

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0360.9
(107 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
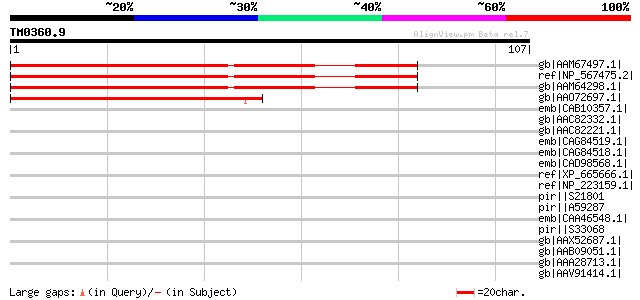
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM67497.1| unknown protein [Arabidopsis thaliana] gi|1817584... 80 2e-14
ref|NP_567475.2| expressed protein [Arabidopsis thaliana] 80 2e-14
gb|AAM64298.1| unknown [Arabidopsis thaliana] 78 5e-14
gb|AAO72697.1| unknown [Oryza sativa (japonica cultivar-group)] 68 6e-11
emb|CAB10357.1| hypothetical protein [Arabidopsis thaliana] gi|7... 37 0.12
gb|AAC82332.1| myosin [Schistosoma japonicum] 35 0.34
gb|AAC82221.1| myosin heavy chain [Schistosoma japonicum] 35 0.34
emb|CAG84519.1| unnamed protein product [Debaryomyces hansenii C... 35 0.58
emb|CAG84518.1| unnamed protein product [Debaryomyces hansenii C... 34 0.76
emb|CAD98568.1| kinesin heavy chain, possible [Cryptosporidium p... 34 0.76
ref|XP_665666.1| kinesin heavy chain [Cryptosporidium hominis] g... 34 0.76
ref|NP_223159.1| hypothetical protein jhp0441 [Helicobacter pylo... 34 0.76
pir||S21801 myosin heavy chain, neuronal [similarity] - rat 34 0.99
pir||A59287 myosin heavy chain - fluke (Schistosoma mansoni) (st... 34 0.99
emb|CAA46548.1| myosin II heavy chain [Schistosoma mansoni] 34 0.99
pir||S33068 myosin heavy chain - fluke (Schistosoma mansoni) (fr... 34 0.99
gb|AAX52687.1| CG15792-PD, isoform D [Drosophila melanogaster] g... 33 1.3
gb|AAB09051.1| nonmuscle myosin-II heavy chain [Drosophila melan... 33 1.3
gb|AAA28713.1| non-muscle myosin heavy chain 33 1.3
gb|AAV91414.1| tropomyosin 1 [Lonomia obliqua] 33 1.3
>gb|AAM67497.1| unknown protein [Arabidopsis thaliana] gi|18175846|gb|AAL59938.1|
unknown protein [Arabidopsis thaliana]
Length = 147
Score = 79.7 bits (195), Expect = 2e-14
Identities = 46/84 (54%), Positives = 61/84 (71%), Gaps = 9/84 (10%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EA 60
ELS LKKTLN+EVEQLR EF+DL+TTL+QQQ+DV+ASL++LGLQD S + S+ E
Sbjct: 73 ELSGLKKTLNLEVEQLREEFKDLKTTLNQQQDDVSASLKSLGLQD-SKEQIDKRSEVTEE 131
Query: 61 KIEEID*KEQQVLPEEDNSKDAEN 84
K+E L +DN+K+AE+
Sbjct: 132 KVE--------ALSTDDNAKEAEH 147
>ref|NP_567475.2| expressed protein [Arabidopsis thaliana]
Length = 191
Score = 79.7 bits (195), Expect = 2e-14
Identities = 46/84 (54%), Positives = 61/84 (71%), Gaps = 9/84 (10%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EA 60
ELS LKKTLN+EVEQLR EF+DL+TTL+QQQ+DV+ASL++LGLQD S + S+ E
Sbjct: 117 ELSGLKKTLNLEVEQLREEFKDLKTTLNQQQDDVSASLKSLGLQD-SKEQIDKRSEVTEE 175
Query: 61 KIEEID*KEQQVLPEEDNSKDAEN 84
K+E L +DN+K+AE+
Sbjct: 176 KVE--------ALSTDDNAKEAEH 191
>gb|AAM64298.1| unknown [Arabidopsis thaliana]
Length = 147
Score = 78.2 bits (191), Expect = 5e-14
Identities = 45/84 (53%), Positives = 61/84 (72%), Gaps = 9/84 (10%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EA 60
ELS LK+TLN+EVEQLR EF+DL+TTL+QQQ+DV+ASL++LGLQD S + S+ E
Sbjct: 73 ELSGLKQTLNLEVEQLREEFKDLKTTLNQQQDDVSASLKSLGLQD-SKEQIDKRSEVTEE 131
Query: 61 KIEEID*KEQQVLPEEDNSKDAEN 84
K+E L +DN+K+AE+
Sbjct: 132 KVE--------ALSTDDNAKEAEH 147
>gb|AAO72697.1| unknown [Oryza sativa (japonica cultivar-group)]
Length = 176
Score = 67.8 bits (164), Expect = 6e-11
Identities = 34/53 (64%), Positives = 44/53 (82%), Gaps = 1/53 (1%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVS-ADGKK 52
ELS++K LN E+EQLR++FQ+LRTTL +QQEDV+ SL+NLGLQD + DG K
Sbjct: 122 ELSDIKTALNSEIEQLRSDFQELRTTLKKQQEDVSNSLKNLGLQDATDNDGNK 174
>emb|CAB10357.1| hypothetical protein [Arabidopsis thaliana]
gi|7268327|emb|CAB78621.1| hypothetical protein
[Arabidopsis thaliana] gi|7485072|pir||C71423
hypothetical protein - Arabidopsis thaliana
Length = 123
Score = 37.0 bits (84), Expect = 0.12
Identities = 24/54 (44%), Positives = 32/54 (58%), Gaps = 4/54 (7%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAE 54
ELS LKKTLN+EVEQLR E QD + + ++ E + L D + K+AE
Sbjct: 73 ELSGLKKTLNLEVEQLR-ELQDSKEQIDKRSEVTEEKVEALSTDD---NAKEAE 122
>gb|AAC82332.1| myosin [Schistosoma japonicum]
Length = 802
Score = 35.4 bits (80), Expect = 0.34
Identities = 24/96 (25%), Positives = 48/96 (50%), Gaps = 3/96 (3%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTL---HQQQEDVTASLRNLGLQDVSADGKKAESQA* 58
LSE+KK ++ E+E+L+ + +DL ++L Q+++ +R L + D +
Sbjct: 259 LSEVKKKMSAEIEELKKDVEDLESSLQKAEQEKQTKDNQIRTLQAEMAQQDETIGKLNKD 318
Query: 59 EAKIEEID*KEQQVLPEEDNSKDAEN*TGFSLEYLL 94
+ +EE + + Q+ L E++ + N LE L
Sbjct: 319 KKNLEEQNKRTQEALQAEEDKVNHLNKLKAKLESTL 354
>gb|AAC82221.1| myosin heavy chain [Schistosoma japonicum]
Length = 528
Score = 35.4 bits (80), Expect = 0.34
Identities = 24/96 (25%), Positives = 48/96 (50%), Gaps = 3/96 (3%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTL---HQQQEDVTASLRNLGLQDVSADGKKAESQA* 58
LSE+KK ++ E+E+L+ + +DL ++L Q+++ +R L + D +
Sbjct: 219 LSEVKKKMSAEIEELKKDVEDLESSLQKAEQEKQTKDNQIRTLQAEMAQQDETIGKLNKD 278
Query: 59 EAKIEEID*KEQQVLPEEDNSKDAEN*TGFSLEYLL 94
+ +EE + + Q+ L E++ + N LE L
Sbjct: 279 KKNLEEQNKRTQEALQAEEDKVNHLNKLKAKLESTL 314
>emb|CAG84519.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50405845|ref|XP_456563.1| unnamed protein product
[Debaryomyces hansenii]
Length = 224
Score = 34.7 bits (78), Expect = 0.58
Identities = 17/49 (34%), Positives = 31/49 (62%), Gaps = 1/49 (2%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSAD 49
E +LK+ LN +V+ L+ EF+DL+ L++ +D+ ++L +DV D
Sbjct: 68 EFKDLKRDLNKDVKDLKIEFKDLKRDLNKDVKDLKIEFKDL-KRDVKRD 115
>emb|CAG84518.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50405843|ref|XP_456562.1| unnamed protein product
[Debaryomyces hansenii]
Length = 78
Score = 34.3 bits (77), Expect = 0.76
Identities = 14/41 (34%), Positives = 27/41 (65%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNL 41
E +LK+ LN +V+ L+ EF+DL+ L++ +D+ ++L
Sbjct: 36 EFKDLKRDLNKDVKDLKIEFKDLKRDLNKDVKDLKGEFKDL 76
>emb|CAD98568.1| kinesin heavy chain, possible [Cryptosporidium parvum]
Length = 757
Score = 34.3 bits (77), Expect = 0.76
Identities = 28/83 (33%), Positives = 46/83 (54%), Gaps = 8/83 (9%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA---- 57
+SE K+ LN+++ L Q+L T +QQQ D ++ +L L++ S G+ +SQ
Sbjct: 492 MSEEKENLNIKLSDL---IQELSQTKYQQQ-DQAETVEHLQLKNKSLIGELEQSQIHIHD 547
Query: 58 *EAKIEEID*KEQQVLPEEDNSK 80
EA+IEE +E Q EE ++
Sbjct: 548 LEARIEEYKSEESQRRNEEKENE 570
>ref|XP_665666.1| kinesin heavy chain [Cryptosporidium hominis]
gi|54656457|gb|EAL35436.1| kinesin heavy chain
[Cryptosporidium hominis]
Length = 757
Score = 34.3 bits (77), Expect = 0.76
Identities = 28/83 (33%), Positives = 46/83 (54%), Gaps = 8/83 (9%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA---- 57
+SE K+ LN+++ L Q+L T +QQQ D ++ +L L++ S G+ +SQ
Sbjct: 492 MSEEKENLNIKLSDL---IQELSQTKYQQQ-DQAETVEHLQLKNKSLIGELEQSQIHIHD 547
Query: 58 *EAKIEEID*KEQQVLPEEDNSK 80
EA+IEE +E Q EE ++
Sbjct: 548 LEARIEEYKSEESQRRNEEKENE 570
>ref|NP_223159.1| hypothetical protein jhp0441 [Helicobacter pylori J99]
gi|4154976|gb|AAD06019.1| putative [Helicobacter pylori
J99] gi|7464724|pir||A71932 hypothetical protein jhp0441
- Helicobacter pylori (strain J99)
Length = 181
Score = 34.3 bits (77), Expect = 0.76
Identities = 20/82 (24%), Positives = 44/82 (53%), Gaps = 10/82 (12%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EAK 61
LS ++L ++++ F + T++H +++VTA+L N +++ ++ KK E+Q E
Sbjct: 81 LSNAGESLKTKMKEYERFFSEFNTSMHANEQEVTATL-NANTENIKSEIKKLENQLIE-- 137
Query: 62 IEEID*KEQQVLPEEDNSKDAE 83
E ++L E++ K +
Sbjct: 138 -------ETRMLLEQETQKSVK 152
>pir||S21801 myosin heavy chain, neuronal [similarity] - rat
Length = 1999
Score = 33.9 bits (76), Expect = 0.99
Identities = 26/72 (36%), Positives = 39/72 (54%), Gaps = 7/72 (9%)
Query: 6 KKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA--*EAKIE 63
K+ L E+E L+TE +DL + QQE LR+ Q+V+ K E +A EA+I+
Sbjct: 1133 KRDLGEELEALKTELEDLTDSTAAQQE-----LRSKREQEVNILKKTLEEEAKTHEAQIQ 1187
Query: 64 EID*KEQQVLPE 75
E+ K Q + E
Sbjct: 1188 EMRQKHSQAVEE 1199
>pir||A59287 myosin heavy chain - fluke (Schistosoma mansoni) (strain Brazilian
LE) gi|161044|gb|AAA29905.1| myosin heavy chain
Length = 1940
Score = 33.9 bits (76), Expect = 0.99
Identities = 23/96 (23%), Positives = 48/96 (49%), Gaps = 3/96 (3%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTL---HQQQEDVTASLRNLGLQDVSADGKKAESQA* 58
L+E+KK ++ E+E+L+ + +DL ++L Q+++ +R L + D +
Sbjct: 922 LTEVKKKMSAEIEELKKDVEDLESSLQKAEQEKQTKDNQIRTLQSEMAQQDEMIGKLNKD 981
Query: 59 EAKIEEID*KEQQVLPEEDNSKDAEN*TGFSLEYLL 94
+ +EE + + Q+ L E++ + N LE L
Sbjct: 982 KKNLEEQNKRTQEALQAEEDKVNHLNKLKAKLESTL 1017
>emb|CAA46548.1| myosin II heavy chain [Schistosoma mansoni]
Length = 528
Score = 33.9 bits (76), Expect = 0.99
Identities = 23/96 (23%), Positives = 48/96 (49%), Gaps = 3/96 (3%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTL---HQQQEDVTASLRNLGLQDVSADGKKAESQA* 58
L+E+KK ++ E+E+L+ + +DL ++L Q+++ +R L + D +
Sbjct: 219 LTEVKKKMSAEIEELKKDVEDLESSLQKAEQEKQTKDNEIRTLQSEMAQQDEMIGKLNKD 278
Query: 59 EAKIEEID*KEQQVLPEEDNSKDAEN*TGFSLEYLL 94
+ +EE + + Q+ L E++ + N LE L
Sbjct: 279 KKNLEEENKRTQEALQAEEDKVNHLNKLKAKLESTL 314
>pir||S33068 myosin heavy chain - fluke (Schistosoma mansoni) (fragment)
Length = 527
Score = 33.9 bits (76), Expect = 0.99
Identities = 23/96 (23%), Positives = 48/96 (49%), Gaps = 3/96 (3%)
Query: 2 LSELKKTLNVEVEQLRTEFQDLRTTL---HQQQEDVTASLRNLGLQDVSADGKKAESQA* 58
L+E+KK ++ E+E+L+ + +DL ++L Q+++ +R L + D +
Sbjct: 219 LTEVKKKMSAEIEELKKDVEDLESSLQKAEQEKQTKDNEIRTLQSEMAQQDEMIGKLNKD 278
Query: 59 EAKIEEID*KEQQVLPEEDNSKDAEN*TGFSLEYLL 94
+ +EE + + Q+ L E++ + N LE L
Sbjct: 279 KKNLEEENKRTQEALQAEEDKVNHLNKLKAKLESTL 314
>gb|AAX52687.1| CG15792-PD, isoform D [Drosophila melanogaster]
gi|62471805|ref|NP_001014552.1| CG15792-PD, isoform D
[Drosophila melanogaster]
Length = 2016
Score = 33.5 bits (75), Expect = 1.3
Identities = 27/80 (33%), Positives = 44/80 (54%), Gaps = 8/80 (10%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EA 60
+++EL+ L E+E+ R+E Q+ T L Q+ E++T L L+ SA K A + E+
Sbjct: 1303 QIAELQVKL-AEIERARSELQEKCTKLQQEAENITNQLEEAELK-ASAAVKSASNM--ES 1358
Query: 61 KIEEID*KEQQVLPEEDNSK 80
++ E QQ+L EE K
Sbjct: 1359 QLTE----AQQLLEEETRQK 1374
>gb|AAB09051.1| nonmuscle myosin-II heavy chain [Drosophila melanogaster]
Length = 1972
Score = 33.5 bits (75), Expect = 1.3
Identities = 27/80 (33%), Positives = 44/80 (54%), Gaps = 8/80 (10%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EA 60
+++EL+ L E+E+ R+E Q+ T L Q+ E++T L L+ SA K A + E+
Sbjct: 1259 QIAELQVKL-AEIERARSELQEKCTKLQQEAENITNQLEEAELK-ASAAVKSASNM--ES 1314
Query: 61 KIEEID*KEQQVLPEEDNSK 80
++ E QQ+L EE K
Sbjct: 1315 QLTE----AQQLLEEETRQK 1330
>gb|AAA28713.1| non-muscle myosin heavy chain
Length = 1972
Score = 33.5 bits (75), Expect = 1.3
Identities = 27/80 (33%), Positives = 44/80 (54%), Gaps = 8/80 (10%)
Query: 1 ELSELKKTLNVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EA 60
+++EL+ L E+E+ R+E Q+ T L Q+ E++T L L+ SA K A + E+
Sbjct: 1259 QIAELQVKL-AEIERARSELQEKCTKLQQEAENITNQLEEAELK-ASAAVKSASNM--ES 1314
Query: 61 KIEEID*KEQQVLPEEDNSK 80
++ E QQ+L EE K
Sbjct: 1315 QLTE----AQQLLEEETRQK 1330
>gb|AAV91414.1| tropomyosin 1 [Lonomia obliqua]
Length = 284
Score = 33.5 bits (75), Expect = 1.3
Identities = 21/74 (28%), Positives = 39/74 (52%), Gaps = 2/74 (2%)
Query: 10 NVEVEQLRTEFQDLRTTLHQQQEDVTASLRNLGLQDVSADGKKAESQA*EAKIEEID*KE 69
N+ E++ E ++L+ L Q +ED+ + L + + K+ + A EA++ ++ K
Sbjct: 33 NLRAEKVNEEVRELQKKLAQVEEDLILNKNKLEQANKDLEEKEKQLTATEAEVAALNRKV 92
Query: 70 QQVLPEEDNSKDAE 83
QQ+ EED K E
Sbjct: 93 QQI--EEDLEKSEE 104
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.331 0.144 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 132,999,265
Number of Sequences: 2540612
Number of extensions: 3973252
Number of successful extensions: 24358
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 88
Number of HSP's that attempted gapping in prelim test: 24207
Number of HSP's gapped (non-prelim): 237
length of query: 107
length of database: 863,360,394
effective HSP length: 83
effective length of query: 24
effective length of database: 652,489,598
effective search space: 15659750352
effective search space used: 15659750352
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0360.9


