKA 232
S G + MRK + L+ EE + +N +L FLL Q R+++SSG++ KA
Sbjct: 146 VTKSNVG-VGGMRKKKVGGTKLQNGNEEPSS---ENVELARFLLNQARNLVSSGDSTHKA 201
Query: 233 LDLALRALKSFEMCAD-GKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDG 291
L+L RA K FE A+ GKP LE +MCLHV A ++C L +YNEAIP+L+RS++IPV+E+G
Sbjct: 202 LELTHRAAKLFEASAENGKPCLEWIMCLHVTAAVHCKLKEYNEAIPVLQRSVEIPVVEEG 261
Query: 292 QDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAH 351
++HALAKFAG MQLGDTYAM+G +E+SI YT GL IQ +VLGE DPR GETCRY+AEA
Sbjct: 262 EEHALAKFAGLMQLGDTYAMVGQLESSISCYTEGLNIQKKVLGENDPRVGETCRYLAEAL 321
Query: 352 VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA 411
VQAL+FDEA+++C+ AL IHR +G S+ E+ADRRLMGLI ++KGD+E ALEH VLA
Sbjct: 322 VQALRFDEAQQVCETALSIHRESG--LPGSIAEAADRRLMGLICETKGDHENALEHLVLA 379
Query: 412 SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYV 471
SMAM+ANGQE +VA VD+SIGD+YL+L+R+DEA+ +YQK+LT K+ KGENHP V SVY+
Sbjct: 380 SMAMAANGQESEVAFVDTSIGDSYLSLSRFDEAICAYQKSLTALKTAKGENHPAVGSVYI 439
Query: 472 RLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLK 531
RLADLYN+ GK +++KSYCENALRI+ I PEEIA+GL D++ I +SMN++E+ +
Sbjct: 440 RLADLYNRTGKVREAKSYCENALRIYESHNLEISPEEIASGLTDISVICESMNEVEQAIT 499
Query: 532 LLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSAL 591
LL+KALKIY +PGQ+ IAGIEAQMGV+YYM+G Y +SYN FKSA++K RA+G+K+S
Sbjct: 500 LLQKALKIYADSPGQKIMIAGIEAQMGVLYYMMGKYMESYNTFKSAISKLRATGKKQSTF 559
Query: 592 FGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRV 651
FGIALNQMGLAC+Q AI EA +LFEEA+ ILE+E G YHP+TLG+YSNLAG YDA+GR+
Sbjct: 560 FGIALNQMGLACIQLDAIEEAVELFEEAKCILEQECGPYHPETLGLYSNLAGAYDAIGRL 619
Query: 652 DDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDAN 709
DDAI++L VVG+REEKLGTANP +DEKRRLA+LLKEAG RK+K SL+TL+D++
Sbjct: 620 DDAIKLLGHVVGVREEKLGTANPVTEDEKRRLAQLLKEAGNVTGRKAK-SLKTLIDSD 676
>dbj|BAC42575.1| unknown protein [Arabidopsis thaliana] gi|29028990|gb|AAO64874.1|
At1g27500 [Arabidopsis thaliana]
Length = 650
Score = 691 bits (1783), Expect = 0.0
Identities = 368/668 (55%), Positives = 484/668 (72%), Gaps = 52/668 (7%)
Query: 55 DGVVDTSIEQLYYNVCEMRSSDQSPSRASFYSYGEESRIDSELGHLVGG------ILEI- 107
D + DT+IE+L N+CE++SS+QSPSR SF SYG+ES+IDS+L HL G ILE
Sbjct: 14 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILEDG 73
Query: 108 --TKEVVTENKEESNGNAAEKDIVSCGKEATKKDNNQSHSPSSRIIEGSAKST---PKSR 162
EV + + N++ D+ ++ K+ ++ + S+ + G K K +
Sbjct: 74 GDEDEVAKPEEFDVKSNSSNLDLEVMPRDMEKQTGKKNVTKSNVGVGGMRKKKVGGTKLQ 133
Query: 163 NLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIATGLDNPDLGPFLLKQTRDM 222
N E PS++ N +L FLL Q R++
Sbjct: 134 NGNEEPSSE------------------------------------NVELARFLLNQARNL 157
Query: 223 ISSGENPQKALDLALRALKSFEMCAD-GKPSLELVMCLHVLATIYCNLGQYNEAIPILER 281
+SSG++ KAL+L RA K FE A+ GKP LE +MCLHV A ++C L +YNEAIP+L+R
Sbjct: 158 VSSGDSTHKALELTHRAAKLFEASAENGKPCLEWIMCLHVTAAVHCKLKEYNEAIPVLQR 217
Query: 282 SIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFG 341
S++IPV+E+G++HALAKFAG MQLGDTYAM+G +E+SI YT GL IQ +VLGE DPR G
Sbjct: 218 SVEIPVVEEGEEHALAKFAGLMQLGDTYAMVGQLESSISCYTEGLNIQKKVLGENDPRVG 277
Query: 342 ETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDY 401
ETCRY+AEA VQAL+FDEA+++C+ AL IHR +G S+ E+ADRRLMGLI ++KGD+
Sbjct: 278 ETCRYLAEALVQALRFDEAQQVCETALSIHRESG--LPGSIAEAADRRLMGLICETKGDH 335
Query: 402 EAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGE 461
E ALEH VLASMAM+ANGQE +VA VD+SIGD+YL+L+R+DEA+ +YQK+LT K+ KGE
Sbjct: 336 ENALEHLVLASMAMAANGQESEVAFVDTSIGDSYLSLSRFDEAICAYQKSLTALKTAKGE 395
Query: 462 NHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQ 521
NHP V SVY+RLADLYN+ GK +++KSYCENALRI+ I PEEIA+GL D++ I +
Sbjct: 396 NHPAVGSVYIRLADLYNRTGKVREAKSYCENALRIYESHNLEISPEEIASGLTDISVICE 455
Query: 522 SMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKF 581
SMN++E+ + LL+KALKIY +PGQ+ IAGIEAQMGV+YYM+G Y +SYN FKSA++K
Sbjct: 456 SMNEVEQAITLLQKALKIYADSPGQKIMIAGIEAQMGVLYYMMGKYMESYNTFKSAISKL 515
Query: 582 RASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNL 641
RA+G+K+S FGIALNQMGLAC+Q AI EA +LFEEA+ ILE+E G YHP+TLG+YSNL
Sbjct: 516 RATGKKQSTFFGIALNQMGLACIQLDAIEEAVELFEEAKCILEQECGPYHPETLGLYSNL 575
Query: 642 AGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRS 701
AG YDA+GR+DDAI++L VVG+REEKLGTANP +DEKRRLA+LLKEAG RK+K S
Sbjct: 576 AGAYDAIGRLDDAIKLLGHVVGVREEKLGTANPVTEDEKRRLAQLLKEAGNVTGRKAK-S 634
Query: 702 LETLLDAN 709
L+TL+D++
Sbjct: 635 LKTLIDSD 642
>gb|AAN41387.1| unknown protein [Arabidopsis thaliana] gi|7267782|emb|CAB81185.1|
putative protein [Arabidopsis thaliana]
gi|4539358|emb|CAB40052.1| putative protein [Arabidopsis
thaliana] gi|18176148|gb|AAL59992.1| unknown protein
[Arabidopsis thaliana] gi|19699309|gb|AAL91265.1|
AT4g10840/F25I24_50 [Arabidopsis thaliana]
gi|3513727|gb|AAC33943.1| contains similarity to TPR
domains (Pfam: TPR.hmm: score: 11.15) and kinesin motor
domains (Pfam: kinesin2.hmm, score: 17.49, 20.52 and
10.94) [Arabidopsis thaliana]
gi|15236944|ref|NP_192822.1| kinesin light chain-related
[Arabidopsis thaliana] gi|7486710|pir||T01892
hypothetical protein F8M12.21 - Arabidopsis thaliana
Length = 609
Score = 645 bits (1664), Expect = 0.0
Identities = 341/573 (59%), Positives = 429/573 (74%), Gaps = 24/573 (4%)
Query: 156 KSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIATGLDNPDLGPFL 215
K TP S R +PS ++ + + P + A+ + + LDNPDLGPFL
Sbjct: 38 KKTPSSTPSRSKPSPNRSTGKKDS-----PTVSSSTAAVIDVDDP----SLDNPDLGPFL 88
Query: 216 LKQTRDMISSGENPQKALDLALRALKSFEMC-----------ADGKPSLELVMCLHVLAT 264
LK RD I+SGE P KALD A+RA KSFE C +DG P L+L M LHVLA
Sbjct: 89 LKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGPVLDLAMSLHVLAA 148
Query: 265 IYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTA 324
IYC+LG+++EA+P LER+I +P G DH+LA F+G MQLGDT +M+G I+ SI Y
Sbjct: 149 IYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSMLGQIDRSIACYEE 208
Query: 325 GLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEE 384
GL+IQ Q LG+TDPR GETCRY+AEA+VQA+QF++AE LC+ LEIHR + AS+EE
Sbjct: 209 GLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIHRAHSE--PASLEE 266
Query: 385 SADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEA 444
+ADRRLM +I ++KGDYE ALEH VLASMAM A+GQE +VAS+D SIG+ Y++L R+DEA
Sbjct: 267 AADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGNIYMSLCRFDEA 326
Query: 445 VFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGI 504
VFSYQKALTVFK++KGE HP VASV+VRLA+LY++ GK ++SKSYCENALRI+ K PG
Sbjct: 327 VFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENALRIYNKPVPGT 386
Query: 505 PPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYML 564
EEIA GL +++AIY+S+++ E+ LKLL+K++K+ PGQQS IAG+EA+MGVMYY +
Sbjct: 387 TVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGLEARMGVMYYTV 446
Query: 565 GNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILE 624
G Y D+ N F+SAV K RA+GE KSA FG+ LNQMGLACVQ + I+EA +LFEEAR ILE
Sbjct: 447 GRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAGELFEEARGILE 505
Query: 625 KEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLA 684
+E G DTLGVYSNLA TYDAMGR++DAIEILE V+ +REEKLGTANPD +DEK+RLA
Sbjct: 506 QERGPCDQDTLGVYSNLAATYDAMGRIEDAIEILEQVLKLREEKLGTANPDFEDEKKRLA 565
Query: 685 ELLKEAGRGRNRKSKRSLETLLDANSRLIKNNS 717
ELLKEAGR RN K+K SL+ L+D N+R K S
Sbjct: 566 ELLKEAGRSRNYKAK-SLQNLIDPNARPPKKES 597
>gb|AAM63491.1| putative kinesin light chain [Arabidopsis thaliana]
Length = 606
Score = 645 bits (1664), Expect = 0.0
Identities = 341/573 (59%), Positives = 429/573 (74%), Gaps = 24/573 (4%)
Query: 156 KSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIATGLDNPDLGPFL 215
K TP S R +PS ++ + + P + A+ + + LDNPDLGPFL
Sbjct: 35 KKTPSSTPSRSKPSPNRSTGKKDS-----PTVSSSTAAVIDVDDP----SLDNPDLGPFL 85
Query: 216 LKQTRDMISSGENPQKALDLALRALKSFEMC-----------ADGKPSLELVMCLHVLAT 264
LK RD I+SGE P KALD A+RA KSFE C +DG P L+L M LHVLA
Sbjct: 86 LKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGPVLDLAMSLHVLAA 145
Query: 265 IYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTA 324
IYC+LG+++EA+P LER+I +P G DH+LA F+G MQLGDT +M+G I+ SI Y
Sbjct: 146 IYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSMLGQIDRSIACYEE 205
Query: 325 GLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEE 384
GL+IQ Q LG+TDPR GETCRY+AEA+VQA+QF++AE LC+ LEIHR + AS+EE
Sbjct: 206 GLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIHRAHSE--PASLEE 263
Query: 385 SADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEA 444
+ADRRLM +I ++KGDYE ALEH VLASMAM A+GQE +VAS+D SIG+ Y++L R+DEA
Sbjct: 264 AADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGNIYMSLCRFDEA 323
Query: 445 VFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGI 504
VFSYQKALTVFK++KGE HP VASV+VRLA+LY++ GK ++SKSYCENALRI+ K PG
Sbjct: 324 VFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENALRIYNKPVPGT 383
Query: 505 PPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYML 564
EEIA GL +++AIY+S+++ E+ LKLL+K++K+ PGQQS IAG+EA+MGVMYY +
Sbjct: 384 TVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGLEARMGVMYYTV 443
Query: 565 GNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILE 624
G Y D+ N F+SAV K RA+GE KSA FG+ LNQMGLACVQ + I+EA +LFEEAR ILE
Sbjct: 444 GRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAGELFEEARGILE 502
Query: 625 KEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLA 684
+E G DTLGVYSNLA TYDAMGR++DAIEILE V+ +REEKLGTANPD +DEK+RLA
Sbjct: 503 QERGPCDQDTLGVYSNLAATYDAMGRIEDAIEILEQVLKLREEKLGTANPDFEDEKKRLA 562
Query: 685 ELLKEAGRGRNRKSKRSLETLLDANSRLIKNNS 717
ELLKEAGR RN K+K SL+ L+D N+R K S
Sbjct: 563 ELLKEAGRSRNYKAK-SLQNLIDPNARPPKKES 594
>ref|XP_463817.1| kinesin light chain-like protein [Oryza sativa (japonica
cultivar-group)] gi|41052919|dbj|BAD07830.1| kinesin
light chain-like protein [Oryza sativa (japonica
cultivar-group)] gi|50252012|dbj|BAD27945.1| kinesin
light chain-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 599
Score = 629 bits (1621), Expect = e-178
Identities = 326/535 (60%), Positives = 412/535 (76%), Gaps = 27/535 (5%)
Query: 206 LDNPDLGPFLLKQTRDMISSGEN--PQKALDLALRALKSFEMCADGKPSLELVMCLHVLA 263
LDNPDLGPFLLKQ RD + SGE +AL+ A RA ++ E +G LEL M LHV A
Sbjct: 58 LDNPDLGPFLLKQARDAMVSGEGGGAARALEFAERAARALERRGEGA-ELELAMSLHVAA 116
Query: 264 TIYCNLGQYNEAIPILERSIDIPV---------------------LEDGQDHALAKFAGC 302
I+C LG++ +AIP+LER++ + + G++ ALA F+G
Sbjct: 117 AIHCGLGRHADAIPVLERAVAVVTPPPPPPPPEGESSAEEQPPEDQQKGEEWALAAFSGW 176
Query: 303 MQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAER 362
MQLGDT+AM+G ++ SI Y GLEIQ LGE DPR ETCRY+AEAHVQALQFDEAE+
Sbjct: 177 MQLGDTHAMLGRMDESIACYGKGLEIQMAALGERDPRVAETCRYLAEAHVQALQFDEAEK 236
Query: 363 LCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQ 422
LC+ ALEIHR + + AS+EE++DRRLM LI D+KGDY+ ALEH VLASM M ANG++
Sbjct: 237 LCRKALEIHREHS--APASLEEASDRRLMALILDAKGDYDGALEHLVLASMTMVANGRDI 294
Query: 423 DVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGK 482
+VA++D +IG+ YLALAR+DEAVFSYQKALTV KS +G++HP+VASV+VRLADLY++ G+
Sbjct: 295 EVATIDVAIGNTYLALARFDEAVFSYQKALTVLKSARGDDHPSVASVFVRLADLYHRTGR 354
Query: 483 FKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGH 542
++SKSYCENALR++ K PG P+E+A GL+++AAIY+++ DL++ LKLL++ALK+
Sbjct: 355 LRESKSYCENALRVYAKPAPGAAPDEVAGGLMEIAAIYEALGDLDEALKLLQRALKLLED 414
Query: 543 APGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLA 602
+PGQ ST+AGIEAQMGV+YYM+G Y+DS N F++AVAK RASGE+KSA FG+ LNQMGLA
Sbjct: 415 SPGQWSTVAGIEAQMGVLYYMVGRYADSRNSFENAVAKLRASGERKSAFFGVLLNQMGLA 474
Query: 603 CVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVV 662
CVQ + I+EAA LFEEAR +LE+E G HPDTLGVYSNLA YDAMGRV+DAIEILE V+
Sbjct: 475 CVQLFKIDEAAQLFEEARAVLEQECGASHPDTLGVYSNLAAIYDAMGRVEDAIEILEHVL 534
Query: 663 GMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRLIKNNS 717
+REEKLGTANPDV+DEK RLAELLKEAGR RNRK K SLE L NS+ +K ++
Sbjct: 535 KVREEKLGTANPDVEDEKLRLAELLKEAGRSRNRKQK-SLENLFVTNSQRVKKDA 588
>ref|NP_974530.1| kinesin light chain-related [Arabidopsis thaliana]
Length = 531
Score = 564 bits (1453), Expect = e-159
Identities = 296/506 (58%), Positives = 374/506 (73%), Gaps = 23/506 (4%)
Query: 156 KSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIATGLDNPDLGPFL 215
K TP S R +PS ++ + + P + A+ + + LDNPDLGPFL
Sbjct: 38 KKTPSSTPSRSKPSPNRSTGKKDS-----PTVSSSTAAVIDVDDP----SLDNPDLGPFL 88
Query: 216 LKQTRDMISSGENPQKALDLALRALKSFEMC-----------ADGKPSLELVMCLHVLAT 264
LK RD I+SGE P KALD A+RA KSFE C +DG P L+L M LHVLA
Sbjct: 89 LKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGPVLDLAMSLHVLAA 148
Query: 265 IYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTA 324
IYC+LG+++EA+P LER+I +P G DH+LA F+G MQLGDT +M+G I+ SI Y
Sbjct: 149 IYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSMLGQIDRSIACYEE 208
Query: 325 GLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEE 384
GL+IQ Q LG+TDPR GETCRY+AEA+VQA+QF++AE LC+ LEIHR + AS+EE
Sbjct: 209 GLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIHRAHSE--PASLEE 266
Query: 385 SADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEA 444
+ADRRLM +I ++KGDYE ALEH VLASMAM A+GQE +VAS+D SIG+ Y++L R+DEA
Sbjct: 267 AADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGNIYMSLCRFDEA 326
Query: 445 VFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGI 504
VFSYQKALTVFK++KGE HP VASV+VRLA+LY++ GK ++SKSYCENALRI+ K PG
Sbjct: 327 VFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENALRIYNKPVPGT 386
Query: 505 PPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYML 564
EEIA GL +++AIY+S+++ E+ LKLL+K++K+ PGQQS IAG+EA+MGVMYY +
Sbjct: 387 TVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGLEARMGVMYYTV 446
Query: 565 GNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILE 624
G Y D+ N F+SAV K RA+GE KSA FG+ LNQMGLACVQ + I+EA +LFEEAR ILE
Sbjct: 447 GRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAGELFEEARGILE 505
Query: 625 KEYGQYHPDTLGVYSNLAGTYDAMGR 650
+E G DTLGVYSNLA TYDAMGR
Sbjct: 506 QERGPCDQDTLGVYSNLAATYDAMGR 531
>dbj|BAC78579.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 309
Score = 405 bits (1041), Expect = e-111
Identities = 199/299 (66%), Positives = 253/299 (84%), Gaps = 1/299 (0%)
Query: 419 GQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYN 478
G++ +VA++D +IG+ YLALAR+DEAVFSYQKALTV KS +G++HP+VASV+VRLADLY+
Sbjct: 1 GRDIEVATIDVAIGNTYLALARFDEAVFSYQKALTVLKSARGDDHPSVASVFVRLADLYH 60
Query: 479 KIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALK 538
+ G+ ++SKSYCENALR++ K PG P+E+A GL+++AAIY+++ DL++ LKLL++ALK
Sbjct: 61 RTGRLRESKSYCENALRVYAKPAPGAAPDEVAGGLMEIAAIYEALGDLDEALKLLQRALK 120
Query: 539 IYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQ 598
+ +PGQ ST+AGIEAQMGV+YYM+G Y+DS N F++AVAK RASGE+KSA FG+ LNQ
Sbjct: 121 LLEDSPGQWSTVAGIEAQMGVLYYMVGRYADSRNSFENAVAKLRASGERKSAFFGVLLNQ 180
Query: 599 MGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEIL 658
MGLACVQ + I+EAA LFEEAR +LE+E G HPDTLGVYSNLA YDAMGRV+DAIEIL
Sbjct: 181 MGLACVQLFKIDEAAQLFEEARAVLEQECGASHPDTLGVYSNLAAIYDAMGRVEDAIEIL 240
Query: 659 EFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRLIKNNS 717
E V+ +REEKLGTANPDV+DEK RLAELLKEAGR RNRK K SLE L NS+ +K ++
Sbjct: 241 EHVLKVREEKLGTANPDVEDEKLRLAELLKEAGRSRNRKQK-SLENLFVTNSQRVKKDA 298
Score = 46.6 bits (109), Expect = 0.003
Identities = 66/288 (22%), Positives = 109/288 (36%), Gaps = 51/288 (17%)
Query: 203 ATGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVL 262
A G D+P + ++ D+ +++ AL+ + A G E+ L +
Sbjct: 40 ARGDDHPSVASVFVRLA-DLYHRTGRLRESKSYCENALRVYAKPAPGAAPDEVAGGLMEI 98
Query: 263 ATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNI---ENSI 319
A IY LG +EA+ +L+R++ + GQ +A Q+G Y M+G NS
Sbjct: 99 AAIYEALGDLDEALKLLQRALKLLEDSPGQWSTVAGIEA--QMGVLYYMVGRYADSRNSF 156
Query: 320 LFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSS 379
A L G+ FG + A VQ + DEA +L + A
Sbjct: 157 ENAVAKLRASGE---RKSAFFGVLLNQMGLACVQLFKIDEAAQLFEEA------------ 201
Query: 380 ASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALA 439
A LE AS D V S++ Y A+
Sbjct: 202 ----------------------RAVLEQECGAS--------HPDTLGVYSNLAAIYDAMG 231
Query: 440 RYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSK 487
R ++A+ + L V + G +P+V +RLA+L + G+ ++ K
Sbjct: 232 RVEDAIEILEHVLKVREEKLGTANPDVEDEKLRLAELLKEAGRSRNRK 279
>gb|AAT07454.1| kinesin-like protein [Mirabilis jalapa]
Length = 194
Score = 297 bits (761), Expect = 7e-79
Identities = 148/196 (75%), Positives = 176/196 (89%), Gaps = 2/196 (1%)
Query: 365 QMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDV 424
QMALEIHR NG S SVEE+ADRRLMGLIY++KGD+E ALEH VLASMAM ANGQE++V
Sbjct: 1 QMALEIHRENG--SPGSVEETADRRLMGLIYETKGDHEKALEHLVLASMAMVANGQEKEV 58
Query: 425 ASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFK 484
ASVD SIGD YL+L+RYDEAVF+Y+KALT FK++KGENHP++ASVYVRLADLYNK GK +
Sbjct: 59 ASVDCSIGDTYLSLSRYDEAVFAYEKALTSFKASKGENHPSIASVYVRLADLYNKTGKLR 118
Query: 485 DSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAP 544
+SKSYCENALRI+ K PGIPPE+IA+G DV+AIY+SMN+LE+ +KLL+KALKIY +AP
Sbjct: 119 ESKSYCENALRIYSKPVPGIPPEDIASGFTDVSAIYESMNELEQAIKLLQKALKIYDNAP 178
Query: 545 GQQSTIAGIEAQMGVM 560
GQQ+TIAGI+AQMGVM
Sbjct: 179 GQQNTIAGIQAQMGVM 194
Score = 46.6 bits (109), Expect = 0.003
Identities = 37/157 (23%), Positives = 74/157 (46%), Gaps = 4/157 (2%)
Query: 519 IYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAV 578
IY++ D EK L+ L A + A GQ+ +A ++ +G Y L Y ++ ++ A+
Sbjct: 28 IYETKGDHEKALEHLVLA-SMAMVANGQEKEVASVDCSIGDTYLSLSRYDEAVFAYEKAL 86
Query: 579 AKFRAS-GEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHP-DTLG 636
F+AS GE ++ + + L + + E+ E A I K P D
Sbjct: 87 TSFKASKGENHPSIASVYVRLADL-YNKTGKLRESKSYCENALRIYSKPVPGIPPEDIAS 145
Query: 637 VYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTAN 673
+++++ Y++M ++ AI++L+ + + + G N
Sbjct: 146 GFTDVSAIYESMNELEQAIKLLQKALKIYDNAPGQQN 182
Score = 38.5 bits (88), Expect = 0.78
Identities = 40/187 (21%), Positives = 83/187 (43%), Gaps = 6/187 (3%)
Query: 248 DGKP-SLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLG 306
+G P S+E ++ IY G + +A+ L + + ++ +GQ+ +A C +G
Sbjct: 10 NGSPGSVEETADRRLMGLIYETKGDHEKALEHLVLA-SMAMVANGQEKEVAS-VDC-SIG 66
Query: 307 DTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQM 366
DTY + + ++ Y L GE P +A+ + + + E++ C+
Sbjct: 67 DTYLSLSRYDEAVFAYEKALTSFKASKGENHPSIASVYVRLADLYNKTGKLRESKSYCEN 126
Query: 367 ALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQDVA 425
AL I+ G + ++ + IY+S + E A++ A + +A GQ+ +A
Sbjct: 127 ALRIYSKPVPGIPPE-DIASGFTDVSAIYESMNELEQAIKLLQKALKIYDNAPGQQNTIA 185
Query: 426 SVDSSIG 432
+ + +G
Sbjct: 186 GIQAQMG 192
>gb|AAL36288.1| putative kinesin light chain [Arabidopsis thaliana]
gi|30684882|ref|NP_850163.1| tetratricopeptide repeat
(TPR)-containing protein [Arabidopsis thaliana]
Length = 617
Score = 170 bits (430), Expect = 2e-40
Identities = 136/491 (27%), Positives = 232/491 (46%), Gaps = 16/491 (3%)
Query: 210 DLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADG-KPSLELVMCLHVLATIYCN 268
+LG LK + GE+P+K L A +ALKSF+ DG KP+L + M ++ +
Sbjct: 116 ELGLSALKLGLHLDREGEDPEKVLSYADKALKSFD--GDGNKPNLLVAMASQLMGSANYG 173
Query: 269 LGQYNEAIPILERS--IDIPVLEDG----QDHALAKFAGCMQLGDTYAMMGNIENSILFY 322
L ++++++ L R+ I + + DG +D A ++L + MG E +I
Sbjct: 174 LKRFSDSLGYLNRANRILVKLEADGDCVVEDVRPVLHAVQLELANVKNAMGRREEAIENL 233
Query: 323 TAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASV 382
LEI+ E G R +A+A+V L F+EA ALEIH+ +SA V
Sbjct: 234 KKSLEIKEMTFDEDSKEMGVANRSLADAYVAVLNFNEALPYALKALEIHKKELGNNSAEV 293
Query: 383 EESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYD 442
+ DRRL+G+IY ++ ALE L+ + G + ++ + + +AL +Y+
Sbjct: 294 AQ--DRRLLGVIYSGLEQHDKALEQNRLSQRVLKNWGMKLELIRAEIDAANMKVALGKYE 351
Query: 443 EAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKP 502
EA+ + +V + T ++ A V++ ++ KF +SK E A I K +
Sbjct: 352 EAIDILK---SVVQQTDKDSEMR-AMVFISMSKALVNQQKFAESKRCLEFACEILEKKET 407
Query: 503 GIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYY 562
+P E +A +VA Y+SMN+ E + LL+K L I P +Q + + A++G +
Sbjct: 408 ALPVE-VAEAYSEVAMQYESMNEFETAISLLQKTLGILEKLPQEQHSEGSVSARIGWLLL 466
Query: 563 MLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTI 622
G S + +SA + + S K G N +G A ++ AA +F A+ I
Sbjct: 467 FSGRVSQAVPYLESAAERLKESFGAKHFGVGYVYNNLGAAYLELGRPQSAAQMFAVAKDI 526
Query: 623 LEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRR 682
++ G H D++ NL+ Y MG A+E + V+ + +A ++ + KR
Sbjct: 527 MDVSLGPNHVDSIDACQNLSKAYAGMGNYSLAVEFQQRVINAWDNHGDSAKDEMKEAKRL 586
Query: 683 LAELLKEAGRG 693
L +L +A G
Sbjct: 587 LEDLRLKARGG 597
>gb|AAD20669.1| putative kinesin light chain [Arabidopsis thaliana]
gi|25408142|pir||C84718 probable kinesin light chain
[imported] - Arabidopsis thaliana
Length = 510
Score = 170 bits (430), Expect = 2e-40
Identities = 136/491 (27%), Positives = 232/491 (46%), Gaps = 16/491 (3%)
Query: 210 DLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADG-KPSLELVMCLHVLATIYCN 268
+LG LK + GE+P+K L A +ALKSF+ DG KP+L + M ++ +
Sbjct: 9 ELGLSALKLGLHLDREGEDPEKVLSYADKALKSFD--GDGNKPNLLVAMASQLMGSANYG 66
Query: 269 LGQYNEAIPILERS--IDIPVLEDG----QDHALAKFAGCMQLGDTYAMMGNIENSILFY 322
L ++++++ L R+ I + + DG +D A ++L + MG E +I
Sbjct: 67 LKRFSDSLGYLNRANRILVKLEADGDCVVEDVRPVLHAVQLELANVKNAMGRREEAIENL 126
Query: 323 TAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASV 382
LEI+ E G R +A+A+V L F+EA ALEIH+ +SA V
Sbjct: 127 KKSLEIKEMTFDEDSKEMGVANRSLADAYVAVLNFNEALPYALKALEIHKKELGNNSAEV 186
Query: 383 EESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYD 442
+ DRRL+G+IY ++ ALE L+ + G + ++ + + +AL +Y+
Sbjct: 187 AQ--DRRLLGVIYSGLEQHDKALEQNRLSQRVLKNWGMKLELIRAEIDAANMKVALGKYE 244
Query: 443 EAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKP 502
EA+ + +V + T ++ A V++ ++ KF +SK E A I K +
Sbjct: 245 EAIDILK---SVVQQTDKDSEMR-AMVFISMSKALVNQQKFAESKRCLEFACEILEKKET 300
Query: 503 GIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYY 562
+P E +A +VA Y+SMN+ E + LL+K L I P +Q + + A++G +
Sbjct: 301 ALPVE-VAEAYSEVAMQYESMNEFETAISLLQKTLGILEKLPQEQHSEGSVSARIGWLLL 359
Query: 563 MLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTI 622
G S + +SA + + S K G N +G A ++ AA +F A+ I
Sbjct: 360 FSGRVSQAVPYLESAAERLKESFGAKHFGVGYVYNNLGAAYLELGRPQSAAQMFAVAKDI 419
Query: 623 LEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRR 682
++ G H D++ NL+ Y MG A+E + V+ + +A ++ + KR
Sbjct: 420 MDVSLGPNHVDSIDACQNLSKAYAGMGNYSLAVEFQQRVINAWDNHGDSAKDEMKEAKRL 479
Query: 683 LAELLKEAGRG 693
L +L +A G
Sbjct: 480 LEDLRLKARGG 490
>ref|XP_468434.1| kinesin light chain-like [Oryza sativa (japonica cultivar-group)]
gi|48716499|dbj|BAD23104.1| kinesin light chain-like
[Oryza sativa (japonica cultivar-group)]
gi|48716364|dbj|BAD22975.1| kinesin light chain-like
[Oryza sativa (japonica cultivar-group)]
Length = 621
Score = 152 bits (384), Expect = 4e-35
Identities = 137/516 (26%), Positives = 235/516 (44%), Gaps = 47/516 (9%)
Query: 224 SSGENPQKALDLALRALKSFEMCADGKPS--------LELVMCLHVLATIYCNLGQYNEA 275
S +P + L LALR+L E + S + L M LH+ + +L ++++A
Sbjct: 106 SGAADPSRVLALALRSLGILEATPNSATSTTASHSDAVSLAMALHLAGSASFDLSRFHDA 165
Query: 276 IPILERSIDI--PVLEDGQDHALAK--------------FAGCMQLGDTYAMMGNIENSI 319
+ L RS+ + P+L A A A +QL + +G E ++
Sbjct: 166 LSFLTRSLRLVSPLLPSSSSAAAASGDDDAQGFDVRPVAHAVRLQLANVKTALGRREEAL 225
Query: 320 LFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSS 379
A L+++ +L G R +AEA+ L F EA LC+ ALE+H+ +S
Sbjct: 226 ADMRACLDLKESILPPGSRELGAAYRDLAEAYSTVLDFKEALPLCEKALELHQSTLGKNS 285
Query: 380 ASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDV---ASVDSSIGDAYL 436
VE + DRRL+G+IY +E AL+ ++ M + G D A +D++ + +
Sbjct: 286 --VEVAHDRRLLGVIYTGLEQHEQALQQNEMSQKVMKSWGVAGDELLHAEIDAA--NIKI 341
Query: 437 ALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRI 496
AL + DEAV + V K + ++ A V++ +A K D+K E A I
Sbjct: 342 ALGKCDEAVTVLRN---VSKQVEKDSEIR-ALVFISMAKALANQEKAGDTKRCLEIACDI 397
Query: 497 FGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQ 556
K K P+++A ++V+++Y+ +N+ +K + LLK++L + P Q + A+
Sbjct: 398 LEK-KELAAPDKVAEAYVEVSSLYEMVNEFDKAISLLKRSLGMLERIPQAQHMEGNVAAR 456
Query: 557 MGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLF 616
+G + + G S++ + AV + + S K G N +G A ++ AA +F
Sbjct: 457 IGWLLLLTGKVSEAVPYLEDAVERMKDSFGPKHYGVGYVYNNLGAAYMEIGRPQSAAQMF 516
Query: 617 EEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDV 676
A+ +++ G +H DT+ +LA Y+AMG A+E + VV + P
Sbjct: 517 ALAKEVMDVSLGPHHSDTIEACQSLANAYNAMGSYALAMEFQKRVV----DSWRNHGPSA 572
Query: 677 DDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRL 712
DE LKEA R + ++L L NS +
Sbjct: 573 RDE-------LKEAIRLYEQIKIKALSCLSPENSAI 601
>ref|ZP_00327784.1| COG0457: FOG: TPR repeat [Trichodesmium erythraeum IMS101]
Length = 1215
Score = 128 bits (322), Expect = 6e-28
Identities = 128/485 (26%), Positives = 216/485 (44%), Gaps = 13/485 (2%)
Query: 213 PFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQY 272
P + T + I +G +K + + S+ + + SL L+ A G+Y
Sbjct: 44 PLTTEATEEQIINGTRNEKGKQGSYKL--SWRQASAKEESLTFATELNDEAFELYKQGKY 101
Query: 273 NEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQV 332
+EA+P+LE+S+ I + G +H L Y G + Y L++ ++
Sbjct: 102 DEAVPLLEQSLKIRLQLLGAEHPDVA-TSLNNLAFLYQRQGRYTEAEPLYIQALDMIKKL 160
Query: 333 LGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMG 392
LG P + +AE + ++ EAE L Q AL++ + V S + +
Sbjct: 161 LGAEHPLVATSLNNLAELYRVQGRYTEAEPLYQQALKMRKKLLGAKHPDVATSLNS--LA 218
Query: 393 LIYDSKGDYEAALEHYVLA-SMAMSANGQEQ-DVASVDSSIGDAYLALARYDEAVFSYQK 450
L+Y +G Y A Y+ A M G E DVAS +++ + Y + RY EA Y +
Sbjct: 219 LLYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAELYRSQGRYTEAEPLYLQ 278
Query: 451 ALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIA 510
AL + K G HP+VAS LA LY G++ +++ AL + K G +A
Sbjct: 279 ALEMTKKLLGAEHPDVASSLNNLAGLYKDQGRYTEAEPLYLQALEM-RKKLLGAEHRYVA 337
Query: 511 NGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNYSD 569
+ L ++A +Y+ + L +AL++ G + +A + ++Y G Y++
Sbjct: 338 SSLNNLALLYRVQGRYTEAEALYIQALEMDKKLLGAEHPDVATSLNNLALLYSDQGRYTE 397
Query: 570 SYNIFKSAVAKFRASGEKKSALFGIALNQM-GLACVQ-RYAINEAADLFEEARTILEKEY 627
+ ++ A+ F+ + +LN + GL VQ RY EA L+ +A + +K
Sbjct: 398 AEPLYIQALEIFKKLLGAEHRYVASSLNNLAGLYRVQGRY--TEAEPLYVQALKMWKKLL 455
Query: 628 GQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELL 687
G HPD +NLA Y GR +A + + + MR++ LG +PDV LA L
Sbjct: 456 GAEHPDVATSLNNLALLYKDQGRYTEAEPLYQQALDMRKKLLGAEHPDVATSLNNLAGLY 515
Query: 688 KEAGR 692
+ GR
Sbjct: 516 RVQGR 520
Score = 125 bits (314), Expect = 5e-27
Identities = 114/442 (25%), Positives = 195/442 (43%), Gaps = 7/442 (1%)
Query: 254 ELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMG 313
++ L+ LA +Y G+Y EA P+ +++D+ G +H L L + Y + G
Sbjct: 125 DVATSLNNLAFLYQRQGRYTEAEPLYIQALDMIKKLLGAEHPLVA-TSLNNLAELYRVQG 183
Query: 314 NIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRG 373
+ Y L+++ ++LG P + +A + ++ EAE L ALE+ +
Sbjct: 184 RYTEAEPLYQQALKMRKKLLGAKHPDVATSLNSLALLYKDQGRYTEAEPLYIQALEMRKK 243
Query: 374 NGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQ-DVASVDSSI 431
V S + + +Y S+G Y A Y+ A M G E DVAS +++
Sbjct: 244 LLGAEHPDVASSLNN--LAELYRSQGRYTEAEPLYLQALEMTKKLLGAEHPDVASSLNNL 301
Query: 432 GDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCE 491
Y RY EA Y +AL + K G H VAS LA LY G++ ++++
Sbjct: 302 AGLYKDQGRYTEAEPLYLQALEMRKKLLGAEHRYVASSLNNLALLYRVQGRYTEAEALYI 361
Query: 492 NALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQST-I 550
AL + K G ++A L ++A +Y + L +AL+I+ G + +
Sbjct: 362 QALEM-DKKLLGAEHPDVATSLNNLALLYSDQGRYTEAEPLYIQALEIFKKLLGAEHRYV 420
Query: 551 AGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAIN 610
A + +Y + G Y+++ ++ A+ ++ + +LN + L +
Sbjct: 421 ASSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPDVATSLNNLALLYKDQGRYT 480
Query: 611 EAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLG 670
EA L+++A + +K G HPD +NLAG Y GR +A + + M ++ LG
Sbjct: 481 EAEPLYQQALDMRKKLLGAEHPDVATSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLG 540
Query: 671 TANPDVDDEKRRLAELLKEAGR 692
+PDV LA L K GR
Sbjct: 541 AEHPDVATSLNNLALLYKAQGR 562
Score = 117 bits (293), Expect = 1e-24
Identities = 109/441 (24%), Positives = 193/441 (43%), Gaps = 7/441 (1%)
Query: 254 ELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMG 313
++ L+ LA +Y + G+Y EA P+ +++++ G +H + L + Y G
Sbjct: 209 DVATSLNSLALLYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVA-SSLNNLAELYRSQG 267
Query: 314 NIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRG 373
+ Y LE+ ++LG P + +A + ++ EAE L ALE+ +
Sbjct: 268 RYTEAEPLYLQALEMTKKLLGAEHPDVASSLNNLAGLYKDQGRYTEAEPLYLQALEMRKK 327
Query: 374 NGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQ-DVASVDSSI 431
V S + + L+Y +G Y A Y+ A M G E DVA+ +++
Sbjct: 328 LLGAEHRYVASSLNN--LALLYRVQGRYTEAEALYIQALEMDKKLLGAEHPDVATSLNNL 385
Query: 432 GDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCE 491
Y RY EA Y +AL +FK G H VAS LA LY G++ +++
Sbjct: 386 ALLYSDQGRYTEAEPLYIQALEIFKKLLGAEHRYVASSLNNLAGLYRVQGRYTEAEPLYV 445
Query: 492 NALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTI 550
AL+++ K G ++A L ++A +Y+ + L ++AL + G + +
Sbjct: 446 QALKMW-KKLLGAEHPDVATSLNNLALLYKDQGRYTEAEPLYQQALDMRKKLLGAEHPDV 504
Query: 551 AGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAIN 610
A + +Y + G Y+++ ++ A+ ++ + +LN + L +
Sbjct: 505 ATSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPDVATSLNNLALLYKAQGRYT 564
Query: 611 EAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLG 670
EA L+ ++ + +K G HP +NLA Y GR DA + + + MR++ LG
Sbjct: 565 EAEPLYIQSLEMRKKLLGAEHPSVAQSLNNLAALYYYQGRYTDAEPLYQQALEMRKKLLG 624
Query: 671 TANPDVDDEKRRLAELLKEAG 691
+PDV LA L G
Sbjct: 625 AEHPDVAISLNNLALLYSAQG 645
Score = 114 bits (284), Expect = 1e-23
Identities = 121/439 (27%), Positives = 195/439 (43%), Gaps = 11/439 (2%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENS 318
L+ LA +Y G+Y EA P+ ++++ + G H L Y G +
Sbjct: 172 LNNLAELYRVQGRYTEAEPLYQQALKMRKKLLGAKHPDVA-TSLNSLALLYKDQGRYTEA 230
Query: 319 ILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGS 378
Y LE++ ++LG P + +AE + ++ EAE L ALE+ +
Sbjct: 231 EPLYIQALEMRKKLLGAEHPDVASSLNNLAELYRSQGRYTEAEPLYLQALEMTKKLLGAE 290
Query: 379 SASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQD-VASVDSSIGDAYL 436
V S + L GL Y +G Y A Y+ A M G E VAS +++ Y
Sbjct: 291 HPDVASSLNN-LAGL-YKDQGRYTEAEPLYLQALEMRKKLLGAEHRYVASSLNNLALLYR 348
Query: 437 ALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRI 496
RY EA Y +AL + K G HP+VA+ LA LY+ G++ +++ AL I
Sbjct: 349 VQGRYTEAEALYIQALEMDKKLLGAEHPDVATSLNNLALLYSDQGRYTEAEPLYIQALEI 408
Query: 497 FGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEA 555
F K G +A+ L ++A +Y+ + L +ALK++ G + +A
Sbjct: 409 F-KKLLGAEHRYVASSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPDVATSLN 467
Query: 556 QMGVMYYMLGNYSDSYNIFKSAV-AKFRASGEKKSALFGIALNQMGLACVQ-RYAINEAA 613
+ ++Y G Y+++ +++ A+ + + G + + N GL VQ RY EA
Sbjct: 468 NLALLYKDQGRYTEAEPLYQQALDMRKKLLGAEHPDVATSLNNLAGLYRVQGRYT--EAE 525
Query: 614 DLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTAN 673
L+ +A + +K G HPD +NLA Y A GR +A + + MR++ LG +
Sbjct: 526 PLYVQALKMWKKLLGAEHPDVATSLNNLALLYKAQGRYTEAEPLYIQSLEMRKKLLGAEH 585
Query: 674 PDVDDEKRRLAELLKEAGR 692
P V LA L GR
Sbjct: 586 PSVAQSLNNLAALYYYQGR 604
Score = 105 bits (261), Expect = 7e-21
Identities = 96/419 (22%), Positives = 185/419 (43%), Gaps = 7/419 (1%)
Query: 254 ELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMG 313
++ L+ LA +Y + G+Y EA P+ +++++ G +H + L Y G
Sbjct: 251 DVASSLNNLAELYRSQGRYTEAEPLYLQALEMTKKLLGAEHPDVA-SSLNNLAGLYKDQG 309
Query: 314 NIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRG 373
+ Y LE++ ++LG + +A + ++ EAE L ALE+ +
Sbjct: 310 RYTEAEPLYLQALEMRKKLLGAEHRYVASSLNNLALLYRVQGRYTEAEALYIQALEMDKK 369
Query: 374 NGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSA--NGQEQDVASVDSSI 431
V S + + L+Y +G Y A Y+ A + + VAS +++
Sbjct: 370 LLGAEHPDVATSLNN--LALLYSDQGRYTEAEPLYIQALEIFKKLLGAEHRYVASSLNNL 427
Query: 432 GDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCE 491
Y RY EA Y +AL ++K G HP+VA+ LA LY G++ +++ +
Sbjct: 428 AGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPDVATSLNNLALLYKDQGRYTEAEPLYQ 487
Query: 492 NALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTI 550
AL + K G ++A L ++A +Y+ + L +ALK++ G + +
Sbjct: 488 QALDM-RKKLLGAEHPDVATSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPDV 546
Query: 551 AGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAIN 610
A + ++Y G Y+++ ++ ++ + + +LN + +
Sbjct: 547 ATSLNNLALLYKAQGRYTEAEPLYIQSLEMRKKLLGAEHPSVAQSLNNLAALYYYQGRYT 606
Query: 611 EAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKL 669
+A L+++A + +K G HPD +NLA Y A G + A++ LE + ++E+ L
Sbjct: 607 DAEPLYQQALEMRKKLLGAEHPDVAISLNNLALLYSAQGNIASAVQYLERGLEVQEKNL 665
Score = 63.9 bits (154), Expect = 2e-08
Identities = 74/312 (23%), Positives = 131/312 (41%), Gaps = 7/312 (2%)
Query: 191 SLALRGIIEERIATGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGK 250
+L ++ + ++ G ++PD+ L + S +A L ++AL+ F+ G
Sbjct: 358 ALYIQALEMDKKLLGAEHPDVATSL-NNLALLYSDQGRYTEAEPLYIQALEIFKKLL-GA 415
Query: 251 PSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYA 310
+ L+ LA +Y G+Y EA P+ +++ + G +H L Y
Sbjct: 416 EHRYVASSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPDVA-TSLNNLALLYK 474
Query: 311 MMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEI 370
G + Y L+++ ++LG P + +A + ++ EAE L AL++
Sbjct: 475 DQGRYTEAEPLYQQALDMRKKLLGAEHPDVATSLNNLAGLYRVQGRYTEAEPLYVQALKM 534
Query: 371 HRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQ-DVASVD 428
+ V S + + L+Y ++G Y A Y+ + M G E VA
Sbjct: 535 WKKLLGAEHPDVATSLNN--LALLYKAQGRYTEAEPLYIQSLEMRKKLLGAEHPSVAQSL 592
Query: 429 SSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKS 488
+++ Y RY +A YQ+AL + K G HP+VA LA LY+ G +
Sbjct: 593 NNLAALYYYQGRYTDAEPLYQQALEMRKKLLGAEHPDVAISLNNLALLYSAQGNIASAVQ 652
Query: 489 YCENALRIFGKS 500
Y E L + K+
Sbjct: 653 YLERGLEVQEKN 664
>ref|XP_470238.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|27261475|gb|AAN87741.1| Hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 556
Score = 127 bits (320), Expect = 1e-27
Identities = 119/482 (24%), Positives = 205/482 (41%), Gaps = 11/482 (2%)
Query: 214 FLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYN 273
F L + + + ++ Q+AL LA++AL+ + + G SL + L + LG+
Sbjct: 65 FALLEAAQLREAADDHQEALALAIKALEPLQ-ASHGGWSLPVARTLRLAGAAASRLGRLT 123
Query: 274 EAIPILERSIDIPVLEDGQDHALAKFAGCM--QLGDTYAMMGNIENSILFYTAGLEIQGQ 331
+++ L + DI + D +A + QL T MG ++ +E++
Sbjct: 124 DSLDSLNAAADIIDSLEAGDAEVAAVGAAVHEQLARTKTAMGRRWDAASDLMRAMELKAV 183
Query: 332 VLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLM 391
L + G + VAEA+ L D+A LC ALEI R + G S V + R+L+
Sbjct: 184 FLEKGSLELGNAYKDVAEAYRGVLACDKALPLCLEALEIARNHFGGDSQEVAKV--RQLL 241
Query: 392 GLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKA 451
IY G E ALE Y + M G + +++ ++ + + L R +EA+ ++
Sbjct: 242 ATIYAGSGRNEEALEQYEIVRMVYERLGLDVELSLAETDVAMVLVLLGRSEEAMDVLKRV 301
Query: 452 LTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIAN 511
+ + G+ A +V +A++ + DSK E A I +K + P ++A
Sbjct: 302 I----NRAGKESEERALAFVAMANILCIQDRKADSKRCLEIAREILD-TKISVSPLQVAQ 356
Query: 512 GLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSY 571
+++ +Y++M + E L L+KK L Q I A+MG + ++
Sbjct: 357 VYAEMSMLYETMIEFEVALCLMKKTLVFLDGVSEMQHIQGSISARMGWLLLKTERVDEAV 416
Query: 572 NIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYH 631
+SA+ K + G A +G A + A F A+ I+ YG H
Sbjct: 417 PYLQSAIEKLKNCFGPLHFGLGFAYKHLGDAYLAMNQSESAIKYFTIAKDIINATYGPKH 476
Query: 632 PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAG 691
DT+ ++A Y MG A++ E V+ E G + E +RL LK
Sbjct: 477 EDTIDTIQSIANAYGVMGSYKQAMDYQEQVID-AYESCGPGAFEELREAQRLRYQLKIKA 535
Query: 692 RG 693
RG
Sbjct: 536 RG 537
>ref|ZP_00324930.1| COG0457: FOG: TPR repeat [Trichodesmium erythraeum IMS101]
Length = 1363
Score = 121 bits (303), Expect = 9e-26
Identities = 118/465 (25%), Positives = 206/465 (43%), Gaps = 8/465 (1%)
Query: 231 KALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLED 290
+A+ L ++LK + A G ++ L+ LA +Y G+Y EA P+ +++++
Sbjct: 217 EAVPLLEQSLKIRQQ-ALGAEHPDVAASLNNLAFLYNAQGRYTEAEPLYIQALEMRKKLL 275
Query: 291 GQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEA 350
G +H + L Y G + Y LE++ ++LG P + +A
Sbjct: 276 GAEHPDVA-SSLNNLAFLYKAQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNLASL 334
Query: 351 HVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVL 410
+ ++ EAE L ALEI + V S + + +Y+++G Y A Y+
Sbjct: 335 YESQGRYTEAEPLYIQALEIFKKLLGAEHPDVATSLNN--LAFLYNAQGRYTEAEPLYIQ 392
Query: 411 A-SMAMSANGQEQ-DVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVAS 468
A M G E VA+ +++ Y RY EA Y +AL + K G HP+VA+
Sbjct: 393 ALDMTKKLLGAEHPSVATSLNNLALLYEDQGRYTEAEPLYIQALEMRKKLLGAEHPDVAT 452
Query: 469 VYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEK 528
LA LYN G++ +++ AL + K G ++A L ++A++Y+S +
Sbjct: 453 SLNNLAGLYNAQGRYTEAEPLYIQALEM-RKKLLGAEHPDVATSLNNLASLYESQGRYTE 511
Query: 529 GLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEK 587
L +AL+I+ G + +A + +Y G Y+++ ++ A+ +
Sbjct: 512 AEPLYIQALEIFKKLLGAEHPDVASSLNNLAGLYKDQGRYTEAEPLYIQALEMRKKLLGA 571
Query: 588 KSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDA 647
+ +LN + + EA L+ +A + +K G HPD +NLAG Y+A
Sbjct: 572 EHPDVASSLNNLAALYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAGLYNA 631
Query: 648 MGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGR 692
GR +A + + MR++ LG +P V LA L GR
Sbjct: 632 QGRYTEAEPLYIQALEMRKKLLGAEHPLVASSLNNLAGLYNAQGR 676
Score = 121 bits (303), Expect = 9e-26
Identities = 109/426 (25%), Positives = 185/426 (42%), Gaps = 7/426 (1%)
Query: 270 GQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQ 329
G+Y+EA+P+LE+S+ I G +H A L Y G + Y LE++
Sbjct: 213 GKYDEAVPLLEQSLKIRQQALGAEHPDVA-ASLNNLAFLYNAQGRYTEAEPLYIQALEMR 271
Query: 330 GQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRR 389
++LG P + +A + ++ EAE L ALE+ + V S +
Sbjct: 272 KKLLGAEHPDVASSLNNLAFLYKAQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNN- 330
Query: 390 LMGLIYDSKGDYEAALEHYV--LASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFS 447
+ +Y+S+G Y A Y+ L + DVA+ +++ Y A RY EA
Sbjct: 331 -LASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVATSLNNLAFLYNAQGRYTEAEPL 389
Query: 448 YQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPE 507
Y +AL + K G HP+VA+ LA LY G++ +++ AL + K G
Sbjct: 390 YIQALDMTKKLLGAEHPSVATSLNNLALLYEDQGRYTEAEPLYIQALEM-RKKLLGAEHP 448
Query: 508 EIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGN 566
++A L ++A +Y + + L +AL++ G + +A + +Y G
Sbjct: 449 DVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNLASLYESQGR 508
Query: 567 YSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKE 626
Y+++ ++ A+ F+ + +LN + + EA L+ +A + +K
Sbjct: 509 YTEAEPLYIQALEIFKKLLGAEHPDVASSLNNLAGLYKDQGRYTEAEPLYIQALEMRKKL 568
Query: 627 YGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAEL 686
G HPD +NLA Y GR +A + + MR++ LG +PDV LA L
Sbjct: 569 LGAEHPDVASSLNNLAALYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAGL 628
Query: 687 LKEAGR 692
GR
Sbjct: 629 YNAQGR 634
Score = 120 bits (301), Expect = 2e-25
Identities = 116/442 (26%), Positives = 197/442 (44%), Gaps = 7/442 (1%)
Query: 254 ELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMG 313
++ L+ LA++Y + G+Y EA P+ ++++I G +H L Y G
Sbjct: 323 DVATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVA-TSLNNLAFLYNAQG 381
Query: 314 NIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRG 373
+ Y L++ ++LG P + +A + ++ EAE L ALE+ +
Sbjct: 382 RYTEAEPLYIQALDMTKKLLGAEHPSVATSLNNLALLYEDQGRYTEAEPLYIQALEMRKK 441
Query: 374 NGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQ-DVASVDSSI 431
V S + L GL Y+++G Y A Y+ A M G E DVA+ +++
Sbjct: 442 LLGAEHPDVATSLNN-LAGL-YNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNL 499
Query: 432 GDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCE 491
Y + RY EA Y +AL +FK G HP+VAS LA LY G++ +++
Sbjct: 500 ASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVASSLNNLAGLYKDQGRYTEAEPLYI 559
Query: 492 NALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTI 550
AL + K G ++A+ L ++AA+Y+ + L +AL++ G + +
Sbjct: 560 QALEM-RKKLLGAEHPDVASSLNNLAALYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDV 618
Query: 551 AGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAIN 610
A + +Y G Y+++ ++ A+ + + L +LN + +
Sbjct: 619 ASSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPLVASSLNNLAGLYNAQGRYT 678
Query: 611 EAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLG 670
EA L+ +A I +K G HP +NLAG Y+A GR +A + + MR++ LG
Sbjct: 679 EAEPLYIQALEIFKKLLGAEHPLVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLG 738
Query: 671 TANPDVDDEKRRLAELLKEAGR 692
+P V LA L GR
Sbjct: 739 AEHPYVATSLNNLALLYYAQGR 760
Score = 119 bits (297), Expect = 5e-25
Identities = 121/490 (24%), Positives = 209/490 (41%), Gaps = 9/490 (1%)
Query: 205 GLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLAT 264
G ++PD+ L + S +A L ++AL+ F+ G ++ L+ LA
Sbjct: 318 GAEHPDVATSL-NNLASLYESQGRYTEAEPLYIQALEIFKKLL-GAEHPDVATSLNNLAF 375
Query: 265 IYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTA 324
+Y G+Y EA P+ +++D+ G +H L Y G + Y
Sbjct: 376 LYNAQGRYTEAEPLYIQALDMTKKLLGAEHPSVA-TSLNNLALLYEDQGRYTEAEPLYIQ 434
Query: 325 GLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEE 384
LE++ ++LG P + +A + ++ EAE L ALE+ + V
Sbjct: 435 ALEMRKKLLGAEHPDVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVAT 494
Query: 385 SADRRLMGLIYDSKGDYEAALEHYVLASMAMSA--NGQEQDVASVDSSIGDAYLALARYD 442
S + + +Y+S+G Y A Y+ A + DVAS +++ Y RY
Sbjct: 495 SLNN--LASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVASSLNNLAGLYKDQGRYT 552
Query: 443 EAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKP 502
EA Y +AL + K G HP+VAS LA LY G++ +++ AL + K
Sbjct: 553 EAEPLYIQALEMRKKLLGAEHPDVASSLNNLAALYKDQGRYTEAEPLYIQALEM-RKKLL 611
Query: 503 GIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMY 561
G ++A+ L ++A +Y + + L +AL++ G + +A + +Y
Sbjct: 612 GAEHPDVASSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPLVASSLNNLAGLY 671
Query: 562 YMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEART 621
G Y+++ ++ A+ F+ + L +LN + + EA L+ +A
Sbjct: 672 NAQGRYTEAEPLYIQALEIFKKLLGAEHPLVATSLNNLAGLYNAQGRYTEAEPLYIQALE 731
Query: 622 ILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKR 681
+ +K G HP +NLA Y A GR +A + + MR++ LG +PDV
Sbjct: 732 MRKKLLGAEHPYVATSLNNLALLYYAQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLN 791
Query: 682 RLAELLKEAG 691
LA L G
Sbjct: 792 NLATLYNVQG 801
Score = 112 bits (279), Expect = 6e-23
Identities = 106/414 (25%), Positives = 182/414 (43%), Gaps = 7/414 (1%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENS 318
L+ LA +Y + G+Y EA P+ +++++ G +H L Y G +
Sbjct: 412 LNNLALLYEDQGRYTEAEPLYIQALEMRKKLLGAEHPDVA-TSLNNLAGLYNAQGRYTEA 470
Query: 319 ILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGS 378
Y LE++ ++LG P + +A + ++ EAE L ALEI +
Sbjct: 471 EPLYIQALEMRKKLLGAEHPDVATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLLGAE 530
Query: 379 SASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQ-DVASVDSSIGDAYL 436
V S + L GL Y +G Y A Y+ A M G E DVAS +++ Y
Sbjct: 531 HPDVASSLNN-LAGL-YKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAALYK 588
Query: 437 ALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRI 496
RY EA Y +AL + K G HP+VAS LA LYN G++ +++ AL +
Sbjct: 589 DQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAGLYNAQGRYTEAEPLYIQALEM 648
Query: 497 FGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQ 556
K G +A+ L ++A +Y + + L +AL+I+ G + +
Sbjct: 649 -RKKLLGAEHPLVASSLNNLAGLYNAQGRYTEAEPLYIQALEIFKKLLGAEHPLVATSLN 707
Query: 557 -MGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADL 615
+ +Y G Y+++ ++ A+ + + +LN + L + EA L
Sbjct: 708 NLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPYVATSLNNLALLYYAQGRYTEAEPL 767
Query: 616 FEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKL 669
+ +A + +K G HPD +NLA Y+ G + A++ L+ + ++E+ L
Sbjct: 768 YIQALEMRKKLLGAEHPDVASSLNNLATLYNVQGDIASAVQYLKRGLEVQEKNL 821
Score = 102 bits (255), Expect = 3e-20
Identities = 86/316 (27%), Positives = 142/316 (44%), Gaps = 3/316 (0%)
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLA 437
S AS E D+ + L K D L L + + DVA+ +++ Y A
Sbjct: 195 SLASATELNDKAIE-LYKQGKYDEAVPLLEQSLKIRQQALGAEHPDVAASLNNLAFLYNA 253
Query: 438 LARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIF 497
RY EA Y +AL + K G HP+VAS LA LY G++ +++ AL +
Sbjct: 254 QGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAFLYKAQGRYTEAEPLYIQALEM- 312
Query: 498 GKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQ 556
K G ++A L ++A++Y+S + L +AL+I+ G + +A
Sbjct: 313 RKKLLGAEHPDVATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVATSLNN 372
Query: 557 MGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLF 616
+ +Y G Y+++ ++ A+ + + +LN + L + EA L+
Sbjct: 373 LAFLYNAQGRYTEAEPLYIQALDMTKKLLGAEHPSVATSLNNLALLYEDQGRYTEAEPLY 432
Query: 617 EEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDV 676
+A + +K G HPD +NLAG Y+A GR +A + + MR++ LG +PDV
Sbjct: 433 IQALEMRKKLLGAEHPDVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDV 492
Query: 677 DDEKRRLAELLKEAGR 692
LA L + GR
Sbjct: 493 ATSLNNLASLYESQGR 508
Score = 73.6 bits (179), Expect = 2e-11
Identities = 70/249 (28%), Positives = 111/249 (44%), Gaps = 5/249 (2%)
Query: 254 ELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMG 313
++ L+ LA +Y + G+Y EA P+ +++++ G +H + L Y G
Sbjct: 575 DVASSLNNLAALYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVA-SSLNNLAGLYNAQG 633
Query: 314 NIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRG 373
+ Y LE++ ++LG P + +A + ++ EAE L ALEI +
Sbjct: 634 RYTEAEPLYIQALEMRKKLLGAEHPLVASSLNNLAGLYNAQGRYTEAEPLYIQALEIFKK 693
Query: 374 NGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA-SMAMSANGQEQD-VASVDSSI 431
V S + L GL Y+++G Y A Y+ A M G E VA+ +++
Sbjct: 694 LLGAEHPLVATSLNN-LAGL-YNAQGRYTEAEPLYIQALEMRKKLLGAEHPYVATSLNNL 751
Query: 432 GDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCE 491
Y A RY EA Y +AL + K G HP+VAS LA LYN G + Y +
Sbjct: 752 ALLYYAQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLATLYNVQGDIASAVQYLK 811
Query: 492 NALRIFGKS 500
L + K+
Sbjct: 812 RGLEVQEKN 820
Score = 57.4 bits (137), Expect = 2e-06
Identities = 35/111 (31%), Positives = 54/111 (48%)
Query: 582 RASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNL 641
+AS + +S LN + ++ +EA L E++ I ++ G HPD +NL
Sbjct: 188 QASAKDQSLASATELNDKAIELYKQGKYDEAVPLLEQSLKIRQQALGAEHPDVAASLNNL 247
Query: 642 AGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGR 692
A Y+A GR +A + + MR++ LG +PDV LA L K GR
Sbjct: 248 AFLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAFLYKAQGR 298
>gb|AAM03641.1| hypothetical protein (multi-domain) [Methanosarcina acetivorans
str. C2A] gi|20089086|ref|NP_615161.1| hypothetical
protein MA0188 [Methanosarcina acetivorans C2A]
Length = 914
Score = 115 bits (288), Expect = 5e-24
Identities = 92/352 (26%), Positives = 168/352 (47%), Gaps = 8/352 (2%)
Query: 230 QKALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLE 289
+KAL + RAL E G ++ L LA +Y ++G Y +A+ + ER+++I
Sbjct: 534 KKALQFSERALAIGETIL-GVQHPDVATSLDNLAGLYESMGNYKQALQLSERALEIYEKV 592
Query: 290 DGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAE 349
G H L Y MG E +++FY +EI+ +VLG F + +A
Sbjct: 593 LGPQHRDVAIT-LDNLAGLYESMGEYEKALIFYQRTIEIKEKVLGPQHSNFATSLDNLAV 651
Query: 350 AHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHY- 408
+ Q ++++A +L Q ALEI+ + + + + L+YDS GDY+ L Y
Sbjct: 652 LYRQMGEYEKALQLSQRALEIYEKVLGPQHPDIATTLNN--IALLYDSMGDYQKTLPLYQ 709
Query: 409 -VLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVA 467
L Q +A+ +++ Y + Y++A+ Q++L + + G HP+VA
Sbjct: 710 RALEINEKVFGPQYLGIATTLNNLAGFYRRVGDYEKALSLSQRSLEIDEKVLGSQHPDVA 769
Query: 468 SVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLE 527
LA +Y IG ++ + ++ + +L I K G ++ L ++A +Y+ M D E
Sbjct: 770 RTLNSLALIYENIGDYEKALAFYQRSLDIREKVL-GPQHPDVGRTLNNLARLYEIMGDHE 828
Query: 528 KGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAV 578
K L L ++ ++I A G Q +A I + ++Y +G Y + +++ A+
Sbjct: 829 KALTLYQRTIEIKEKALGPQHPDVATILNNLAGLHYRIGEYKKALPLYQRAL 880
Score = 107 bits (267), Expect = 1e-21
Identities = 96/399 (24%), Positives = 178/399 (44%), Gaps = 15/399 (3%)
Query: 249 GKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDT 308
G ++V L++L +Y +G+Y +A+ ER++ I G H L
Sbjct: 510 GPEHKDIVTTLNILDVLYYKMGEYKKALQFSERALAIGETILGVQHPDVA-TSLDNLAGL 568
Query: 309 YAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMAL 368
Y MGN + ++ LEI +VLG T +A + ++++A Q +
Sbjct: 569 YESMGNYKQALQLSERALEIYEKVLGPQHRDVAITLDNLAGLYESMGEYEKALIFYQRTI 628
Query: 369 EIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALE--HYVLASMAMSANGQEQDVAS 426
EI ++ S D + ++Y G+YE AL+ L Q D+A+
Sbjct: 629 EIKEKVLGPQHSNFATSLDN--LAVLYRQMGEYEKALQLSQRALEIYEKVLGPQHPDIAT 686
Query: 427 VDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDS 486
++I Y ++ Y + + YQ+AL + + G + +A+ LA Y ++G ++ +
Sbjct: 687 TLNNIALLYDSMGDYQKTLPLYQRALEINEKVFGPQYLGIATTLNNLAGFYRRVGDYEKA 746
Query: 487 KSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQ 546
S + +L I K G ++A L +A IY+++ D EK L +++L I G
Sbjct: 747 LSLSQRSLEIDEKVL-GSQHPDVARTLNSLALIYENIGDYEKALAFYQRSLDIREKVLGP 805
Query: 547 QSTIAGIEA-QMGVMYYMLGNYSDSYNIFKSAV-AKFRASGEKKSALFGIALNQMGLACV 604
Q G + +Y ++G++ + +++ + K +A G + + I N GL
Sbjct: 806 QHPDVGRTLNNLARLYEIMGDHEKALTLYQRTIEIKEKALGPQHPDVATILNNLAGL--- 862
Query: 605 QRYAINE---AADLFEEARTILEKEYGQYHPDTLGVYSN 640
Y I E A L++ A I+EK+ GQ P+++ + +N
Sbjct: 863 -HYRIGEYKKALPLYQRALDIVEKKLGQNQPNSVVIKNN 900
Score = 107 bits (267), Expect = 1e-21
Identities = 101/429 (23%), Positives = 187/429 (43%), Gaps = 19/429 (4%)
Query: 277 PILERSIDIPVLEDGQDHALAKFAGCMQLGDT-YAMMGNIENSILFYTAGLEIQGQVLGE 335
P+ E + I + G +H + + D Y MG + ++ F L I +LG
Sbjct: 496 PMYEEMLQILEAKHGPEHK--DIVTTLNILDVLYYKMGEYKKALQFSERALAIGETILGV 553
Query: 336 TDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIY 395
P + +A + + +A +L + ALEI+ V + D L GL Y
Sbjct: 554 QHPDVATSLDNLAGLYESMGNYKQALQLSERALEIYEKVLGPQHRDVAITLDN-LAGL-Y 611
Query: 396 DSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALT 453
+S G+YE AL Y + Q + A+ ++ Y + Y++A+ Q+AL
Sbjct: 612 ESMGEYEKALIFYQRTIEIKEKVLGPQHSNFATSLDNLAVLYRQMGEYEKALQLSQRALE 671
Query: 454 VFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEEI 509
+++ G HP++A+ +A LY+ +G ++ + + AL ++FG GI
Sbjct: 672 IYEKVLGPQHPDIATTLNNIALLYDSMGDYQKTLPLYQRALEINEKVFGPQYLGI----- 726
Query: 510 ANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQST-IAGIEAQMGVMYYMLGNYS 568
A L ++A Y+ + D EK L L +++L+I G Q +A + ++Y +G+Y
Sbjct: 727 ATTLNNLAGFYRRVGDYEKALSLSQRSLEIDEKVLGSQHPDVARTLNSLALIYENIGDYE 786
Query: 569 DSYNIFKSAV-AKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEY 627
+ ++ ++ + + G + + G LN + +A L++ I EK
Sbjct: 787 KALAFYQRSLDIREKVLGPQHPDV-GRTLNNLARLYEIMGDHEKALTLYQRTIEIKEKAL 845
Query: 628 GQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELL 687
G HPD + +NLAG + +G A+ + + + + E+KLG P+ K LL
Sbjct: 846 GPQHPDVATILNNLAGLHYRIGEYKKALPLYQRALDIVEKKLGQNQPNSVVIKNNYNRLL 905
Query: 688 KEAGRGRNR 696
+ +
Sbjct: 906 SKMSENEKK 914
Score = 106 bits (265), Expect = 2e-21
Identities = 103/387 (26%), Positives = 172/387 (43%), Gaps = 20/387 (5%)
Query: 346 YVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAAL 405
Y + +ALQF E AL I V S D L GL Y+S G+Y+ AL
Sbjct: 528 YKMGEYKKALQFSER------ALAIGETILGVQHPDVATSLDN-LAGL-YESMGNYKQAL 579
Query: 406 E--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENH 463
+ L Q +DVA ++ Y ++ Y++A+ YQ+ + + + G H
Sbjct: 580 QLSERALEIYEKVLGPQHRDVAITLDNLAGLYESMGEYEKALIFYQRTIEIKEKVLGPQH 639
Query: 464 PNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSM 523
N A+ LA LY ++G+++ + + AL I+ K G +IA L ++A +Y SM
Sbjct: 640 SNFATSLDNLAVLYRQMGEYEKALQLSQRALEIYEKVL-GPQHPDIATTLNNIALLYDSM 698
Query: 524 NDLEKGLKLLKKALKIYGHAPGQQST-IAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFR 582
D +K L L ++AL+I G Q IA + Y +G+Y + ++ + ++
Sbjct: 699 GDYQKTLPLYQRALEINEKVFGPQYLGIATTLNNLAGFYRRVGDYEKALSLSQRSLEIDE 758
Query: 583 ASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLA 642
+ LN + L +A ++ + I EK G HPD +NLA
Sbjct: 759 KVLGSQHPDVARTLNSLALIYENIGDYEKALAFYQRSLDIREKVLGPQHPDVGRTLNNLA 818
Query: 643 GTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKS--KR 700
Y+ MG + A+ + + + ++E+ LG +PDV LA L G + +R
Sbjct: 819 RLYEIMGDHEKALTLYQRTIEIKEKALGPQHPDVATILNNLAGLHYRIGEYKKALPLYQR 878
Query: 701 SLETL------LDANSRLIKNNSIKVL 721
+L+ + NS +IKNN ++L
Sbjct: 879 ALDIVEKKLGQNQPNSVVIKNNYNRLL 905
Score = 37.7 bits (86), Expect = 1.3
Identities = 29/96 (30%), Positives = 53/96 (55%), Gaps = 3/96 (3%)
Query: 198 IEERIATGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVM 257
I E++ G +PD+G L R G++ +KAL L R ++ E A G ++
Sbjct: 798 IREKVL-GPQHPDVGRTLNNLARLYEIMGDH-EKALTLYQRTIEIKEK-ALGPQHPDVAT 854
Query: 258 CLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQD 293
L+ LA ++ +G+Y +A+P+ +R++DI + GQ+
Sbjct: 855 ILNNLAGLHYRIGEYKKALPLYQRALDIVEKKLGQN 890
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.132 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,159,185,433
Number of Sequences: 2540612
Number of extensions: 48018403
Number of successful extensions: 124537
Number of sequences better than 10.0: 1046
Number of HSP's better than 10.0 without gapping: 326
Number of HSP's successfully gapped in prelim test: 750
Number of HSP's that attempted gapping in prelim test: 118214
Number of HSP's gapped (non-prelim): 3948
length of query: 721
length of database: 863,360,394
effective HSP length: 135
effective length of query: 586
effective length of database: 520,377,774
effective search space: 304941375564
effective search space used: 304941375564
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0400a.8


