

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0370.9
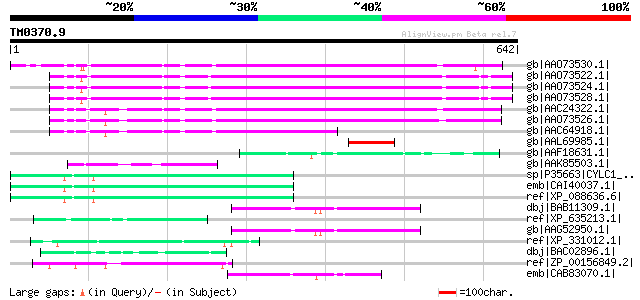
(642 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO73530.1| envelope-like protein [Glycine max] 375 e-102
gb|AAO73522.1| envelope-like protein [Glycine max] 363 1e-98
gb|AAO73524.1| envelope-like protein [Glycine max] 361 5e-98
gb|AAO73528.1| envelope-like protein [Glycine max] 360 8e-98
gb|AAC24322.1| envelope-like [Glycine max] gi|7488670|pir||T0889... 355 2e-96
gb|AAO73526.1| envelope-like protein [Glycine max] 350 6e-95
gb|AAC64918.1| envelope-like [Glycine max] 204 9e-51
gb|AAL69985.1| putative envelope protein [Vicia faba] 70 3e-10
gb|AAF18631.1| F5J5.2 [Arabidopsis thaliana] 63 3e-08
gb|AAK85503.1| Hypothetical protein Y55B1BR.3 [Caenorhabditis el... 59 6e-07
sp|P35663|CYLC1_HUMAN Cylicin-1 (Cylicin I) (Multiple-band polyp... 57 2e-06
emb|CAI40037.1| cylicin, basic protein of sperm head cytoskeleto... 57 2e-06
ref|XP_088636.6| PREDICTED: cylicin, basic protein of sperm head... 57 2e-06
dbj|BAB11309.1| unnamed protein product [Arabidopsis thaliana] 55 5e-06
ref|XP_635213.1| hypothetical protein DDB0183896 [Dictyostelium ... 55 5e-06
gb|AAG52950.1| putative envelope protein [Arabidopsis thaliana] 55 5e-06
ref|XP_331012.1| predicted protein [Neurospora crassa] gi|289209... 55 5e-06
dbj|BAC02896.1| tobacco nucleolin [Nicotiana tabacum] 55 5e-06
ref|ZP_00156849.2| COG3468: Type V secretory pathway, adhesin Ai... 53 3e-05
emb|CAB83070.1| putative protein [Arabidopsis thaliana] gi|11357... 53 3e-05
>gb|AAO73530.1| envelope-like protein [Glycine max]
Length = 685
Score = 375 bits (964), Expect = e-102
Identities = 238/645 (36%), Positives = 353/645 (53%), Gaps = 52/645 (8%)
Query: 1 MSQESGKQSAPGLVPVKNAHGVRFLGLKSPSSHSTRVTRSSSGIAAQSSSPGHSPTPKKM 60
M+ + SAPG V ++ K+PS+ R I PG +P P+K+
Sbjct: 69 MATSPKETSAPGSPSVPSSPS----STKAPSNQE----RPEFNIQPIQMIPGQAPVPEKL 120
Query: 61 RTKQVVTKKKTLRFSPPQDTSPISESDTE----------NV---ADIPDLDIVHAEDLLD 107
K+ K + +P TSP E DTE N+ +PD D +D+
Sbjct: 121 VPKRQQGVK--ISENPSLATSP-REVDTEMDKKIRTIVSNILKDVSVPDAD----KDVPT 173
Query: 108 LASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPETSSRKGKEHV 167
++ VS P + +V S++ + + + ++ T AP G
Sbjct: 174 SSTPNVSVPDAEKDVPTSSTPNAEVLSSPSKEDSTEEEDQATEETPAPRAPEPAPGDLID 233
Query: 168 ILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST--IRTPKDSKAAGSVRKSKGETTS 225
+ + + D + I RL+ + K TP+K + I+T K+ TTS
Sbjct: 234 LEEVESDEEPIANKLAPGIAERLQSR-KGKTPIKRSGRIKTMAQKKSTPIT-----PTTS 287
Query: 226 KGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVADVPLDNVSFHY 285
+ + + +KKRK +S +SD D E +PDI R + GK+VP NV D PLDN+SFH
Sbjct: 288 RRSKVAIPSKKRKEISSSDSDDDVELDVPDIK---RAKTSGKKVPGNVPDAPLDNISFHS 344
Query: 286 ADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLVREFIV 345
DN +RWKFV RR+A+EREL +AL+ KEI+DL+ A L+KTV +G CYE LVREFIV
Sbjct: 345 IDNVERWKFVYQRRLALERELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIV 404
Query: 346 NISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDLNEIAKVISGKQ 405
NI SD + S E+ KVF R KCV SP VIN++LGR T V + ++IAK I+ KQ
Sbjct: 405 NIPSDITNRKSDEYQKVFVRGKCVRFSPAVINKYLGRPTDGVVDITVSEHQIAKEITAKQ 464
Query: 406 VMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWIGTGAMMDFGQ 465
V WPKKG L +GKL+ K+ +L +IGAANW+ TNHTS + L K ++ +GT + +FG
Sbjct: 465 VQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGN 524
Query: 466 HVFEHTFKHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPLNFDYRLFAGT 525
++F+ T KH S+AVKL I FP ++ ++L QHP+IL ++ K+ PL+ Y+LF GT
Sbjct: 525 YIFDQTVKHSESFAVKLPIAFPTVLCGIMLSQHPNILNYTDSVMKRESPLSLHYKLFEGT 584
Query: 526 HVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKVRKHNVDQLIK 585
HVP+IV + K A S + KD ++AELK+ K L+ATI+A+ +K +++LIK
Sbjct: 585 HVPDIVSTSGKAAASG-------AVSKDALIAELKDTCKVLEATIKATTEKKMELERLIK 637
Query: 586 MLA------APADDEVSDPADDEDAEADLDEEDDDEALSDDDNVE 624
L+ A +E + A++E+ A+ +E+ ++ SDDD+ E
Sbjct: 638 RLSDSGIDDGEAAEEEEEAAEEEEEAAEEEEDAAEDTESDDDDSE 682
>gb|AAO73522.1| envelope-like protein [Glycine max]
Length = 680
Score = 363 bits (931), Expect = 1e-98
Identities = 218/601 (36%), Positives = 343/601 (56%), Gaps = 39/601 (6%)
Query: 51 PGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTE-------------NVADIPDL 97
PG + P+K+ ++ K + +P TSP E DTE A +P+
Sbjct: 103 PGQASVPEKLVPRRPQGVK--IAENPSPATSP-REVDTEMDKKIRSIVSSILKDASVPEA 159
Query: 98 DIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPE 157
D ED+ ++ VS P V+ +V S++ + +++ + ++ AP
Sbjct: 160 D----EDVPTSSNPNVSVPDVKKDVPTSSAPNAEALPSPSEEGSTEEDDQAAEETPAPRA 215
Query: 158 TSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST--IRTPKDSKAAGS 215
G + + + D + I RL+ + K TP+K + I+T K+
Sbjct: 216 PEPAPGDLIDLEEVESDEEPIANRLAPGIAERLQSR-KGKTPIKRSGRIKTMAQKKSTPI 274
Query: 216 VRKSKGETTSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVAD 275
TS+ + + +KKRK +S +SD+D E D+ST+ + + GK+VP NV D
Sbjct: 275 T-----PATSRRSKVAIPSKKRKEISSSDSDKDVEL---DVSTSKKAKTSGKKVPGNVPD 326
Query: 276 VPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRC 335
PLDN+SFH N ++WK+V RR+A+EREL +AL+ KEI+DL+ A L+KTV +G C
Sbjct: 327 APLDNISFHSIGNVEKWKYVYQRRLAVERELGRDALDCKEIMDLIKAAGLLKTVSKLGDC 386
Query: 336 YEKLVREFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDLN 395
YE LVREFIVNI SD + S E+ KVF R KCV SP VIN++LGR T V + + +
Sbjct: 387 YEGLVREFIVNIPSDISNRKSDEYQKVFVRGKCVKFSPAVINKYLGRPTDGVIDIDVSEH 446
Query: 396 EIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWI 455
+IAK I+ K+V WPKKG L +GKL+ K+ +L +IGAANW+ TNHTS + L K ++ +
Sbjct: 447 QIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAV 506
Query: 456 GTGAMMDFGQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPL 515
GT + +FG ++F+ T KH S+A+KL I FP ++ ++L QHP++L ++ K+ PL
Sbjct: 507 GTKSKFNFGNYIFDQTVKHSESFAIKLPIAFPTVLCGIMLSQHPNMLNYTDSVMKRESPL 566
Query: 516 NFDYRLFAGTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKV 575
+ Y+LF GTHVP+IV + + A+ + V+ KD ++AELK+ K L+ATI+A+
Sbjct: 567 SLHYKLFEGTHVPDIVSTSVSTSGKAAASGAVS---KDALIAELKDTCKVLEATIKATTE 623
Query: 576 RKHNVDQLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADS 635
+K ++ LIK L+ D+ + A +E+ EA E+++EA ++++ EE E D+++
Sbjct: 624 KKMELELLIKRLSESGIDD--EEAAEEEGEA---AEEEEEAAEEEEDAAEETESDDDSEA 678
Query: 636 T 636
T
Sbjct: 679 T 679
>gb|AAO73524.1| envelope-like protein [Glycine max]
Length = 682
Score = 361 bits (926), Expect = 5e-98
Identities = 217/601 (36%), Positives = 342/601 (56%), Gaps = 39/601 (6%)
Query: 51 PGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTE-------------NVADIPDL 97
PG + P+K+ ++ K + +P TSP E DTE A +P+
Sbjct: 105 PGQASVPEKLVPRRPQGVK--IAENPSPATSP-REVDTEMDKKIRSIVSSILKDASVPEA 161
Query: 98 DIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPE 157
D ED+ ++ VS P V+ +V S++ + +++ + ++ AP
Sbjct: 162 D----EDVPTSSNPNVSVPDVKKDVPTSSAPNAEALPSPGEEGSTEEDDQAAEETPAPRA 217
Query: 158 TSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST--IRTPKDSKAAGS 215
G + + + D + I RL+ + K TP+K + I+T K+
Sbjct: 218 PEPAPGDLIDLEEVESDEEPIANRLAPGIAERLQSR-KGKTPIKRSGRIKTMAQKKSTPI 276
Query: 216 VRKSKGETTSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVAD 275
TS+ + + +KKRK +S +SD+D E D+ST+ + + GK+VP NV D
Sbjct: 277 T-----PATSRRSKVAIPSKKRKEISSSDSDKDVEL---DVSTSKKAKTSGKKVPGNVPD 328
Query: 276 VPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRC 335
PLDN+SFH N ++WK+V RR+A+EREL +AL+ KEI+DL+ L+KTV +G C
Sbjct: 329 APLDNISFHSIGNVEKWKYVYQRRLAVERELGRDALDCKEIMDLIKAGGLLKTVSKLGDC 388
Query: 336 YEKLVREFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDLN 395
YE LVREFIVNI SD + S E+ KVF R KCV SP VIN++LGR T V + + +
Sbjct: 389 YEGLVREFIVNIPSDISNRKSDEYQKVFVRGKCVKFSPAVINKYLGRPTDGVIDIDVSEH 448
Query: 396 EIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWI 455
+IAK I+ K+V WPKKG L +GKL+ K+ +L +IGAANW+ TNHTS + L K ++ +
Sbjct: 449 QIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAV 508
Query: 456 GTGAMMDFGQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPL 515
GT + +FG ++F+ T KH S+A+KL I FP ++ ++L QHP++L ++ K+ PL
Sbjct: 509 GTKSKFNFGNYIFDQTVKHSESFAIKLPIAFPTVLCGIMLSQHPNMLNYTDSVMKRESPL 568
Query: 516 NFDYRLFAGTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKV 575
+ Y+LF GTHVP+IV + + A+ + V+ KD ++AELK+ K L+ATI+A+
Sbjct: 569 SLHYKLFEGTHVPDIVSTSVSTSGKAAASGAVS---KDALIAELKDTCKVLEATIKATTE 625
Query: 576 RKHNVDQLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADS 635
+K ++ LIK L+ D+ + A +E+ EA E+++EA ++++ EE E D+++
Sbjct: 626 KKMELELLIKRLSESGIDD--EEAAEEEGEA---AEEEEEAAEEEEDAAEETESDDDSEA 680
Query: 636 T 636
T
Sbjct: 681 T 681
>gb|AAO73528.1| envelope-like protein [Glycine max]
Length = 680
Score = 360 bits (924), Expect = 8e-98
Identities = 216/601 (35%), Positives = 343/601 (56%), Gaps = 39/601 (6%)
Query: 51 PGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTE-------------NVADIPDL 97
PG + P+K+ ++ K + +P TSP E DTE A +P+
Sbjct: 103 PGQASVPEKLVPRRPQGVK--IAENPSPATSP-REVDTEMDKKIRSIVSSILKDASVPEA 159
Query: 98 DIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPE 157
D ED+ ++ VS P V+ +V S++ + +++ + ++ AP
Sbjct: 160 D----EDVPTSSNPDVSVPDVKKDVPTSSAPNAEALPSPSEEGSTEEDDQAAEETPAPRA 215
Query: 158 TSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST--IRTPKDSKAAGS 215
G + + + D + I RL+ + K TP+K + I+T K+
Sbjct: 216 PEPAPGDLIDLEEVESDEEPIANRLAPGIAERLQSR-KGKTPIKRSGRIKTMAQKKSTPI 274
Query: 216 VRKSKGETTSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVAD 275
TS+ + + +KKRK +S +SD+D E D+ST+ + + GK+VP NV D
Sbjct: 275 T-----PATSRRSKVAIPSKKRKEISSSDSDKDVEL---DVSTSKKAKTSGKKVPGNVPD 326
Query: 276 VPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRC 335
PLDN+SFH N ++WK+V RR+A+EREL +AL+ KEI+DL+ A L+KTV +G C
Sbjct: 327 APLDNISFHSIGNVEKWKYVYQRRLAVERELGRDALDCKEIMDLIKAAGLLKTVSKLGDC 386
Query: 336 YEKLVREFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDLN 395
YE LVREFIVNI SD + S ++ +VF R KCV SP VIN++LGR T V + + +
Sbjct: 387 YEGLVREFIVNIPSDISNRKSDDYQRVFVRGKCVRFSPAVINKYLGRPTDGVIDIDVSEH 446
Query: 396 EIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWI 455
+IAK I+ K+V WPKKG L +GKL+ K+ +L +IGAANW+ TNHTS + L K ++ +
Sbjct: 447 QIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAV 506
Query: 456 GTGAMMDFGQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPL 515
GT + +FG ++F+ T KH S+A+KL I FP ++ ++L QHP++L ++ K+ PL
Sbjct: 507 GTKSKFNFGNYIFDQTVKHSESFAIKLPIAFPTVLCGIMLSQHPNMLNYTDSVMKRESPL 566
Query: 516 NFDYRLFAGTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKV 575
+ Y+LF GTHVP+IV + + A+ + V+ KD ++AELK+ K L+ATI+A+
Sbjct: 567 SLHYKLFEGTHVPDIVSTSVSTSGKAAASGAVS---KDALIAELKDTCKVLEATIKATTE 623
Query: 576 RKHNVDQLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADS 635
+K ++ LIK L+ D+ + A +E+ EA E+++EA ++++ EE E D+++
Sbjct: 624 KKMELELLIKRLSESGIDD--EEAAEEEGEA---AEEEEEAAEEEEDAAEETESDDDSEA 678
Query: 636 T 636
T
Sbjct: 679 T 679
>gb|AAC24322.1| envelope-like [Glycine max] gi|7488670|pir||T08898 envelope-like -
soybean (fragment)
Length = 648
Score = 355 bits (912), Expect = 2e-96
Identities = 223/579 (38%), Positives = 327/579 (55%), Gaps = 40/579 (6%)
Query: 51 PGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTENVADIPDLDIVHAEDLLDLAS 110
PG +P P+K+ K+ K + +P TSP E DTE I + +L AS
Sbjct: 98 PGQAPVPEKLVPKRQQGVK--ISENPSIATSP-REVDTEMDKKIRSI----VSSILKNAS 150
Query: 111 ARVSEPPVQS----NVEK-STSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPETSSRKGKE 165
++ V + N E S+S E+S + + A T AP G
Sbjct: 151 VPDADKDVPTSSTPNAEVLSSSSKEESTEEEEQA---------TEETPAPRAPEPAPGDL 201
Query: 166 HVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST--IRTPKDSKAAGSVRKSKGET 223
+ + + D + I RL+ + K TP+ + I+T K+ T
Sbjct: 202 IDLEEVESDEEPIANKLAPGIAERLQSR-KGKTPITRSGRIKTMAQKKSTPIT-----PT 255
Query: 224 TSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVADVPLDNVSF 283
TS+ + + +KKRK S +SD D E +PDI R + GK+VP NV D PLDN+SF
Sbjct: 256 TSRWSKVAIPSKKRKEFSSSDSDDDVELDVPDIK---RAKKSGKKVPGNVPDAPLDNISF 312
Query: 284 HYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLVREF 343
H N +RWKFV RR+A+EREL +AL+ KEI+DL+ A L+KTV +G CYE LVREF
Sbjct: 313 HSIGNVERWKFVYQRRLALERELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREF 372
Query: 344 IVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDLNEIAKVISG 403
IVNI SD + S E+ KVF R KCV SP VIN++LGR T V + ++IAK I+
Sbjct: 373 IVNIPSDITNRKSDEYQKVFVRGKCVRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITA 432
Query: 404 KQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWIGTGAMMDF 463
KQV WPKKG L +GKL+ K+ +L +IGAANW+ TNHTS + L K ++ +GT + +F
Sbjct: 433 KQVQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNF 492
Query: 464 GQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPLNFDYRLFA 523
G+++F+ T KH S+AVKL I FP ++ ++L QHP+IL ++ K+ L+ Y+LF
Sbjct: 493 GKYIFDQTVKHSESFAVKLPIAFPTVLCGIMLSQHPNILNNIDSVMKRESALSLHYKLFE 552
Query: 524 GTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKVRKHNVDQL 583
GTHVP+IV + K A S + KD ++AELK+ K L+ATI+A+ +K +++L
Sbjct: 553 GTHVPDIVSTSGKAAASG-------AVSKDALIAELKDTCKVLEATIKATTEKKMELERL 605
Query: 584 IKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDN 622
IK L+ D+ + A++E+ A+ +++ ++ SDDD+
Sbjct: 606 IKRLSDSGIDD-GEAAEEEEEAAEEEKDAAEDTESDDDD 643
>gb|AAO73526.1| envelope-like protein [Glycine max]
Length = 656
Score = 350 bits (899), Expect = 6e-95
Identities = 223/579 (38%), Positives = 324/579 (55%), Gaps = 40/579 (6%)
Query: 51 PGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTENVADIPDLDIVHAEDLLDLAS 110
PG +P P+K+ K+ K + +P TSP E DTE I + +L AS
Sbjct: 106 PGPAPVPEKLVPKRQQGVK--ISENPSLATSP-REVDTEMDKKIRSI----VSSILKNAS 158
Query: 111 ARVSEPPVQS----NVEK-STSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPETSSRKGKE 165
++ V + N E S+S E S + D A T AP G
Sbjct: 159 VPDADKDVPTSSTPNAEVLSSSSKEKSTEEEDQA---------TEETPAPRAPEPAPGDL 209
Query: 166 HVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST--IRTPKDSKAAGSVRKSKGET 223
+ + + D V I RL+ + K TP+ + I+T K+ T
Sbjct: 210 IDLEEVESDEEPIVKKLALGIAERLQSR-KGKTPITRSGRIKTIAQKKSTPIT-----PT 263
Query: 224 TSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVADVPLDNVSF 283
TS+ + + +KKRK +S +SD D E +PDI R + GK+VP NV D PLDN+SF
Sbjct: 264 TSRWSKVAIPSKKRKEISSSDSDDDVELDVPDIK---RAKKSGKKVPGNVPDAPLDNISF 320
Query: 284 HYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLVREF 343
H N +RWKFV RR+A+EREL +AL+ KEI+DL+ A L+KTV +G CYE LVREF
Sbjct: 321 HSIGNVERWKFVYQRRLALERELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREF 380
Query: 344 IVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDLNEIAKVISG 403
IVNI SD + S E+ VF R K + SP VIN++LGR T V + ++IAK I+
Sbjct: 381 IVNIPSDITNRKSDEYQTVFVRGKGIRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITA 440
Query: 404 KQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWIGTGAMMDF 463
KQV WPKKG L +GKL+ K+ +L +IG ANW+ TNHTS + L K ++ +GT + +F
Sbjct: 441 KQVQHWPKKGKLSAGKLSVKYAILHRIGTANWVPTNHTSTVATGLGKFLYAVGTKSKFNF 500
Query: 464 GQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPLNFDYRLFA 523
G ++F+ T KH S+AVKL I FP ++ ++L QHP+IL ++ K+ L+ Y+LF
Sbjct: 501 GNYIFDQTVKHSESFAVKLPIAFPTVLCGIMLSQHPNILNNIDSVKKRESALSLHYKLFE 560
Query: 524 GTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKVRKHNVDQL 583
GTHVP+IV + K A S + T KD ++AELK+ K L+ATI+A+ +K +++L
Sbjct: 561 GTHVPDIVSTSGKAAASGAVT-------KDALIAELKDTCKVLEATIKATTEKKMELERL 613
Query: 584 IKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDN 622
IK L+ D+ + A++E+ A+ +E+ ++ SDDD+
Sbjct: 614 IKRLSDSGIDD-GEAAEEEEEAAEEEEDAAEDTESDDDD 651
>gb|AAC64918.1| envelope-like [Glycine max]
Length = 444
Score = 204 bits (518), Expect = 9e-51
Identities = 143/372 (38%), Positives = 194/372 (51%), Gaps = 32/372 (8%)
Query: 51 PGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTENVADIPDLDIVHAEDLLDLAS 110
PG +P P+K+ K+ K + +P TSP E DTE I + +L AS
Sbjct: 98 PGQAPVPEKLVPKRQQGVK--ISENPSIATSP-REVDTEMDKKIRSI----VSSILKNAS 150
Query: 111 ARVSEPPVQS----NVEK-STSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPETSSRKGKE 165
++ V + N E S+S E+S + + A T AP G
Sbjct: 151 VPDADKDVPTSSTPNAEVLSSSSKEESTEEEEQA---------TEETPAPRAPEPAPGDL 201
Query: 166 HVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST--IRTPKDSKAAGSVRKSKGET 223
+ + + D + I RL+ + K TP+ + I+T K+ T
Sbjct: 202 IDLEEVESDEEPIANKLAPGIAERLQSR-KGKTPITRSGRIKTMAQKKSTPIT-----PT 255
Query: 224 TSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVADVPLDNVSF 283
TS+ + + +KKRK S +SD D E +PDI R + GK+VP NV D PLDN+SF
Sbjct: 256 TSRWSKVAIPSKKRKEFSSSDSDDDVELDVPDIK---RAKKSGKKVPGNVPDAPLDNISF 312
Query: 284 HYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLVREF 343
H N +RWKFV RR+A+EREL +AL+ KEI+DL+ A L+KTV +G CYE LVREF
Sbjct: 313 HSIGNVERWKFVYQRRLALERELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREF 372
Query: 344 IVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDLNEIAKVISG 403
IVNI SD + S E+ KVF R KCV SP VIN++LGR T V + ++IAK I+
Sbjct: 373 IVNIPSDITNRKSDEYQKVFVRGKCVRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITA 432
Query: 404 KQVMQWPKKGLL 415
KQV WPKKG L
Sbjct: 433 KQVQHWPKKGKL 444
>gb|AAL69985.1| putative envelope protein [Vicia faba]
Length = 59
Score = 69.7 bits (169), Expect = 3e-10
Identities = 31/58 (53%), Positives = 41/58 (70%)
Query: 430 IGAANWLATNHTSGITIPLAKLIFWIGTGAMMDFGQHVFEHTFKHVGSYAVKLSIMFP 487
IGAANW+ TNH S I+ L +LI+ +GT D+G+++F+ T KH GSYAVK I FP
Sbjct: 1 IGAANWVPTNHKSTISTGLGRLIYAVGTRTEFDYGRYIFDQTLKHAGSYAVKGPIAFP 58
>gb|AAF18631.1| F5J5.2 [Arabidopsis thaliana]
Length = 463
Score = 62.8 bits (151), Expect = 3e-08
Identities = 77/342 (22%), Positives = 133/342 (38%), Gaps = 57/342 (16%)
Query: 291 RWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLVREFIVNISSD 350
R+K + +R R L + LL L+ TV I ++V EF N+
Sbjct: 166 RYKVIGNRCFNDMRFLPLDGNNTASTQQLLFNGGLLPTVTEIDSYVHEVVMEFYANLPDG 225
Query: 351 YDDAGSAEFHKVFFREKCVHLSPVVINEFL----------GRSTTAVAEGESDLNEIAKV 400
+ G + VF R S +IN+ G S V E ++E+A
Sbjct: 226 --EEGDNLAYSVFVRRNMYEFSLAIINQMFQLPNPSYPLDGMSEIQVPES---MDEVAIA 280
Query: 401 ISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWIGTGAM 460
+S + W +L S L+ + +L+KI NW T + S + L++ +
Sbjct: 281 LSNGKANSWK---MLTSRLLSPELALLNKICCHNWSPTVNRSVLKPERMTLLYMVAKALP 337
Query: 461 MDFGQHVFEHTFK--HVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPLNFD 518
+FG+ +F+ + + L ++ P LI +++ Q I+++DE + PL F
Sbjct: 338 FNFGKLIFDENLGVFEFANPSSTLCLVLPNLIDQMLRFQR--IVRSDEGDTTSSAPLKFT 395
Query: 519 YRLFAGTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKVRKH 578
+ VP + ++P L +TL A++ VR
Sbjct: 396 MEV---KPVPLLQSDSP----------------------TLDAALETLIASLTTMHVR-- 428
Query: 579 NVDQLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDD 620
LA +VS D+ + D++EEDDD+ L D+
Sbjct: 429 --------LAGGEYSDVSYSVPDDVGDEDVEEEDDDDELDDN 462
>gb|AAK85503.1| Hypothetical protein Y55B1BR.3 [Caenorhabditis elegans]
gi|17556242|ref|NP_497198.1| chromo domain containing
protein (77.9 kD) (3B49) [Caenorhabditis elegans]
Length = 679
Score = 58.5 bits (140), Expect = 6e-07
Identities = 45/190 (23%), Positives = 82/190 (42%), Gaps = 27/190 (14%)
Query: 74 FSPPQDTSPISESDTENVADIPDLDIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSV 133
F P + + + EN ++ LD + E +DL+ +++PP +S + ++ SE+S
Sbjct: 141 FPDPVSNTEMWGEELENCQEL--LDAYNEEHDIDLSKTVMNKPPKKSKHQDTSDDSEESE 198
Query: 134 DVHDDAPPMKKSVIKTGSAKAPPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKK 193
D + K+P + SS+K + + DSD D+ D D +KK
Sbjct: 199 DEEE---------------KSPRKRSSKKRRAASVSDSDSDSDSDFD----------KKK 233
Query: 194 MKHATPVKSTIRTPKDSKAAGSVRKSKGETTSKGKGLNVSNKKRKHVSDHESDQDGEPTM 253
K +S D + R+ K + + K K S K+++ V+D SD + E
Sbjct: 234 WKKNKAKRSKRDDSSDDDSEMERRRKKSKKSKKSKKFKKSEKRKRAVNDSSSDDEDEEEK 293
Query: 254 PDISTTTRKR 263
P+ + K+
Sbjct: 294 PEKRSKKSKK 303
Score = 37.7 bits (86), Expect = 1.2
Identities = 55/251 (21%), Positives = 100/251 (38%), Gaps = 36/251 (14%)
Query: 44 IAAQSSSPGHSPTPKKMRTKQ------------VVTKKKTLRFSPPQDTSPISESDTENV 91
+ + +S TPKK K+ VV KKK+ + + + S + E V
Sbjct: 344 VEVKKNSKSPKKTPKKTAVKEESEESSGDEEEEVVKKKKSSKINK-RKAKESSSDEEEEV 402
Query: 92 ADIPDLDIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGS 151
+ P S++ S +S E S + E+ VD P KK S
Sbjct: 403 EESPKKKTKSPRK-----SSKKSAAKEESEEESSDNEEEEEVDYS----PKKKVKSPKKS 453
Query: 152 AKAPPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSK 211
+K P + K + D++ + + + +K ++ + K A V+ST +
Sbjct: 454 SKKP----AAKVESEEPSDNEEEEEEVEESPIKKDKTPRKYSRKSAAKVEST-------E 502
Query: 212 AAGSVRKSKGETTSKGKGLN--VSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRV 269
++G+ + + E + K KG S+KK V + +++++ P T+ RK K KR
Sbjct: 503 SSGNEEEEEVEESPKKKGKTPRKSSKKSAAVEESDNEEEDVEESPKKRTSPRKSSK-KRA 561
Query: 270 PMNVADVPLDN 280
++ DN
Sbjct: 562 AKEESEESSDN 572
>sp|P35663|CYLC1_HUMAN Cylicin-1 (Cylicin I) (Multiple-band polypeptide I)
gi|396105|emb|CAA80457.1| cylicin [Homo sapiens]
Length = 598
Score = 56.6 bits (135), Expect = 2e-06
Identities = 79/386 (20%), Positives = 147/386 (37%), Gaps = 28/386 (7%)
Query: 1 MSQESGKQSAPGLVPVKNAHGVRFLGLKSPSSHSTRVTRSSSGIAAQSSSPGH--SPTPK 58
+ +++ +Q+ P+K++H SS + +++S ++ S S K
Sbjct: 95 VEEKTKRQNEADKTPLKSSHENEQSKKSKSSSETNPESQNSKTVSKNCSQKDKKDSKNSK 154
Query: 59 KMRTKQVVTK---KKTLRFSPPQDTSPISESDTENVADIPDLDIVHAED----------- 104
K T+ + TK KK L+ S + PISE +EN ++ L +V D
Sbjct: 155 KTNTEFLHTKNNPKKDLKRSKTSN-DPISEICSENSLNVDFLMLVGQSDDESINFDAWLR 213
Query: 105 --------LLDLASARVSEPPVQSNVEKSTSF-SEDSVDVHDDAPPMKKSVIKTGSAK-A 154
L + ++ + N +KS+ SEDS D D+ +KK+V K K
Sbjct: 214 NYSQNNSKNYSLKYTKYTKKDTKKNAKKSSDAESEDSKDAKKDSKKVKKNVKKDDKKKDV 273
Query: 155 PPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSKAAG 214
+T S + D DT D K+ + + KK +T +S + +
Sbjct: 274 KKDTESTDAESGDSKDERKDTKKDKKKLKKDDKKKDTKKYPESTDTESGDAKDARNDSRN 333
Query: 215 SVRKSKGETTSK-GKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNV 273
+ SK + K K + S + ES +D + D T +K VK +
Sbjct: 334 LKKASKNDDKKKDAKKITFSTDSESELESKESQKDEKKDKKDSKTDNKKSVKNDEESTDA 393
Query: 274 ADVPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIG 333
P + D + K + + + E+ E + ++L + K KG
Sbjct: 394 DSEPKGDSKKGKKDEKKGKKDSKKDDKKKDAKKNAESTEMESDLELKKDKKHSKEKKGSK 453
Query: 334 RCYEKLVREFIVNISSDYDDAGSAEF 359
+ +K R+ + +++D++ F
Sbjct: 454 KDIKKDARKDTESTDAEFDESSKTGF 479
>emb|CAI40037.1| cylicin, basic protein of sperm head cytoskeleton 1 [Homo sapiens]
Length = 651
Score = 56.6 bits (135), Expect = 2e-06
Identities = 79/386 (20%), Positives = 147/386 (37%), Gaps = 28/386 (7%)
Query: 1 MSQESGKQSAPGLVPVKNAHGVRFLGLKSPSSHSTRVTRSSSGIAAQSSSPGH--SPTPK 58
+ +++ +Q+ P+K++H SS + +++S ++ S S K
Sbjct: 148 VEEKTKRQNEADKTPLKSSHENEQSKKSKSSSETNPESQNSKTVSKNCSQKDKKDSKNSK 207
Query: 59 KMRTKQVVTK---KKTLRFSPPQDTSPISESDTENVADIPDLDIVHAED----------- 104
K T+ + TK KK L+ S + PISE +EN ++ L +V D
Sbjct: 208 KTNTEFLHTKNNPKKDLKRSKTSN-DPISEICSENSLNVDFLMLVGQSDDESINFDAWLR 266
Query: 105 --------LLDLASARVSEPPVQSNVEKSTSF-SEDSVDVHDDAPPMKKSVIKTGSAK-A 154
L + ++ + N +KS+ SEDS D D+ +KK+V K K
Sbjct: 267 NYSQNNSKNYSLKYTKYTKKDTKKNAKKSSDAESEDSKDAKKDSKKVKKNVKKDDKKKDV 326
Query: 155 PPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSKAAG 214
+T S + D DT D K+ + + KK +T +S + +
Sbjct: 327 KKDTESTDAESGDSKDERKDTKKDKKKLKKDDKKKDTKKYPESTDTESGDAKDARNDSRN 386
Query: 215 SVRKSKGETTSK-GKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNV 273
+ SK + K K + S + ES +D + D T +K VK +
Sbjct: 387 LKKASKNDDKKKDAKKITFSTDSESELESKESQKDEKKDKKDSKTDNKKSVKNDEESTDA 446
Query: 274 ADVPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIG 333
P + D + K + + + E+ E + ++L + K KG
Sbjct: 447 DSEPKGDSKKGKKDEKKGKKDSKKDDKKKDAKKNAESTEMESDLELKKDKKHSKEKKGSK 506
Query: 334 RCYEKLVREFIVNISSDYDDAGSAEF 359
+ +K R+ + +++D++ F
Sbjct: 507 KDIKKDARKDTESTDAEFDESSKTGF 532
>ref|XP_088636.6| PREDICTED: cylicin, basic protein of sperm head cytoskeleton 1
[Homo sapiens]
Length = 650
Score = 56.6 bits (135), Expect = 2e-06
Identities = 79/386 (20%), Positives = 147/386 (37%), Gaps = 28/386 (7%)
Query: 1 MSQESGKQSAPGLVPVKNAHGVRFLGLKSPSSHSTRVTRSSSGIAAQSSSPGH--SPTPK 58
+ +++ +Q+ P+K++H SS + +++S ++ S S K
Sbjct: 147 VEEKTKRQNEADKTPLKSSHENEQSKKSKSSSETNPESQNSKTVSKNCSQKDKKDSKNSK 206
Query: 59 KMRTKQVVTK---KKTLRFSPPQDTSPISESDTENVADIPDLDIVHAED----------- 104
K T+ + TK KK L+ S + PISE +EN ++ L +V D
Sbjct: 207 KTNTEFLHTKNNPKKDLKRSKTSN-DPISEICSENSLNVDFLMLVGQSDDESINFDAWLR 265
Query: 105 --------LLDLASARVSEPPVQSNVEKSTSF-SEDSVDVHDDAPPMKKSVIKTGSAK-A 154
L + ++ + N +KS+ SEDS D D+ +KK+V K K
Sbjct: 266 NYSQNNSKNYSLKYTKYTKKDTKKNAKKSSDAESEDSKDAKKDSKKVKKNVKKDDKKKDV 325
Query: 155 PPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSKAAG 214
+T S + D DT D K+ + + KK +T +S + +
Sbjct: 326 KKDTESTDAESGDSKDERKDTKKDKKKLKKDDKKKDTKKYPESTDTESGDAKDARNDSRN 385
Query: 215 SVRKSKGETTSK-GKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNV 273
+ SK + K K + S + ES +D + D T +K VK +
Sbjct: 386 LKKASKNDDKKKDAKKITFSTDSESELESKESQKDEKKDKKDSKTDNKKSVKNDEESTDA 445
Query: 274 ADVPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIG 333
P + D + K + + + E+ E + ++L + K KG
Sbjct: 446 DSEPKGDSKKGKKDEKKGKKDSKKDDKKKDAKKNAESTEMESDLELKKDKKHSKEKKGSK 505
Query: 334 RCYEKLVREFIVNISSDYDDAGSAEF 359
+ +K R+ + +++D++ F
Sbjct: 506 KDIKKDARKDTESTDAEFDESSKTGF 531
>dbj|BAB11309.1| unnamed protein product [Arabidopsis thaliana]
Length = 515
Score = 55.5 bits (132), Expect = 5e-06
Identities = 57/247 (23%), Positives = 105/247 (42%), Gaps = 13/247 (5%)
Query: 281 VSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLV 340
+ F + +R+K R +R + E ++++ ++ L+ TV I ++++
Sbjct: 170 IQFLTSQGVERYKDFKLRDPYEQRYVPRGPPEMRDMMKVIEDNHLLDTVAEIDPFVKEVI 229
Query: 341 REFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTT---AVAEGES---DL 394
EF N ++ + KVF R K SP +IN+ A AE E
Sbjct: 230 WEFYANFHIFNEETKKS---KVFVRGKMYEFSPQMINKTFRSPAIKWHAKAEFERVTMSK 286
Query: 395 NEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFW 454
+ +AK +S +V+ W L + + L I NW+ + + S +T+ AK+++
Sbjct: 287 HALAKYLSDGKVVTWKD---LRTPVMPTHLAGLYLICCINWIPSKNNSNVTVDRAKMLYL 343
Query: 455 IGTGAMMDFGQHVFEHTFKHVGSYAVKLSIM-FPCLITELILQQHPSILQADEAPSKKGL 513
+ +FG+ V++ K S +I+ FP LI +++ Q L+ E P
Sbjct: 344 LNERIPFNFGKLVYDQVEKAARSPGQIQNILPFPNLIYRVLMFQRVITLENYEKPGPHPK 403
Query: 514 PLNFDYR 520
N D R
Sbjct: 404 LFNPDVR 410
>ref|XP_635213.1| hypothetical protein DDB0183896 [Dictyostelium discoideum]
gi|60463521|gb|EAL61706.1| hypothetical protein
DDB0183896 [Dictyostelium discoideum]
Length = 447
Score = 55.5 bits (132), Expect = 5e-06
Identities = 56/220 (25%), Positives = 86/220 (38%), Gaps = 10/220 (4%)
Query: 31 SSHSTRVTRSSSGIAAQSSSPGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTEN 90
SS +V + SS + S S S + + K++ KKT +D+S SESDTE+
Sbjct: 72 SSKKHKVEKDSSDSSDSSDSESDSSDSEDEKEKEMKVDKKT------EDSSSSSESDTES 125
Query: 91 VADIPDLDIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTG 150
+ D E D +S+ SE S+ +S S SE S ++ KK K
Sbjct: 126 SSSSESEDEKKKEKESDDSSSSESESDSSSSESESES-SESSSSSESESEDEKK---KES 181
Query: 151 SAKAPPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDS 210
S + E+ K K+ DS S+ D + E +KK++ + S + S
Sbjct: 182 SDSSDSESEDEKKKKVESSDSSSSESESSDSSDSESEDEKKKKVESSDSSSSESESSSSS 241
Query: 211 KAAGSVRKSKGETTSKGKGLNVSNKKRKHVSDHESDQDGE 250
+ S K K + S+ K D D E
Sbjct: 242 SSDSSDSSDSESEDEKKKKVEKSSDKETSSESESEDSDDE 281
Score = 45.1 bits (105), Expect = 0.007
Identities = 52/235 (22%), Positives = 93/235 (39%), Gaps = 17/235 (7%)
Query: 40 SSSGIAAQSSSPGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTENVADIPDLDI 99
S S + SSS K+ + + + S + S SES + + ++ D
Sbjct: 119 SESDTESSSSSESEDEKKKEKESDDSSSSESESDSSSSESESESSESSSSSESESEDEKK 178
Query: 100 VHAEDLLDLASARVSEPPVQSNVEKST-SFSEDSVDVHDDAPPMKKSVIKTGSAKAPPET 158
+ D D S + V+S+ S+ S S DS D + KK V + S+ + E+
Sbjct: 179 KESSDSSDSESEDEKKKKVESSDSSSSESESSDSSD-SESEDEKKKKVESSDSSSSESES 237
Query: 159 SSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSKAAGSVRK 218
SS + SD S+ D K +E K + S+ +DS +K
Sbjct: 238 SSSSSSD----SSDSSDSESEDEKKKKVE-------KSSDKETSSESESEDSDDEKDKKK 286
Query: 219 SKGETTSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNV 273
SK E + K +++K + +SD+D + M + ++ +K ++ V
Sbjct: 287 SKKEDKKENK----KQEEKKKPIEKDSDEDSDDEMKVVEQPKKQEIKQQQQQQTV 337
>gb|AAG52950.1| putative envelope protein [Arabidopsis thaliana]
Length = 461
Score = 55.5 bits (132), Expect = 5e-06
Identities = 57/247 (23%), Positives = 105/247 (42%), Gaps = 13/247 (5%)
Query: 281 VSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLV 340
+ F + +R+K R +R + E ++++ ++ L+ TV I ++++
Sbjct: 170 IQFLTSQGVERYKDFKLRDPYEQRYVPRGPPEMRDMMKVIEDNHLLDTVAEIDPFVKEVI 229
Query: 341 REFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTT---AVAEGES---DL 394
EF N ++ + KVF R K SP +IN+ A AE E
Sbjct: 230 WEFYANFHIFNEETKKS---KVFVRGKMYEFSPQMINKTFRSPAIKWHAKAEFERVTMSK 286
Query: 395 NEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFW 454
+ +AK +S +V+ W L + + L I NW+ + + S +T+ AK+++
Sbjct: 287 HALAKYLSDGKVVTWKD---LRTPVMPTHLAGLYLICCINWIPSKNNSNVTVDRAKMLYL 343
Query: 455 IGTGAMMDFGQHVFEHTFKHVGSYAVKLSIM-FPCLITELILQQHPSILQADEAPSKKGL 513
+ +FG+ V++ K S +I+ FP LI +++ Q L+ E P
Sbjct: 344 LNERIPFNFGKLVYDQVEKAARSPGQIQNILPFPNLIYRVLMFQRVITLENYEKPGPHPK 403
Query: 514 PLNFDYR 520
N D R
Sbjct: 404 LFNPDVR 410
>ref|XP_331012.1| predicted protein [Neurospora crassa] gi|28920929|gb|EAA30259.1|
predicted protein [Neurospora crassa]
Length = 577
Score = 55.5 bits (132), Expect = 5e-06
Identities = 71/312 (22%), Positives = 121/312 (38%), Gaps = 36/312 (11%)
Query: 27 LKSPSSHSTRVTRSSSGIAAQSSSPGHSPTPKK-----MRTKQVVTKKKTLRFSPPQDTS 81
LK + T+ T++++ SP PKK ++ K V TKK+ S +++
Sbjct: 80 LKKTNMAQTKATKAAA-----------SPAPKKAKAEPVKAKPVATKKEESESSSDSESA 128
Query: 82 PISESDTENVADIPDLD--IVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDA 139
SES++E+ +++ D + AE+ A A V + + +S E+S D++
Sbjct: 129 SESESESESESEVEDKKKPVKKAEEKKPAAPA-VKKAAKKDESSSESSSEEESGSGSDES 187
Query: 140 PPMKKSVIKTGSAKAPPETSSRKGKEHVILDSDGDTSDDVDVT-VKNIESRLRKKMKHAT 198
+ K P+T+ K K+ +S++ + ++ E K A
Sbjct: 188 SSDDEEETKPAPKATTPKTAPAKTKQQTAKQPTPSSSEEESSSESESDEEPAPKANTSAK 247
Query: 199 PVKSTIRTPKDSKAAGSVRKSKGETTSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDIST 258
K +TP+ KS ET SK + S + + S+ ES D E
Sbjct: 248 LAKPASKTPEAKPVVNGTSKS-NETVSKSDDESSSEEDEESGSEEESSSDEEMADAPQPK 306
Query: 259 TTRKRVKGKRVP--------MNVADVPLD------NVSFHYADNAQRWKFVSHRRIAIER 304
T K+ G ++P M AD D ++ Q W F + I IE
Sbjct: 307 TAPKKTSGPQLPQVIAPNFHMRKADANADAKDVARTFQKAQSEGKQIWYFTAPASIPIE- 365
Query: 305 ELSEEALEFKEI 316
+ E AL +I
Sbjct: 366 VIQEHALPLSKI 377
>dbj|BAC02896.1| tobacco nucleolin [Nicotiana tabacum]
Length = 620
Score = 55.5 bits (132), Expect = 5e-06
Identities = 65/235 (27%), Positives = 93/235 (38%), Gaps = 28/235 (11%)
Query: 40 SSSGIAAQSSSPGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTENVADIPDLDI 99
+SS + SS +P PKK+ ++ D S SE D+ + D+P
Sbjct: 103 ASSSDDSDSSDEDDAPPPKKVVAASAKNGVAAMKKDESSDDSS-SEDDSSSEEDVPAAKK 161
Query: 100 VHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPETS 159
A SA VS+ S+ S SED DDA KK+ K G A A +
Sbjct: 162 PAAN-----GSAAVSKKDESSD----QSSSEDDSSSEDDAVA-KKTAPKKGLAAAASK-- 209
Query: 160 SRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSKAAGSVRKS 219
KE DS D+S D D + + A P K+ +T S+ S
Sbjct: 210 ----KEESSEDSSDDSSSDDDEPASKVAA--------AQPAKAAKKTGDGSEETDSDEDD 257
Query: 220 KGETTSKGKGLNVSNKKRKHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVA 274
KGK VS KK + SD SD++ E + D + +K++K P V+
Sbjct: 258 SDSDVDKGKAAAVSKKKVESSSD-SSDEESEES--DEEGSQKKKIKPSSTPATVS 309
>ref|ZP_00156849.2| COG3468: Type V secretory pathway, adhesin AidA [Haemophilus
influenzae R2866]
Length = 1781
Score = 53.1 bits (126), Expect = 3e-05
Identities = 61/278 (21%), Positives = 115/278 (40%), Gaps = 42/278 (15%)
Query: 29 SPSSHSTRVTRSSSGIAAQS----------SSPGHSPTPKKMRTKQVV-TKKKTLRFSPP 77
SP+S T+SS G A+ S P + K+ +K+ T K + SP
Sbjct: 1234 SPNSKPAEETQSSEGTNAKPVTPVVSENKVSQPTETEKTAKVESKEAQETPKVASQASPK 1293
Query: 78 QDTS----PISESDTENVADIPDLDIVHAEDLLDLASARVSEPPVQ---SNVEKSTSFSE 130
Q+ S P +E ++E V + + + V A+ ++ +E P + SNVE+
Sbjct: 1294 QEQSETVQPQAELESEKVPTVKNAEEVQAQPQTQPSAIVSTEQPAKETSSNVEQ------ 1347
Query: 131 DSVDVHDDAPPMKKSVIKTGSAKAPPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRL 190
P ++S TGSA A ET+ + K + ++ D + + N +
Sbjct: 1348 ----------PEQESSTNTGSATAMTETAEKSDKPQMETVTENDRQPEANTVADNSVANN 1397
Query: 191 RKKMKHATPVKSTIRTPKDSKAAGSVRKSKGETT--SKGKGLNVSNKKRKHVSDHESDQD 248
+ + + + ++ PK++ + S +TT K + + R+ VS +
Sbjct: 1398 SESSESKSRRRRSVSQPKETSTEETTVTSTDKTTVADNSKSSKPNRRHRRSVSQPQETST 1457
Query: 249 GEPTMPDISTTTRKRVKGKR----VPMNVADVPLDNVS 282
+ T+ D S ++ + + +R P NV PL N++
Sbjct: 1458 EKTTVADNSKRSKPKRRSRRDVSSAPHNVE--PLHNLT 1493
>emb|CAB83070.1| putative protein [Arabidopsis thaliana] gi|11357591|pir||T47405
hypothetical protein F23N14.70 - Arabidopsis thaliana
Length = 539
Score = 52.8 bits (125), Expect = 3e-05
Identities = 48/203 (23%), Positives = 86/203 (41%), Gaps = 13/203 (6%)
Query: 276 VPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRC 335
VP F A A+R+K ++ R ++ L + L + + +TV + +
Sbjct: 235 VPPHTKRFLSAAAAERYKHIAKRDFIFQKTLPLDPEVLTATKYFLEHSGMAQTVVAVEQF 294
Query: 336 YEKLVREFIVNISS-DYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTA------VA 388
++VREF N+ +Y + G V+ R K SP +IN +A V
Sbjct: 295 VPEVVREFYANLPEMEYRECG---LDLVYVRGKMYEFSPALINHMFSIDDSALDPEAPVT 351
Query: 389 EGESDLNEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPL 448
+ +++A +++G +W + L P+ L +L K+ NW T +TS + +
Sbjct: 352 LSTASRDDLALMMTGGTTRRWLR--LQPADHLDT-MKMLHKVCCGNWFPTTNTSTLRVDR 408
Query: 449 AKLIFWIGTGAMMDFGQHVFEHT 471
+LI G + G+ V HT
Sbjct: 409 LRLIDMGTHGKSFNLGKLVVTHT 431
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.310 0.128 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,072,551,154
Number of Sequences: 2540612
Number of extensions: 46957261
Number of successful extensions: 346627
Number of sequences better than 10.0: 2657
Number of HSP's better than 10.0 without gapping: 950
Number of HSP's successfully gapped in prelim test: 1797
Number of HSP's that attempted gapping in prelim test: 285734
Number of HSP's gapped (non-prelim): 27922
length of query: 642
length of database: 863,360,394
effective HSP length: 134
effective length of query: 508
effective length of database: 522,918,386
effective search space: 265642540088
effective search space used: 265642540088
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0370.9


