

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0399.7
(167 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
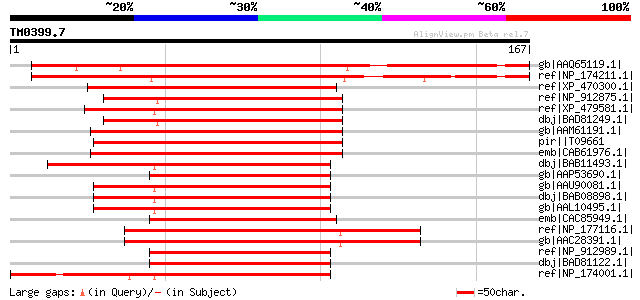
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ65119.1| At2g34140 [Arabidopsis thaliana] gi|62319965|dbj|... 170 2e-41
ref|NP_174211.1| Dof-type zinc finger domain-containing protein ... 166 3e-40
ref|XP_470300.1| putative zinc finger DNA-binding protein [Oryza... 131 6e-30
ref|NP_912875.1| unnamed protein product [Oryza sativa (japonica... 131 8e-30
ref|XP_479581.1| putative ascorbate oxidase promoter-binding pro... 131 8e-30
dbj|BAD81249.1| putative ascorbate oxidase promoter-binding prot... 131 8e-30
gb|AAM61191.1| H-protein promoter binding factor-2a [Arabidopsis... 130 1e-29
pir||T09661 ascorbate oxidase promoter-binding protein AOBP - wi... 130 1e-29
emb|CAB61976.1| H-protein promoter binding factor-2a [Arabidopsi... 129 2e-29
dbj|BAB11493.1| H-protein promoter binding factor-like protein [... 128 5e-29
gb|AAP53690.1| putative H-protein promoter binding factor-2a [Or... 127 1e-28
gb|AAU90081.1| At5g39660 [Arabidopsis thaliana] gi|18421891|ref|... 127 1e-28
dbj|BAB08898.1| unnamed protein product [Arabidopsis thaliana] 127 1e-28
gb|AAL10495.1| AT5g39660/MIJ24_130 [Arabidopsis thaliana] 127 1e-28
emb|CAC85949.1| dof zinc finger protein [Hordeum vulgare subsp. ... 127 2e-28
ref|NP_177116.1| Dof-type zinc finger domain-containing protein ... 126 3e-28
gb|AAC28391.1| H-protein promoter binding factor-2b [Arabidopsis... 126 3e-28
ref|NP_912989.1| unnamed protein product [Oryza sativa (japonica... 125 6e-28
dbj|BAD81122.1| ascorbate oxidase promoter-binding protein AOBP ... 125 6e-28
ref|NP_174001.1| Dof-type zinc finger domain-containing protein ... 124 8e-28
>gb|AAQ65119.1| At2g34140 [Arabidopsis thaliana] gi|62319965|dbj|BAD94068.1|
putative DOF zinc finger protein [Arabidopsis thaliana]
gi|2342734|gb|AAB67632.1| putative DOF zinc finger
protein [Arabidopsis thaliana] gi|25408331|pir||H84752
probable DOF zinc finger protein [imported] -
Arabidopsis thaliana gi|15226224|ref|NP_180961.1|
Dof-type zinc finger domain-containing protein
[Arabidopsis thaliana] gi|55583789|sp|O22967|DOF23_ARATH
Dof zinc finger protein DOF2.3 (AtDOF2.3)
Length = 170
Score = 170 bits (430), Expect = 2e-41
Identities = 96/174 (55%), Positives = 105/174 (60%), Gaps = 21/174 (12%)
Query: 8 QDSEGIKLFGTTI-----TLHGGRETRERSED-----KKGSGEERDQEKKPDKIIPCPRC 57
QDS+GIKLFG TI T+ ET ++ + S + EK+PDKII CPRC
Sbjct: 4 QDSQGIKLFGKTIAFNTRTIKNEEETHPPEQEATIAVRSSSSSDLTAEKRPDKIIACPRC 63
Query: 58 KSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRKMRPS----CDELLD 113
KSMETKFCYFNNYNVNQPRHFCKGC RYWTAGGALRNVPVGAGRRK +P L D
Sbjct: 64 KSMETKFCYFNNYNVNQPRHFCKGCHRYWTAGGALRNVPVGAGRRKSKPPGRVVVGMLGD 123
Query: 114 GGGSICDASATVEQLGLGTHREWHEAAAMFHGDFRQLFPSKRPRTTAGSGGQPC 167
G G VE + EW AAA HG FR FP KR R S GQ C
Sbjct: 124 GNG-----VRQVELINGLLVEEWQHAAAAAHGSFRHDFPMKRLR--CYSDGQSC 170
>ref|NP_174211.1| Dof-type zinc finger domain-containing protein [Arabidopsis
thaliana] gi|55583799|sp|P68350|DOF15_ARATH Dof zinc
finger protein DOF1.5 (AtDOF1.5)
Length = 175
Score = 166 bits (419), Expect = 3e-40
Identities = 96/181 (53%), Positives = 109/181 (60%), Gaps = 30/181 (16%)
Query: 8 QDSEGIKLFGTTITLHGGRETRERSEDKKGSGEERDQ--------------EKKPDKIIP 53
QDS+GIKLFG TIT + + E+++ + Q EK+PDKIIP
Sbjct: 4 QDSQGIKLFGKTITFNANITQTIKKEEQQQQQQPELQATTAVRSPSSDLTAEKRPDKIIP 63
Query: 54 CPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRKMRP-----SC 108
CPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK +P
Sbjct: 64 CPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRKSKPPGRVGGF 123
Query: 109 DELLDGGGSICDASATVEQLGLGT--HREWHEAAAMFHGDFRQLFPSKRPRTTAGSGGQP 166
ELL A+ V+Q+ L EW A A HG FR FP KR R + GQ
Sbjct: 124 AELLGA------ATGAVDQVELDALLVEEWRAATAS-HGGFRHDFPVKRLR--CYTDGQS 174
Query: 167 C 167
C
Sbjct: 175 C 175
>ref|XP_470300.1| putative zinc finger DNA-binding protein [Oryza sativa (japonica
cultivar-group)] gi|29367351|gb|AAO72548.1| putative
H-protein promoter binding factor-2a [Oryza sativa
(japonica cultivar-group)] gi|19071625|gb|AAL84292.1|
putative zinc finger DNA-binding protein [Oryza sativa
(japonica cultivar-group)]
Length = 440
Score = 131 bits (330), Expect = 6e-30
Identities = 52/80 (65%), Positives = 67/80 (83%)
Query: 26 RETRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRY 85
+ + E + + + ++++ KKPDKI+PCPRC SM+TKFCY+NNYN+NQPRHFCK CQRY
Sbjct: 80 KNSSENQQQQGDTANQKEKLKKPDKILPCPRCSSMDTKFCYYNNYNINQPRHFCKNCQRY 139
Query: 86 WTAGGALRNVPVGAGRRKMR 105
WTAGGA+RNVPVGAGRRK +
Sbjct: 140 WTAGGAMRNVPVGAGRRKSK 159
>ref|NP_912875.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 420
Score = 131 bits (329), Expect = 8e-30
Identities = 56/80 (70%), Positives = 67/80 (83%), Gaps = 3/80 (3%)
Query: 31 RSEDKKGSGEERDQEK---KPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWT 87
+ E++K + +E Q+K KPDKI+PCPRC SM+TKFCY+NNYNVNQPRHFCK CQRYWT
Sbjct: 13 KDENEKTANDESGQDKVLKKPDKILPCPRCNSMDTKFCYYNNYNVNQPRHFCKNCQRYWT 72
Query: 88 AGGALRNVPVGAGRRKMRPS 107
AGG +RNVPVGAGRRK + S
Sbjct: 73 AGGTMRNVPVGAGRRKSKSS 92
>ref|XP_479581.1| putative ascorbate oxidase promoter-binding protein AOBP [Oryza
sativa (japonica cultivar-group)]
gi|51963658|ref|XP_506580.1| PREDICTED OSJNBa0060O17.31
gene product [Oryza sativa (japonica cultivar-group)]
gi|29367353|gb|AAO72549.1| AOBP-like protein [Oryza
sativa (japonica cultivar-group)]
gi|34394527|dbj|BAC83814.1| putative ascorbate oxidase
promoter-binding protein AOBP [Oryza sativa (japonica
cultivar-group)]
Length = 476
Score = 131 bits (329), Expect = 8e-30
Identities = 54/84 (64%), Positives = 68/84 (80%), Gaps = 1/84 (1%)
Query: 25 GRETRERSEDKKGSGEERDQE-KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQ 83
G E++ + +G G ++ KKPDKI+PCPRC SM+TKFCY+NNYN+NQPRHFCK CQ
Sbjct: 80 GSESKSEAAQTEGGGSSEEKVLKKPDKILPCPRCNSMDTKFCYYNNYNINQPRHFCKSCQ 139
Query: 84 RYWTAGGALRNVPVGAGRRKMRPS 107
RYWTAGG++RN+PVGAGRRK + S
Sbjct: 140 RYWTAGGSMRNLPVGAGRRKSKSS 163
>dbj|BAD81249.1| putative ascorbate oxidase promoter-binding protein AOBP [Oryza
sativa (japonica cultivar-group)]
Length = 496
Score = 131 bits (329), Expect = 8e-30
Identities = 56/80 (70%), Positives = 67/80 (83%), Gaps = 3/80 (3%)
Query: 31 RSEDKKGSGEERDQEK---KPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWT 87
+ E++K + +E Q+K KPDKI+PCPRC SM+TKFCY+NNYNVNQPRHFCK CQRYWT
Sbjct: 81 KDENEKTANDESGQDKVLKKPDKILPCPRCNSMDTKFCYYNNYNVNQPRHFCKNCQRYWT 140
Query: 88 AGGALRNVPVGAGRRKMRPS 107
AGG +RNVPVGAGRRK + S
Sbjct: 141 AGGTMRNVPVGAGRRKSKSS 160
>gb|AAM61191.1| H-protein promoter binding factor-2a [Arabidopsis thaliana]
Length = 447
Score = 130 bits (328), Expect = 1e-29
Identities = 56/81 (69%), Positives = 65/81 (80%)
Query: 27 ETRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYW 86
E+ E ED + + + KKP KI+PCPRCKSMETKFCY+NNYN+NQPRHFCK CQRYW
Sbjct: 85 ESSETPEDNQQTTPDGKTLKKPTKILPCPRCKSMETKFCYYNNYNINQPRHFCKACQRYW 144
Query: 87 TAGGALRNVPVGAGRRKMRPS 107
TAGG +RNVPVGAGRRK + S
Sbjct: 145 TAGGTMRNVPVGAGRRKNKSS 165
>pir||T09661 ascorbate oxidase promoter-binding protein AOBP - winter squash
gi|1669341|dbj|BAA08094.1| AOBP (ascorbate oxidase
promoter-binding protein) [Cucurbita maxima]
Length = 380
Score = 130 bits (328), Expect = 1e-29
Identities = 54/80 (67%), Positives = 66/80 (82%)
Query: 28 TRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWT 87
TR+ +++ + +++ KKPDKI+PCPRC SMETKFCY+NNYNVNQPRHFCK CQRYWT
Sbjct: 16 TRKTEKEQNDAPNSKEKLKKPDKILPCPRCNSMETKFCYYNNYNVNQPRHFCKACQRYWT 75
Query: 88 AGGALRNVPVGAGRRKMRPS 107
GG +RNVPVGAGRRK + S
Sbjct: 76 EGGTIRNVPVGAGRRKNKNS 95
>emb|CAB61976.1| H-protein promoter binding factor-2a [Arabidopsis thaliana]
gi|22136158|gb|AAM91157.1| H-protein promoter binding
factor-2a [Arabidopsis thaliana]
gi|17473786|gb|AAL38328.1| H-protein promoter binding
factor-2a [Arabidopsis thaliana]
gi|3386546|gb|AAC28390.1| H-protein promoter binding
factor-2a [Arabidopsis thaliana]
gi|15232818|ref|NP_190334.1| Dof-type zinc finger
domain-containing protein [Arabidopsis thaliana]
gi|11357324|pir||T45710 H-protein promoter binding
factor-2a [imported] - Arabidopsis thaliana
gi|55584038|sp|Q8LFV3|DOF33_ARATH Dof zinc finger
protein DOF3.3 (AtDOF3.3) (H-protein promoter binding
factor-2a)
Length = 448
Score = 129 bits (325), Expect = 2e-29
Identities = 55/81 (67%), Positives = 65/81 (79%)
Query: 27 ETRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYW 86
E+ + ED + + + KKP KI+PCPRCKSMETKFCY+NNYN+NQPRHFCK CQRYW
Sbjct: 85 ESSDTPEDNQQTTPDGKTLKKPTKILPCPRCKSMETKFCYYNNYNINQPRHFCKACQRYW 144
Query: 87 TAGGALRNVPVGAGRRKMRPS 107
TAGG +RNVPVGAGRRK + S
Sbjct: 145 TAGGTMRNVPVGAGRRKNKSS 165
>dbj|BAB11493.1| H-protein promoter binding factor-like protein [Arabidopsis
thaliana] gi|55583963|sp|Q8W1E3|DOF55_ARATH Dof zinc
finger protein DOF5.5 (AtDOF5.5)
Length = 298
Score = 128 bits (322), Expect = 5e-29
Identities = 59/97 (60%), Positives = 68/97 (69%), Gaps = 6/97 (6%)
Query: 13 IKLFGTTITLHGGRETRERSEDKKGSGEERDQE------KKPDKIIPCPRCKSMETKFCY 66
IKLFG I E + E+K + DQ KKP KI+PCPRC SMETKFCY
Sbjct: 9 IKLFGMKIPFPTVLEVADEEEEKNQNKTLTDQSEKDKTLKKPTKILPCPRCNSMETKFCY 68
Query: 67 FNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
+NNYNVNQPRHFCK CQRYWT+GG +R+VP+GAGRRK
Sbjct: 69 YNNYNVNQPRHFCKACQRYWTSGGTMRSVPIGAGRRK 105
>gb|AAP53690.1| putative H-protein promoter binding factor-2a [Oryza sativa
(japonica cultivar-group)] gi|37534202|ref|NP_921403.1|
putative H-protein promoter binding factor-2a [Oryza
sativa (japonica cultivar-group)]
gi|15451553|gb|AAK98677.1| Putative H-protein promoter
binding factor-2a [Oryza sativa]
Length = 486
Score = 127 bits (319), Expect = 1e-28
Identities = 53/58 (91%), Positives = 56/58 (96%)
Query: 46 KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
KKPDKI+PCPRC SM+TKFCYFNNYNVNQPRHFCK CQRYWTAGGA+RNVPVGAGRRK
Sbjct: 138 KKPDKILPCPRCSSMDTKFCYFNNYNVNQPRHFCKHCQRYWTAGGAMRNVPVGAGRRK 195
>gb|AAU90081.1| At5g39660 [Arabidopsis thaliana] gi|18421891|ref|NP_568567.1|
Dof-type zinc finger domain-containing protein
[Arabidopsis thaliana] gi|30693448|ref|NP_851106.1|
Dof-type zinc finger domain-containing protein
[Arabidopsis thaliana] gi|55583973|sp|Q93ZL5|DOF52_ARATH
Dof zinc finger protein DOF5.2 (AtDOF5.2)
Length = 457
Score = 127 bits (319), Expect = 1e-28
Identities = 56/79 (70%), Positives = 63/79 (78%), Gaps = 3/79 (3%)
Query: 28 TRERSEDKKGSGEERDQE---KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQR 84
T+ +++ G QE KKPDKI+PCPRC SMETKFCY+NNYNVNQPRHFCK CQR
Sbjct: 111 TKAAKTNEESGGTACSQEGKLKKPDKILPCPRCNSMETKFCYYNNYNVNQPRHFCKKCQR 170
Query: 85 YWTAGGALRNVPVGAGRRK 103
YWTAGG +RNVPVGAGRRK
Sbjct: 171 YWTAGGTMRNVPVGAGRRK 189
>dbj|BAB08898.1| unnamed protein product [Arabidopsis thaliana]
Length = 515
Score = 127 bits (319), Expect = 1e-28
Identities = 56/79 (70%), Positives = 63/79 (78%), Gaps = 3/79 (3%)
Query: 28 TRERSEDKKGSGEERDQE---KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQR 84
T+ +++ G QE KKPDKI+PCPRC SMETKFCY+NNYNVNQPRHFCK CQR
Sbjct: 111 TKAAKTNEESGGTACSQEGKLKKPDKILPCPRCNSMETKFCYYNNYNVNQPRHFCKKCQR 170
Query: 85 YWTAGGALRNVPVGAGRRK 103
YWTAGG +RNVPVGAGRRK
Sbjct: 171 YWTAGGTMRNVPVGAGRRK 189
>gb|AAL10495.1| AT5g39660/MIJ24_130 [Arabidopsis thaliana]
Length = 457
Score = 127 bits (319), Expect = 1e-28
Identities = 56/79 (70%), Positives = 63/79 (78%), Gaps = 3/79 (3%)
Query: 28 TRERSEDKKGSGEERDQE---KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQR 84
T+ +++ G QE KKPDKI+PCPRC SMETKFCY+NNYNVNQPRHFCK CQR
Sbjct: 111 TKAAKTNEESGGTACSQEGKLKKPDKILPCPRCNSMETKFCYYNNYNVNQPRHFCKKCQR 170
Query: 85 YWTAGGALRNVPVGAGRRK 103
YWTAGG +RNVPVGAGRRK
Sbjct: 171 YWTAGGTMRNVPVGAGRRK 189
>emb|CAC85949.1| dof zinc finger protein [Hordeum vulgare subsp. vulgare]
Length = 270
Score = 127 bits (318), Expect = 2e-28
Identities = 51/60 (85%), Positives = 57/60 (95%)
Query: 46 KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRKMR 105
KKPDKI+PCPRC SM+TKFCY+NNYN+NQPRHFCK CQRYWTAGGA+RNVPVGAGRRK +
Sbjct: 2 KKPDKILPCPRCSSMDTKFCYYNNYNINQPRHFCKNCQRYWTAGGAMRNVPVGAGRRKSK 61
>ref|NP_177116.1| Dof-type zinc finger domain-containing protein [Arabidopsis
thaliana] gi|12597792|gb|AAG60104.1| H-protein promoter
binding factor-2b [Arabidopsis thaliana]
gi|25404815|pir||D96717 hypothetical protein F24J1.25
[imported] - Arabidopsis thaliana
gi|51970834|dbj|BAD44109.1| putative H-protein promoter
binding factor-2b [Arabidopsis thaliana]
gi|6692253|gb|AAF24604.1| H-protein promoter binding
factor-2b, putative; 37606-39065 [Arabidopsis thaliana]
gi|55584043|sp|Q9SEZ3|DOF1A_ARATH Dof zinc finger
protein DOF1.10 (AtDOF1.10) (H-protein promoter binding
factor-2b)
Length = 399
Score = 126 bits (316), Expect = 3e-28
Identities = 57/97 (58%), Positives = 71/97 (72%), Gaps = 2/97 (2%)
Query: 38 SGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPV 97
SGE+ KKPDK+IPCPRC+S TKFCY+NNYNVNQPR+FC+ CQRYWTAGG++RNVPV
Sbjct: 118 SGEKSTALKKPDKLIPCPRCESANTKFCYYNNYNVNQPRYFCRNCQRYWTAGGSMRNVPV 177
Query: 98 GAGRRKMR--PSCDELLDGGGSICDASATVEQLGLGT 132
G+GRRK + PS + L CD + + L G+
Sbjct: 178 GSGRRKNKGWPSSNHYLQVTSEDCDNNNSGTILSFGS 214
>gb|AAC28391.1| H-protein promoter binding factor-2b [Arabidopsis thaliana]
gi|25453561|pir||T51953 H-protein promoter binding
factor-2b [imported] - Arabidopsis thaliana
Length = 400
Score = 126 bits (316), Expect = 3e-28
Identities = 57/97 (58%), Positives = 71/97 (72%), Gaps = 2/97 (2%)
Query: 38 SGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPV 97
SGE+ KKPDK+IPCPRC+S TKFCY+NNYNVNQPR+FC+ CQRYWTAGG++RNVPV
Sbjct: 119 SGEKSTALKKPDKLIPCPRCESANTKFCYYNNYNVNQPRYFCRNCQRYWTAGGSMRNVPV 178
Query: 98 GAGRRKMR--PSCDELLDGGGSICDASATVEQLGLGT 132
G+GRRK + PS + L CD + + L G+
Sbjct: 179 GSGRRKNKGWPSSNHYLQVTSEDCDNNNSGTILSFGS 215
>ref|NP_912989.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 454
Score = 125 bits (313), Expect = 6e-28
Identities = 51/58 (87%), Positives = 56/58 (95%)
Query: 46 KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
KKPDKI+PCPRC SMETKFCYFNNYNV+QPRHFC+ CQRYWTAGGA+RNVPVGAGRR+
Sbjct: 72 KKPDKILPCPRCNSMETKFCYFNNYNVHQPRHFCRNCQRYWTAGGAMRNVPVGAGRRR 129
>dbj|BAD81122.1| ascorbate oxidase promoter-binding protein AOBP -like [Oryza sativa
(japonica cultivar-group)]
Length = 490
Score = 125 bits (313), Expect = 6e-28
Identities = 51/58 (87%), Positives = 56/58 (95%)
Query: 46 KKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
KKPDKI+PCPRC SMETKFCYFNNYNV+QPRHFC+ CQRYWTAGGA+RNVPVGAGRR+
Sbjct: 108 KKPDKILPCPRCNSMETKFCYFNNYNVHQPRHFCRNCQRYWTAGGAMRNVPVGAGRRR 165
>ref|NP_174001.1| Dof-type zinc finger domain-containing protein [Arabidopsis
thaliana]
Length = 396
Score = 124 bits (312), Expect = 8e-28
Identities = 62/110 (56%), Positives = 78/110 (70%), Gaps = 9/110 (8%)
Query: 1 MAQVQSGQDSEGIKLFGTTITLHGGRETRERSEDKKG------SGEERDQE-KKPDKIIP 53
M QS D+ +KL ++ + +ET E S+D+ S EE+ E KKPDKI+P
Sbjct: 79 MMNNQSVTDNTSLKL--SSNLNNESKETSENSDDQHSEITTITSEEEKTTELKKPDKILP 136
Query: 54 CPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
CPRC S +TKFCY+NNYNVNQPRHFC+ CQRYWTAGG++R VPVG+GRRK
Sbjct: 137 CPRCNSADTKFCYYNNYNVNQPRHFCRKCQRYWTAGGSMRIVPVGSGRRK 186
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.136 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 323,180,537
Number of Sequences: 2540612
Number of extensions: 14173881
Number of successful extensions: 46498
Number of sequences better than 10.0: 164
Number of HSP's better than 10.0 without gapping: 150
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 46322
Number of HSP's gapped (non-prelim): 185
length of query: 167
length of database: 863,360,394
effective HSP length: 118
effective length of query: 49
effective length of database: 563,568,178
effective search space: 27614840722
effective search space used: 27614840722
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0399.7


