

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0349b.8
(340 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
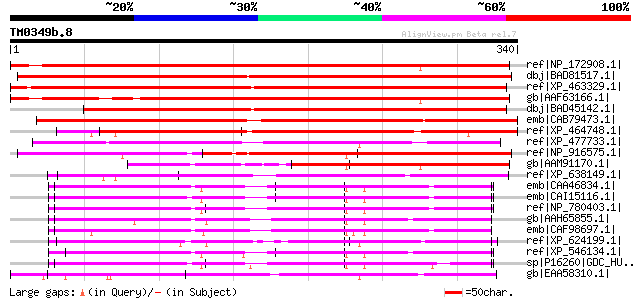
ref|NP_172908.1| mitochondrial substrate carrier family protein ... 442 e-123
dbj|BAD81517.1| Graves disease mitochondrial solute carrier prot... 418 e-115
ref|XP_463329.1| putative mitochondrial carrier [Oryza sativa (j... 414 e-114
gb|AAF63166.1| T5E21.6 [Arabidopsis thaliana] 400 e-110
dbj|BAD45142.1| mitochondrial carrier protein-like [Oryza sativa... 374 e-102
emb|CAB79473.1| putative mitochondrial carrier protein [Arabidop... 355 1e-96
ref|XP_464748.1| putative mitochondrial solute carrier protein [... 291 3e-77
ref|XP_477733.1| putative mitochondrial carrier protein [Oryza s... 259 6e-68
ref|NP_916575.1| P0456F08.3 [Oryza sativa (japonica cultivar-gro... 223 5e-57
gb|AAM91170.1| putative mitochondrial carrier protein [Arabidops... 221 2e-56
ref|XP_638149.1| hypothetical protein DDB0186597 [Dictyostelium ... 218 2e-55
emb|CAA46834.1| Graves disease carrier protein from bovine heart... 215 2e-54
emb|CAI15116.1| solute carrier family 25 (mitochondrial carrier\... 214 3e-54
ref|NP_780403.1| solute carrier family 25 (mitochondrial carrier... 213 7e-54
gb|AAH65855.1| Unknown (protein for MGC:77742) [Danio rerio] gi|... 213 9e-54
emb|CAF98697.1| unnamed protein product [Tetraodon nigroviridis] 212 1e-53
ref|XP_624199.1| PREDICTED: similar to ENSANGP00000008222, parti... 210 4e-53
ref|XP_546134.1| PREDICTED: similar to solute carrier family 25,... 205 2e-51
sp|P16260|GDC_HUMAN Grave's disease carrier protein (GDC) (Grave... 201 2e-50
gb|EAA58310.1| hypothetical protein AN5801.2 [Aspergillus nidula... 201 3e-50
>ref|NP_172908.1| mitochondrial substrate carrier family protein [Arabidopsis
thaliana]
Length = 331
Score = 442 bits (1136), Expect = e-123
Identities = 220/337 (65%), Positives = 262/337 (77%), Gaps = 10/337 (2%)
Query: 1 MDSSQGSTLAGLVDNASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQ 60
M SSQGSTL+ V S D +PV K L+AGG AGA++KTAVAPLER+KIL Q
Sbjct: 1 MGSSQGSTLSADV--------MSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQ 52
Query: 61 TRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNC 120
TRT F TLGV QSL K+++ +G LG YKGNGAS +RI+PYAALH+MTYE Y+ WIL
Sbjct: 53 TRTNDFKTLGVSQSLKKVLQFDGPLGFYKGNGASVIRIIPYAALHYMTYEVYRDWILEKN 112
Query: 121 PALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHI 180
LGSGP +DL+AGSAAGGT+VLCTYPLDLARTKLAYQV DTR +++ G G QP +
Sbjct: 113 LPLGSGPIVDLVAGSAAGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYS 172
Query: 181 GIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLS 240
GIK VL Y+EGG RGLYRG+GPT+ GILPYAGLKFY YE+LK HVPEEHQ S+ M L
Sbjct: 173 GIKEVLAMAYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLP 232
Query: 241 CGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHE--DTRYRNTLDGLRKIVRNQGWRQLF 298
CGALAGLFGQT+TYPLDVV+RQMQV +LQ E + RY+NT DGL IVR QGW+QLF
Sbjct: 233 CGALAGLFGQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLF 292
Query: 299 AGVSINYIRIVPSAAISFTAYDTMKAWLGIPPQQKSR 335
AG+SINYI+IVPS AI FT Y++MK+W+ IPP+++S+
Sbjct: 293 AGLSINYIKIVPSVAIGFTVYESMKSWMRIPPRERSK 329
>dbj|BAD81517.1| Graves disease mitochondrial solute carrier protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 337
Score = 418 bits (1074), Expect = e-115
Identities = 211/332 (63%), Positives = 253/332 (75%), Gaps = 2/332 (0%)
Query: 6 GSTLAGLVDNASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGG 65
G+ G A+ D D VPV+ KE++AGGVAGA SKTA+APLER+KIL QTRT
Sbjct: 2 GTPSQGSAAVAAAGVDLCVLDLVPVFAKEMIAGGVAGAFSKTAIAPLERLKILLQTRTNE 61
Query: 66 FHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGS 125
F +LGV +SL KL +H+G LG YKGNGAS +RIVPYAALH+M YERY+ WILNNCP+LG+
Sbjct: 62 FSSLGVLKSLKKLKQHDGILGFYKGNGASVLRIVPYAALHYMAYERYRCWILNNCPSLGT 121
Query: 126 GPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSV 185
GP +DLLAGSA+GGT+VLCTYPLDLARTKLA+QV ++ I G+K QP + GIK V
Sbjct: 122 GPLVDLLAGSASGGTAVLCTYPLDLARTKLAFQV-NSSDQISSGLKRTNFQPKYGGIKDV 180
Query: 186 LTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCGALA 245
VY EGG R LYRGVGPT+ GILPYAGLKFY YE LK HVPE+++ S+ ++LSCGA A
Sbjct: 181 FRGVYSEGGVRALYRGVGPTLMGILPYAGLKFYIYEGLKAHVPEDYKNSVTLKLSCGAAA 240
Query: 246 GLFGQTLTYPLDVVKRQMQVGSLQ-NAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSIN 304
GLFGQTLTYPLDVV+RQMQV S Q + + R T GL I + QGWRQLFAG+S+N
Sbjct: 241 GLFGQTLTYPLDVVRRQMQVQSQQYHDKFGGPQIRGTFQGLMIIKQTQGWRQLFAGLSLN 300
Query: 305 YIRIVPSAAISFTAYDTMKAWLGIPPQQKSRS 336
YI++VPS AI FTAYDTMK+ L IPP++K S
Sbjct: 301 YIKVVPSVAIGFTAYDTMKSLLKIPPREKKMS 332
>ref|XP_463329.1| putative mitochondrial carrier [Oryza sativa (japonica
cultivar-group)] gi|20161078|dbj|BAB90009.1|
mitochondrial carrier protein-like [Oryza sativa
(japonica cultivar-group)]
Length = 340
Score = 414 bits (1064), Expect = e-114
Identities = 211/335 (62%), Positives = 261/335 (76%), Gaps = 5/335 (1%)
Query: 1 MDSSQGSTLAGLVDNASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQ 60
M SSQ ST + + + Q + D +PVY KEL+AGG AGA +KTAVAPLERVKIL Q
Sbjct: 1 MGSSQESTFSSA--STAAQVNACALDLLPVYAKELIAGGAAGAFAKTAVAPLERVKILLQ 58
Query: 61 TRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNN- 119
TRT GF +LG+ QSL KL ++EG G YKGNGAS +RIVPYAALH+MTYE+Y+ WILNN
Sbjct: 59 TRTHGFQSLGILQSLRKLWQYEGIRGFYKGNGASVLRIVPYAALHYMTYEQYRCWILNNF 118
Query: 120 CPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAH 179
P++G+GP +DLLAGSAAGGT+VLCTYPLDLARTKLAYQV + G + + QPA+
Sbjct: 119 APSVGTGPVVDLLAGSAAGGTAVLCTYPLDLARTKLAYQVSNV-GQPGNALGNAGRQPAY 177
Query: 180 IGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRL 239
GIK V +VY+EGGAR LYRGVGPT+ GILPYAGLKFY YE LK VPE++++S++++L
Sbjct: 178 GGIKDVFKTVYKEGGARALYRGVGPTLIGILPYAGLKFYIYEDLKSRVPEDYKRSVVLKL 237
Query: 240 SCGALAGLFGQTLTYPLDVVKRQMQVGSLQ-NAAHEDTRYRNTLDGLRKIVRNQGWRQLF 298
SCGALAGLFGQTLTYPLDVV+RQMQV + Q + A++ R R T GL I+R QGWRQLF
Sbjct: 238 SCGALAGLFGQTLTYPLDVVRRQMQVQNKQPHNANDAFRIRGTFQGLALIIRCQGWRQLF 297
Query: 299 AGVSINYIRIVPSAAISFTAYDTMKAWLGIPPQQK 333
AG+S+NY+++VPS AI FT YD MK L +PP+++
Sbjct: 298 AGLSLNYVKVVPSVAIGFTTYDMMKNLLRVPPRER 332
>gb|AAF63166.1| T5E21.6 [Arabidopsis thaliana]
Length = 319
Score = 400 bits (1029), Expect = e-110
Identities = 207/337 (61%), Positives = 246/337 (72%), Gaps = 22/337 (6%)
Query: 1 MDSSQGSTLAGLVDNASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQ 60
M SSQGSTL+ V S D +PV K L+AGG AGA++KTAVAPLER+KIL Q
Sbjct: 1 MGSSQGSTLSADV--------MSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQ 52
Query: 61 TRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNC 120
L LN H +GNGAS +RI+PYAALH+MTYE Y+ WIL
Sbjct: 53 --------LSSTTLLNSCDVHNS----RRGNGASVIRIIPYAALHYMTYEVYRDWILEKN 100
Query: 121 PALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHI 180
LGSGP +DL+AGSAAGGT+VLCTYPLDLARTKLAYQV DTR +++ G G QP +
Sbjct: 101 LPLGSGPIVDLVAGSAAGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYS 160
Query: 181 GIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLS 240
GIK VL Y+EGG RGLYRG+GPT+ GILPYAGLKFY YE+LK HVPEEHQ S+ M L
Sbjct: 161 GIKEVLAMAYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLP 220
Query: 241 CGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHE--DTRYRNTLDGLRKIVRNQGWRQLF 298
CGALAGLFGQT+TYPLDVV+RQMQV +LQ E + RY+NT DGL IVR QGW+QLF
Sbjct: 221 CGALAGLFGQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLF 280
Query: 299 AGVSINYIRIVPSAAISFTAYDTMKAWLGIPPQQKSR 335
AG+SINYI+IVPS AI FT Y++MK+W+ IPP+++S+
Sbjct: 281 AGLSINYIKIVPSVAIGFTVYESMKSWMRIPPRERSK 317
>dbj|BAD45142.1| mitochondrial carrier protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 330
Score = 374 bits (960), Expect = e-102
Identities = 186/286 (65%), Positives = 230/286 (80%), Gaps = 3/286 (1%)
Query: 50 APLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTY 109
+PLERVKIL QTRT GF +LG+ QSL KL ++EG G YKGNGAS +RIVPYAALH+MTY
Sbjct: 38 SPLERVKILLQTRTHGFQSLGILQSLRKLWQYEGIRGFYKGNGASVLRIVPYAALHYMTY 97
Query: 110 ERYKSWILNN-CPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKD 168
E+Y+ WILNN P++G+GP +DLLAGSAAGGT+VLCTYPLDLARTKLAYQV + G +
Sbjct: 98 EQYRCWILNNFAPSVGTGPVVDLLAGSAAGGTAVLCTYPLDLARTKLAYQVSNV-GQPGN 156
Query: 169 GIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVP 228
+ QPA+ GIK V +VY+EGGAR LYRGVGPT+ GILPYAGLKFY YE LK VP
Sbjct: 157 ALGNAGRQPAYGGIKDVFKTVYKEGGARALYRGVGPTLIGILPYAGLKFYIYEDLKSRVP 216
Query: 229 EEHQKSILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQ-NAAHEDTRYRNTLDGLRK 287
E++++S++++LSCGALAGLFGQTLTYPLDVV+RQMQV + Q + A++ R R T GL
Sbjct: 217 EDYKRSVVLKLSCGALAGLFGQTLTYPLDVVRRQMQVQNKQPHNANDAFRIRGTFQGLAL 276
Query: 288 IVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAWLGIPPQQK 333
I+R QGWRQLFAG+S+NY+++VPS AI FT YD MK L +PP+++
Sbjct: 277 IIRCQGWRQLFAGLSLNYVKVVPSVAIGFTTYDMMKNLLRVPPRER 322
>emb|CAB79473.1| putative mitochondrial carrier protein [Arabidopsis thaliana]
gi|4538947|emb|CAB39683.1| putative mitochondrial
carrier protein [Arabidopsis thaliana]
gi|15236140|ref|NP_194348.1| mitochondrial substrate
carrier family protein [Arabidopsis thaliana]
gi|7441723|pir||T04273 hypothetical protein F20B18.290 -
Arabidopsis thaliana
Length = 325
Score = 355 bits (910), Expect = 1e-96
Identities = 176/323 (54%), Positives = 229/323 (70%), Gaps = 11/323 (3%)
Query: 19 QKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKL 78
++ D +P++ KEL+AGGV G ++KTAVAPLER+KIL+QTR F +G+ S+NK+
Sbjct: 5 EEKNGIIDSIPLFAKELIAGGVTGGIAKTAVAPLERIKILFQTRRDEFKRIGLVGSINKI 64
Query: 79 VKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAG 138
K EG +G Y+GNGAS RIVPYAALH+M YE Y+ WI+ P GP +DL+AGS AG
Sbjct: 65 GKTEGLMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGFPDTTRGPLLDLVAGSFAG 124
Query: 139 GTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVR-PQPAHIGIKSVLTSVYREGGARG 197
GT+VL TYPLDL RTKLAYQ +K + Q + GI + YRE GARG
Sbjct: 125 GTAVLFTYPLDLVRTKLAYQT---------QVKAIPVEQIIYRGIVDCFSRTYRESGARG 175
Query: 198 LYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCGALAGLFGQTLTYPLD 257
LYRGV P++ GI PYAGLKFY YE++K HVP EH++ I ++L CG++AGL GQTLTYPLD
Sbjct: 176 LYRGVAPSLYGIFPYAGLKFYFYEEMKRHVPPEHKQDISLKLVCGSVAGLLGQTLTYPLD 235
Query: 258 VVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFT 317
VV+RQMQV L +A E+TR R T+ L KI R +GW+QLF+G+SINY+++VPS AI FT
Sbjct: 236 VVRRQMQVERLYSAVKEETR-RGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFT 294
Query: 318 AYDTMKAWLGIPPQQKSRSVSAT 340
YD MK L +PP+++ + + T
Sbjct: 295 VYDIMKLHLRVPPREEPEAEAVT 317
>ref|XP_464748.1| putative mitochondrial solute carrier protein [Oryza sativa
(japonica cultivar-group)] gi|49388534|dbj|BAD25656.1|
putative mitochondrial solute carrier protein [Oryza
sativa (japonica cultivar-group)]
Length = 426
Score = 291 bits (744), Expect = 3e-77
Identities = 144/301 (47%), Positives = 197/301 (64%), Gaps = 28/301 (9%)
Query: 61 TRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNC 120
TR FH G+ S + + EG LG Y+GNGAS RIVPYAALH+M YE Y+ WI+
Sbjct: 121 TRRAEFHGSGLIGSFRTISRTEGLLGFYRGNGASVARIVPYAALHYMAYEEYRRWIILGF 180
Query: 121 PALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRP-QPAH 179
P + GP +DL+AGS AGGT+V+CTYPLDL RTKLAYQV +G +K ++ +P + +
Sbjct: 181 PNVEQGPILDLVAGSIAGGTAVICTYPLDLVRTKLAYQV---KGAVKLSLREYKPSEQVY 237
Query: 180 IGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRL 239
GI + ++YR+ G RGLYRG+ P++ GI PY+GLKFY YE +K +VPEEH+K I+ +L
Sbjct: 238 KGILDCVKTIYRQNGLRGLYRGMAPSLYGIFPYSGLKFYFYETMKTYVPEEHRKDIIAKL 297
Query: 240 SCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFA 299
+CG++AGL GQT+TYPLDVV+RQMQ S N + + T + I ++QGWRQLF+
Sbjct: 298 ACGSVAGLLGQTITYPLDVVRRQMQAFSSSNL----EKGKGTFGSIAMIAKHQGWRQLFS 353
Query: 300 GVSINYI--------------------RIVPSAAISFTAYDTMKAWLGIPPQQKSRSVSA 339
G+SINY+ ++VPS AI FT YD+MK WL +P ++ + +
Sbjct: 354 GLSINYLKELYQLDTSVCAFIYVQCGEKVVPSVAIGFTVYDSMKVWLKVPSREDTAIAAL 413
Query: 340 T 340
T
Sbjct: 414 T 414
Score = 53.9 bits (128), Expect = 7e-06
Identities = 41/138 (29%), Positives = 62/138 (44%), Gaps = 18/138 (13%)
Query: 32 VKELVAGGVAGALSKTAVAPLE--RVKILWQTRTGGFHTL-----------GVCQSLNKL 78
+ +LVAG +AG + PL+ R K+ +Q + +L G+ + +
Sbjct: 188 ILDLVAGSIAGGTAVICTYPLDLVRTKLAYQVKGAVKLSLREYKPSEQVYKGILDCVKTI 247
Query: 79 VKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLA-GSAA 137
+ G GLY+G S I PY+ L F YE K+++ P I LA GS A
Sbjct: 248 YRQNGLRGLYRGMAPSLYGIFPYSGLKFYFYETMKTYV----PEEHRKDIIAKLACGSVA 303
Query: 138 GGTSVLCTYPLDLARTKL 155
G TYPLD+ R ++
Sbjct: 304 GLLGQTITYPLDVVRRQM 321
Score = 33.5 bits (75), Expect = 9.4
Identities = 27/110 (24%), Positives = 45/110 (40%), Gaps = 28/110 (25%)
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQTRTG-----GFHTLGVCQSLNKLVKHEGFLG 86
+ +L G VAG L +T PL+ V+ Q + G T G S+ + KH+G+
Sbjct: 294 IAKLACGSVAGLLGQTITYPLDVVRRQMQAFSSSNLEKGKGTFG---SIAMIAKHQGWRQ 350
Query: 87 LYKGNGASAV--------------------RIVPYAALHFMTYERYKSWI 116
L+ G + + ++VP A+ F Y+ K W+
Sbjct: 351 LFSGLSINYLKELYQLDTSVCAFIYVQCGEKVVPSVAIGFTVYDSMKVWL 400
>ref|XP_477733.1| putative mitochondrial carrier protein [Oryza sativa (japonica
cultivar-group)] gi|34394749|dbj|BAC84113.1| putative
mitochondrial carrier protein [Oryza sativa (japonica
cultivar-group)]
Length = 333
Score = 259 bits (663), Expect = 6e-68
Identities = 145/321 (45%), Positives = 193/321 (59%), Gaps = 30/321 (9%)
Query: 16 ASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSL 75
A++ + + P + +E+VAGGVAG ++KTAVAPLERV ++ Q G Q L
Sbjct: 26 ANLSRTATAICSTPAFAREMVAGGVAGVVAKTAVAPLERVNLMRQVGAAP-RGAGAVQML 84
Query: 76 NKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGS 135
++ + EG GL++GNGA+A+R+ ALHFM YERYK ++L P+LG GP +DLLAGS
Sbjct: 85 REIGRGEGVAGLFRGNGANALRVFHTKALHFMAYERYKRFLLGAAPSLGDGPVVDLLAGS 144
Query: 136 AAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYRE-GG 194
AAGGT+VL TYPLDLART+LA P A G+ VL S YRE GG
Sbjct: 145 AAGGTAVLATYPLDLARTRLA-------------CAAAPPGAAAAGMSGVLRSAYREGGG 191
Query: 195 ARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQK------SILMRLSCGALAGLF 248
RG+YRG+ P++ +LP +GL F YE LK +P E ++ +++CG AGL
Sbjct: 192 VRGVYRGLCPSLARVLPMSGLNFCVYEALKAQIPREEEEHGARGWRRAAKVACGVAAGLV 251
Query: 249 GQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRI 308
T TYPLDVV+RQ+Q+G TL R IVR QG RQL+AG+ I Y++
Sbjct: 252 ASTATYPLDVVRRQIQLGGGGG---------GTLQAFRAIVRAQGARQLYAGLGITYVKK 302
Query: 309 VPSAAISFTAYDTMKAWLGIP 329
VPS A+ AYD MK+ L +P
Sbjct: 303 VPSTAVGLVAYDYMKSLLMLP 323
>ref|NP_916575.1| P0456F08.3 [Oryza sativa (japonica cultivar-group)]
Length = 239
Score = 223 bits (569), Expect = 5e-57
Identities = 118/208 (56%), Positives = 145/208 (68%), Gaps = 4/208 (1%)
Query: 130 DLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSV 189
+++AG AG S PL+ R K+ QV ++ I G+K QP + GIK V V
Sbjct: 30 EMIAGGVAGAFSKTAIAPLE--RLKILLQV-NSSDQISSGLKRTNFQPKYGGIKDVFRGV 86
Query: 190 YREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCGALAGLFG 249
Y EGG R LYRGVGPT+ GILPYAGLKFY YE LK HVPE+++ S+ ++LSCGA AGLFG
Sbjct: 87 YSEGGVRALYRGVGPTLMGILPYAGLKFYIYEGLKAHVPEDYKNSVTLKLSCGAAAGLFG 146
Query: 250 QTLTYPLDVVKRQMQVGSLQ-NAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRI 308
QTLTYPLDVV+RQMQV S Q + + R T GL I + QGWRQLFAG+S+NYI++
Sbjct: 147 QTLTYPLDVVRRQMQVQSQQYHDKFGGPQIRGTFQGLMIIKQTQGWRQLFAGLSLNYIKV 206
Query: 309 VPSAAISFTAYDTMKAWLGIPPQQKSRS 336
VPS AI FTAYDTMK+ L IPP++K S
Sbjct: 207 VPSVAIGFTAYDTMKSLLKIPPREKKMS 234
Score = 91.3 bits (225), Expect = 4e-17
Identities = 73/242 (30%), Positives = 113/242 (46%), Gaps = 25/242 (10%)
Query: 6 GSTLAGLVDNASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGG 65
G+ G A+ D D VPV+ KE++AGGVAGA SKTA+APLER+KIL Q +
Sbjct: 2 GTPSQGSAAVAAAGVDLCVLDLVPVFAKEMIAGGVAGAFSKTAIAPLERLKILLQVNSSD 61
Query: 66 FHTLGVCQS------------LNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYK 113
+ G+ ++ + G LY+G G + + I+PYA L F YE K
Sbjct: 62 QISSGLKRTNFQPKYGGIKDVFRGVYSEGGVRALYRGVGPTLMGILPYAGLKFYIYEGLK 121
Query: 114 SWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGV 173
+ + + + + L G+AAG TYPLD+ R ++ QV + K G +
Sbjct: 122 AHVPED---YKNSVTLKLSCGAAAGLFGQTLTYPLDVVRRQM--QVQSQQYHDKFGGPQI 176
Query: 174 RPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK--MHVPEEH 231
R G L + + G R L+ G+ ++P + F Y+ +K + +P
Sbjct: 177 R------GTFQGLMIIKQTQGWRQLFAGLSLNYIKVVPSVAIGFTAYDTMKSLLKIPPRE 230
Query: 232 QK 233
+K
Sbjct: 231 KK 232
>gb|AAM91170.1| putative mitochondrial carrier protein [Arabidopsis thaliana]
gi|20260324|gb|AAM13060.1| putative mitochondrial
carrier protein [Arabidopsis thaliana]
Length = 152
Score = 221 bits (564), Expect = 2e-56
Identities = 103/148 (69%), Positives = 124/148 (83%), Gaps = 2/148 (1%)
Query: 190 YREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCGALAGLFG 249
Y+EGG RGLYRG+GPT+ GILPYAGLKFY YE+LK HVPEEHQ S+ M L CGALAGLFG
Sbjct: 3 YKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLFG 62
Query: 250 QTLTYPLDVVKRQMQVGSLQNAAHE--DTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIR 307
QT+TYPLDVV+RQMQV +LQ E + RY+NT DGL IVR QGW+QLFAG+SINYI+
Sbjct: 63 QTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYIK 122
Query: 308 IVPSAAISFTAYDTMKAWLGIPPQQKSR 335
IVPS AI FT Y++MK+W+ IPP+++S+
Sbjct: 123 IVPSVAIGFTVYESMKSWMRIPPRERSK 150
Score = 51.2 bits (121), Expect = 4e-05
Identities = 44/151 (29%), Positives = 68/151 (44%), Gaps = 12/151 (7%)
Query: 80 KHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGG 139
K G GLY+G G + + I+PYA L F YE K + + + L G+ AG
Sbjct: 4 KEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQ---NSVRMHLPCGALAGL 60
Query: 140 TSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLY 199
TYPLD+ R ++ QV + + +G R + G L ++ R G + L+
Sbjct: 61 FGQTITYPLDVVRRQM--QVENLQPMTSEG-NNKRYKNTFDG----LNTIVRTQGWKQLF 113
Query: 200 RGVGPTITGILPYAGLKFYTYEKLK--MHVP 228
G+ I+P + F YE +K M +P
Sbjct: 114 AGLSINYIKIVPSVAIGFTVYESMKSWMRIP 144
Score = 40.4 bits (93), Expect = 0.077
Identities = 25/90 (27%), Positives = 39/90 (42%), Gaps = 8/90 (8%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRT--------GGFHTLGVCQSLNKLVKHEGFLG 86
L G +AG +T PL+ V+ Q LN +V+ +G+
Sbjct: 52 LPCGALAGLFGQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQ 111
Query: 87 LYKGNGASAVRIVPYAALHFMTYERYKSWI 116
L+ G + ++IVP A+ F YE KSW+
Sbjct: 112 LFAGLSINYIKIVPSVAIGFTVYESMKSWM 141
>ref|XP_638149.1| hypothetical protein DDB0186597 [Dictyostelium discoideum]
gi|60466585|gb|EAL64637.1| hypothetical protein
DDB0186597 [Dictyostelium discoideum]
Length = 434
Score = 218 bits (555), Expect = 2e-55
Identities = 118/310 (38%), Positives = 182/310 (58%), Gaps = 26/310 (8%)
Query: 33 KELVAGGVAGALSKTAVAPLERVKILWQT-------RTGGFHTLGVCQSLNKLVKHEGFL 85
K L++GGVAGA+S+T +PLER+KIL Q + G+ QSL + EGF+
Sbjct: 140 KLLLSGGVAGAVSRTCTSPLERLKILNQVGHMNLEQNAPKYKGRGIIQSLKTMYTTEGFI 199
Query: 86 GLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCT 145
G +KGNG + +RI PY+A+ F++YE+YK+++LNN + +L G AAG TS+LCT
Sbjct: 200 GFFKGNGTNVIRIAPYSAIQFLSYEKYKNFLLNNNDQTHLTTYENLFVGGAAGVTSLLCT 259
Query: 146 YPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPT 205
YPLDL R++L QV + + GI + RE G GLY+G+ +
Sbjct: 260 YPLDLIRSRLTVQVFGNK---------------YNGIADTCKMIIREEGVAGLYKGLFAS 304
Query: 206 ITGILPYAGLKFYTYEKL-KMHVPEEHQKSILMRLSCGALAGLFGQTLTYPLDVVKRQMQ 264
G+ PY + F TYE L K +P++ +++ L+ GA++G QTLTYP+D+++R++Q
Sbjct: 305 ALGVAPYVAINFTTYENLKKTFIPKDTTPTVVQSLTFGAISGATAQTLTYPIDLIRRRLQ 364
Query: 265 VGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKA 324
V Q +D Y T D RKI+R++G L+ G+ Y++++P+ +ISF Y+ MK
Sbjct: 365 V---QGIGGKDILYNGTFDAFRKIIRDEGVLGLYNGMIPCYLKVIPAISISFCVYEVMKK 421
Query: 325 WLGIPPQQKS 334
L I ++ S
Sbjct: 422 ILKIDSKKIS 431
Score = 48.5 bits (114), Expect = 3e-04
Identities = 27/91 (29%), Positives = 47/91 (50%), Gaps = 3/91 (3%)
Query: 26 DGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTR-TGGFHTL--GVCQSLNKLVKHE 82
D P V+ L G ++GA ++T P++ ++ Q + GG L G + K+++ E
Sbjct: 330 DTTPTVVQSLTFGAISGATAQTLTYPIDLIRRRLQVQGIGGKDILYNGTFDAFRKIIRDE 389
Query: 83 GFLGLYKGNGASAVRIVPYAALHFMTYERYK 113
G LGLY G ++++P ++ F YE K
Sbjct: 390 GVLGLYNGMIPCYLKVIPAISISFCVYEVMK 420
>emb|CAA46834.1| Graves disease carrier protein from bovine heart mitochondria [Bos
taurus] gi|266574|sp|Q01888|GDC_BOVIN Grave's disease
carrier protein (GDC) (Mitochondrial solute carrier
protein homolog) gi|27807213|ref|NP_777097.1| solute
carrier family 25, member 16 [Bos taurus]
Length = 330
Score = 215 bits (547), Expect = 2e-54
Identities = 123/310 (39%), Positives = 180/310 (57%), Gaps = 36/310 (11%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKG 90
+++ +AGG+AG +KT VAPL+RVK+L Q + LGV +L + K EG+LGLYKG
Sbjct: 34 WLRSFLAGGIAGCCAKTTVAPLDRVKVLLQAHNHHYRHLGVFSTLRAVPKKEGYLGLYKG 93
Query: 91 NGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDL 150
NGA +RI PY A+ FM +E YK+ I SG L+AGS AG T+V+CTYPLD+
Sbjct: 94 NGAMMIRIFPYGAIQFMAFEHYKTLITTKLGV--SGHVHRLMAGSMAGMTAVICTYPLDM 151
Query: 151 ARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVY-REGGARGLYRGVGPTITGI 209
R +LA+Q V+ + + GI ++Y +EGG G YRG+ PTI G+
Sbjct: 152 VRVRLAFQ--------------VKGEHTYTGIIHAFKTIYAKEGGFLGFYRGLMPTILGM 197
Query: 210 LPYAGLKFYTYEKLK----------MHVPEEHQKSIL-----MRLSCGALAGLFGQTLTY 254
PYAG+ F+T+ LK + P ++L + L CG +AG QT++Y
Sbjct: 198 APYAGVSFFTFGTLKSVGLSYAPTLLGRPSSDNPNVLVLKTHINLLCGGVAGAIAQTISY 257
Query: 255 PLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQ-LFAGVSINYIRIVPSAA 313
P DV +R+MQ+G++ + R T ++ + + G R+ L+ G+S+NYIR VPS A
Sbjct: 258 PFDVTRRRMQLGAVLPEFEKCLTMRET---MKYVYGHHGIRKGLYRGLSLNYIRCVPSQA 314
Query: 314 ISFTAYDTMK 323
++FT Y+ MK
Sbjct: 315 VAFTTYELMK 324
Score = 74.7 bits (182), Expect = 4e-12
Identities = 60/214 (28%), Positives = 94/214 (43%), Gaps = 29/214 (13%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-LGVCQSLNKL-VKHEGF 84
GV +V L+AG +AG + PL+ V++ + G HT G+ + + K GF
Sbjct: 124 GVSGHVHRLMAGSMAGMTAVICTYPLDMVRVRLAFQVKGEHTYTGIIHAFKTIYAKEGGF 183
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGP------------FIDLL 132
LG Y+G + + + PYA + F T+ KS L+ P L P I+LL
Sbjct: 184 LGFYRGLMPTILGMAPYAGVSFFTFGTLKSVGLSYAPTLLGRPSSDNPNVLVLKTHINLL 243
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQ-PAHIGIKSVLTSVYR 191
G AG + +YP D+ R ++ + V P+ + ++ + VY
Sbjct: 244 CGGVAGAIAQTISYPFDVTRRRMQ-------------LGAVLPEFEKCLTMRETMKYVYG 290
Query: 192 EGGAR-GLYRGVGPTITGILPYAGLKFYTYEKLK 224
G R GLYRG+ +P + F TYE +K
Sbjct: 291 HHGIRKGLYRGLSLNYIRCVPSQAVAFTTYELMK 324
Score = 60.8 bits (146), Expect = 6e-08
Identities = 39/148 (26%), Positives = 72/148 (48%), Gaps = 7/148 (4%)
Query: 179 HIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKS-ILM 237
H+G+ S L +V ++ G GLY+G G + I PY ++F +E K + + S +
Sbjct: 71 HLGVFSTLRAVPKKEGYLGLYKGNGAMMIRIFPYGAIQFMAFEHYKTLITTKLGVSGHVH 130
Query: 238 RLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQG-WRQ 296
RL G++AG+ TYPLD+V+ ++ + Y + + I +G +
Sbjct: 131 RLMAGSMAGMTAVICTYPLDMVRVRLAF-----QVKGEHTYTGIIHAFKTIYAKEGGFLG 185
Query: 297 LFAGVSINYIRIVPSAAISFTAYDTMKA 324
+ G+ + + P A +SF + T+K+
Sbjct: 186 FYRGLMPTILGMAPYAGVSFFTFGTLKS 213
Score = 37.4 bits (85), Expect = 0.65
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 6/86 (6%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVK----HEGFL-GLYK 89
L+ GGVAGA+++T P + + Q C ++ + +K H G GLY+
Sbjct: 242 LLCGGVAGAIAQTISYPFDVTRRRMQLGAV-LPEFEKCLTMRETMKYVYGHHGIRKGLYR 300
Query: 90 GNGASAVRIVPYAALHFMTYERYKSW 115
G + +R VP A+ F TYE K +
Sbjct: 301 GLSLNYIRCVPSQAVAFTTYELMKQF 326
>emb|CAI15116.1| solute carrier family 25 (mitochondrial carrier\; Graves disease
autoantigen), member 16 [Homo sapiens]
gi|20988432|gb|AAH30266.1| Solute carrier family 25,
member 16 [Homo sapiens] gi|27544933|ref|NP_689920.1|
solute carrier family 25, member 16 [Homo sapiens]
Length = 332
Score = 214 bits (545), Expect = 3e-54
Identities = 124/311 (39%), Positives = 184/311 (58%), Gaps = 38/311 (12%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKG 90
+++ +AGG+AG +KT VAPL+RVK+L Q + LGV +L + + EGFLGLYKG
Sbjct: 36 WLRSFLAGGIAGCCAKTTVAPLDRVKVLLQAHNHHYKHLGVFSALRAVPQKEGFLGLYKG 95
Query: 91 NGASAVRIVPYAALHFMTYERYKSWILNNCPALG-SGPFIDLLAGSAAGGTSVLCTYPLD 149
NGA +RI PY A+ FM +E YK+ I LG SG L+AGS AG T+V+CTYPLD
Sbjct: 96 NGAMMIRIFPYGAIQFMAFEHYKTLITTK---LGISGHVHRLMAGSMAGMTAVICTYPLD 152
Query: 150 LARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVY-REGGARGLYRGVGPTITG 208
+ R +LA+Q V+ + ++ GI ++Y +EGG G YRG+ PTI G
Sbjct: 153 MVRVRLAFQ--------------VKGEHSYTGIIHAFKTIYAKEGGFFGFYRGLMPTILG 198
Query: 209 ILPYAGLKFYTYEKLK----------MHVPEEHQKSIL-----MRLSCGALAGLFGQTLT 253
+ PYAG+ F+T+ LK + P ++L + L CG +AG QT++
Sbjct: 199 MAPYAGVSFFTFGTLKSVGLSHAPTLLGRPSSDNPNVLVLKTHVNLLCGGVAGAIAQTIS 258
Query: 254 YPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQ-LFAGVSINYIRIVPSA 312
YP DV +R+MQ+G++ + R+T ++ + + G R+ L+ G+S+NYIR +PS
Sbjct: 259 YPFDVTRRRMQLGTVLPEFEKCLTMRDT---MKYVYGHHGIRKGLYRGLSLNYIRCIPSQ 315
Query: 313 AISFTAYDTMK 323
A++FT Y+ MK
Sbjct: 316 AVAFTTYELMK 326
Score = 73.2 bits (178), Expect = 1e-11
Identities = 56/213 (26%), Positives = 94/213 (43%), Gaps = 27/213 (12%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-LGVCQSLNKL-VKHEGF 84
G+ +V L+AG +AG + PL+ V++ + G H+ G+ + + K GF
Sbjct: 126 GISGHVHRLMAGSMAGMTAVICTYPLDMVRVRLAFQVKGEHSYTGIIHAFKTIYAKEGGF 185
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGP------------FIDLL 132
G Y+G + + + PYA + F T+ KS L++ P L P ++LL
Sbjct: 186 FGFYRGLMPTILGMAPYAGVSFFTFGTLKSVGLSHAPTLLGRPSSDNPNVLVLKTHVNLL 245
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYRE 192
G AG + +YP D+ R ++ GT+ + + ++ + VY
Sbjct: 246 CGGVAGAIAQTISYPFDVTRRRMQL------GTVLPEFEKC------LTMRDTMKYVYGH 293
Query: 193 GGAR-GLYRGVGPTITGILPYAGLKFYTYEKLK 224
G R GLYRG+ +P + F TYE +K
Sbjct: 294 HGIRKGLYRGLSLNYIRCIPSQAVAFTTYELMK 326
Score = 59.7 bits (143), Expect = 1e-07
Identities = 39/148 (26%), Positives = 72/148 (48%), Gaps = 7/148 (4%)
Query: 179 HIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKS-ILM 237
H+G+ S L +V ++ G GLY+G G + I PY ++F +E K + + S +
Sbjct: 73 HLGVFSALRAVPQKEGFLGLYKGNGAMMIRIFPYGAIQFMAFEHYKTLITTKLGISGHVH 132
Query: 238 RLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQG-WRQ 296
RL G++AG+ TYPLD+V+ ++ + Y + + I +G +
Sbjct: 133 RLMAGSMAGMTAVICTYPLDMVRVRLAF-----QVKGEHSYTGIIHAFKTIYAKEGGFFG 187
Query: 297 LFAGVSINYIRIVPSAAISFTAYDTMKA 324
+ G+ + + P A +SF + T+K+
Sbjct: 188 FYRGLMPTILGMAPYAGVSFFTFGTLKS 215
Score = 38.1 bits (87), Expect = 0.38
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 6/86 (6%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVK----HEGFL-GLYK 89
L+ GGVAGA+++T P + + Q T C ++ +K H G GLY+
Sbjct: 244 LLCGGVAGAIAQTISYPFDVTRRRMQLGTV-LPEFEKCLTMRDTMKYVYGHHGIRKGLYR 302
Query: 90 GNGASAVRIVPYAALHFMTYERYKSW 115
G + +R +P A+ F TYE K +
Sbjct: 303 GLSLNYIRCIPSQAVAFTTYELMKQF 328
>ref|NP_780403.1| solute carrier family 25 (mitochondrial carrier, Graves disease
autoantigen), member 16 [Mus musculus]
gi|26326839|dbj|BAC27163.1| unnamed protein product [Mus
musculus]
Length = 332
Score = 213 bits (542), Expect = 7e-54
Identities = 121/310 (39%), Positives = 180/310 (58%), Gaps = 36/310 (11%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKG 90
+++ +AGG+AG +KT VAPL+RVK+L Q + LGV +L + + EG+LGLYKG
Sbjct: 36 WLRSFLAGGIAGCCAKTTVAPLDRVKVLLQAHNRHYKHLGVLSTLRAVPQKEGYLGLYKG 95
Query: 91 NGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDL 150
NGA +RI PY A+ FM +E YK++I SG L+AGS AG T+V+CTYPLD+
Sbjct: 96 NGAMMIRIFPYGAIQFMAFEHYKTFITTKLGV--SGHVHRLMAGSMAGMTAVICTYPLDV 153
Query: 151 ARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVY-REGGARGLYRGVGPTITGI 209
R +LA+Q V+ + + GI ++Y +EGG G YRG+ PTI G+
Sbjct: 154 VRVRLAFQ--------------VKGEHTYSGIIHAFKTIYAKEGGFLGFYRGLMPTILGM 199
Query: 210 LPYAGLKFYTYEKLK----------MHVPEEHQKSIL-----MRLSCGALAGLFGQTLTY 254
PYAG+ F+T+ LK + P ++L + L CG +AG QT++Y
Sbjct: 200 APYAGVSFFTFGTLKSVGLSYAPALLGRPSSDNPNVLVLKTHINLLCGGVAGAIAQTISY 259
Query: 255 PLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQ-LFAGVSINYIRIVPSAA 313
P DV +R+MQ+G++ + R T ++ + G R+ L+ G+S+NYIR +PS A
Sbjct: 260 PFDVTRRRMQLGAVLPEFEKCLTMRET---MKYVYGQHGIRRGLYRGLSLNYIRCIPSQA 316
Query: 314 ISFTAYDTMK 323
++FT Y+ MK
Sbjct: 317 VAFTTYELMK 326
Score = 76.6 bits (187), Expect = 1e-12
Identities = 61/214 (28%), Positives = 96/214 (44%), Gaps = 29/214 (13%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTL-GVCQSLNKL-VKHEGF 84
GV +V L+AG +AG + PL+ V++ + G HT G+ + + K GF
Sbjct: 126 GVSGHVHRLMAGSMAGMTAVICTYPLDVVRVRLAFQVKGEHTYSGIIHAFKTIYAKEGGF 185
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGP------------FIDLL 132
LG Y+G + + + PYA + F T+ KS L+ PAL P I+LL
Sbjct: 186 LGFYRGLMPTILGMAPYAGVSFFTFGTLKSVGLSYAPALLGRPSSDNPNVLVLKTHINLL 245
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQ-PAHIGIKSVLTSVYR 191
G AG + +YP D+ R ++ + V P+ + ++ + VY
Sbjct: 246 CGGVAGAIAQTISYPFDVTRRRMQ-------------LGAVLPEFEKCLTMRETMKYVYG 292
Query: 192 EGG-ARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
+ G RGLYRG+ +P + F TYE +K
Sbjct: 293 QHGIRRGLYRGLSLNYIRCIPSQAVAFTTYELMK 326
Score = 63.2 bits (152), Expect = 1e-08
Identities = 51/195 (26%), Positives = 86/195 (43%), Gaps = 22/195 (11%)
Query: 132 LAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYR 191
LAG AG + PLD R K+ Q + H+G+ S L +V +
Sbjct: 41 LAGGIAGCCAKTTVAPLD--RVKVLLQAHNRHYK-------------HLGVLSTLRAVPQ 85
Query: 192 EGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKS-ILMRLSCGALAGLFGQ 250
+ G GLY+G G + I PY ++F +E K + + S + RL G++AG+
Sbjct: 86 KEGYLGLYKGNGAMMIRIFPYGAIQFMAFEHYKTFITTKLGVSGHVHRLMAGSMAGMTAV 145
Query: 251 TLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQG-WRQLFAGVSINYIRIV 309
TYPLDVV+ ++ + Y + + I +G + + G+ + +
Sbjct: 146 ICTYPLDVVRVRLAF-----QVKGEHTYSGIIHAFKTIYAKEGGFLGFYRGLMPTILGMA 200
Query: 310 PSAAISFTAYDTMKA 324
P A +SF + T+K+
Sbjct: 201 PYAGVSFFTFGTLKS 215
Score = 35.4 bits (80), Expect = 2.5
Identities = 25/86 (29%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVK-----HEGFLGLYK 89
L+ GGVAGA+++T P + + Q C ++ + +K H GLY+
Sbjct: 244 LLCGGVAGAIAQTISYPFDVTRRRMQLGAV-LPEFEKCLTMRETMKYVYGQHGIRRGLYR 302
Query: 90 GNGASAVRIVPYAALHFMTYERYKSW 115
G + +R +P A+ F TYE K +
Sbjct: 303 GLSLNYIRCIPSQAVAFTTYELMKQF 328
>gb|AAH65855.1| Unknown (protein for MGC:77742) [Danio rerio]
gi|28277902|gb|AAH45977.1| Unknown (protein for
MGC:77742) [Danio rerio] gi|45387539|ref|NP_991112.1|
solute carrier family 25, member 16 [Danio rerio]
Length = 321
Score = 213 bits (541), Expect = 9e-54
Identities = 124/310 (40%), Positives = 179/310 (57%), Gaps = 36/310 (11%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKG 90
+++ AGGVAG +K+ +APL+RVKIL Q + + LGV +L + K EGFLGLYKG
Sbjct: 25 WLRSFTAGGVAGCCAKSTIAPLDRVKILLQAQNPHYKHLGVFATLKAVPKKEGFLGLYKG 84
Query: 91 NGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDL 150
NGA +RI PY A+ FM ++ YK ++ SG L+AGS AG T+V+CTYPLD+
Sbjct: 85 NGAMMIRIFPYGAIQFMAFDNYKKFLHTKVGI--SGHVHRLMAGSMAGMTAVICTYPLDV 142
Query: 151 ARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVY-REGGARGLYRGVGPTITGI 209
R +LA+QV + GI+ ++Y +EGG G YRG+ PTI G+
Sbjct: 143 IRARLAFQVTG--------------HHRYSGIRHAFQTIYHKEGGISGFYRGLIPTIIGM 188
Query: 210 LPYAGLKFYTYEKLK----MHVPEEHQKSIL-----------MRLSCGALAGLFGQTLTY 254
PYAG F+T+ LK H PE+ K L + L CG +AG QT++Y
Sbjct: 189 APYAGFSFFTFGTLKTLGLTHFPEQLGKPSLDNPDVLVLKTQVNLLCGGVAGAIAQTISY 248
Query: 255 PLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQ-LFAGVSINYIRIVPSAA 313
PLDV +R+MQ+G+ + + + + L+ + G ++ L+ G+S+NYIR VPS A
Sbjct: 249 PLDVARRRMQLGA---SLPDHDKCCSLTKTLKHVYSQYGVKKGLYRGLSLNYIRCVPSQA 305
Query: 314 ISFTAYDTMK 323
++FT Y+ MK
Sbjct: 306 VAFTTYEFMK 315
Score = 65.1 bits (157), Expect = 3e-09
Identities = 54/213 (25%), Positives = 89/213 (41%), Gaps = 27/213 (12%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHE--GF 84
G+ +V L+AG +AG + PL+ ++ + G H + + + H+ G
Sbjct: 115 GISGHVHRLMAGSMAGMTAVICTYPLDVIRARLAFQVTGHHRYSGIRHAFQTIYHKEGGI 174
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFID------------LL 132
G Y+G + + + PYA F T+ K+ L + P P +D LL
Sbjct: 175 SGFYRGLIPTIIGMAPYAGFSFFTFGTLKTLGLTHFPEQLGKPSLDNPDVLVLKTQVNLL 234
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYRE 192
G AG + +YPLD+AR ++ ++ D K + L VY +
Sbjct: 235 CGGVAGAIAQTISYPLDVARRRM-----QLGASLPDHDK-------CCSLTKTLKHVYSQ 282
Query: 193 GGA-RGLYRGVGPTITGILPYAGLKFYTYEKLK 224
G +GLYRG+ +P + F TYE +K
Sbjct: 283 YGVKKGLYRGLSLNYIRCVPSQAVAFTTYEFMK 315
Score = 39.3 bits (90), Expect = 0.17
Identities = 26/91 (28%), Positives = 42/91 (45%), Gaps = 6/91 (6%)
Query: 236 LMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWR 295
L + G +AG ++ PLD VK +Q A + ++ L+ + + +G+
Sbjct: 26 LRSFTAGGVAGCCAKSTIAPLDRVKILLQ------AQNPHYKHLGVFATLKAVPKKEGFL 79
Query: 296 QLFAGVSINYIRIVPSAAISFTAYDTMKAWL 326
L+ G IRI P AI F A+D K +L
Sbjct: 80 GLYKGNGAMMIRIFPYGAIQFMAFDNYKKFL 110
>emb|CAF98697.1| unnamed protein product [Tetraodon nigroviridis]
Length = 313
Score = 212 bits (539), Expect = 1e-53
Identities = 124/310 (40%), Positives = 180/310 (58%), Gaps = 36/310 (11%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKG 90
+++ VAGGVAG +KT +APL+R+KIL Q + + LGV +L + + EGFLGLYKG
Sbjct: 17 WLRSFVAGGVAGCCAKTTIAPLDRIKILLQAQNPHYKHLGVFATLRAVPQKEGFLGLYKG 76
Query: 91 NGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDL 150
NGA VRI PY A+ FM ++ YK + SG L+AGS AG T+V+CTYPLD+
Sbjct: 77 NGAMMVRIFPYGAIQFMAFDNYKKLLSTQIGI--SGHIHRLMAGSMAGMTAVICTYPLDV 134
Query: 151 ARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVY-REGGARGLYRGVGPTITGI 209
R +LA+QV + + GI + ++Y +EGG G YRG+ PT+ G+
Sbjct: 135 VRARLAFQVTG--------------EHRYTGIANAFHTIYLKEGGVLGFYRGLTPTLIGM 180
Query: 210 LPYAGLKFYTYEKLK----MHVPE------EHQKSIL-----MRLSCGALAGLFGQTLTY 254
PYAG F+T+ LK H PE ++L + L CG +AG QT++Y
Sbjct: 181 APYAGFSFFTFGTLKSLGLKHFPELLGRPSSDNPNVLVLKPQVNLLCGGMAGAVAQTISY 240
Query: 255 PLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQ-LFAGVSINYIRIVPSAA 313
PLDV +R+MQ+G++ + + T L + + G ++ L+ G+S+NYIR VPS A
Sbjct: 241 PLDVARRRMQLGAVLPDSDKCVSLSKT---LTYVYKQYGIKKGLYRGLSLNYIRCVPSQA 297
Query: 314 ISFTAYDTMK 323
++FT Y+ MK
Sbjct: 298 MAFTTYEFMK 307
Score = 77.8 bits (190), Expect = 4e-13
Identities = 60/214 (28%), Positives = 98/214 (45%), Gaps = 29/214 (13%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLE--RVKILWQTRTGGFHTLGVCQSLNKL-VKHEG 83
G+ ++ L+AG +AG + PL+ R ++ +Q TG G+ + + + +K G
Sbjct: 107 GISGHIHRLMAGSMAGMTAVICTYPLDVVRARLAFQV-TGEHRYTGIANAFHTIYLKEGG 165
Query: 84 FLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPF------------IDL 131
LG Y+G + + + PYA F T+ KS L + P L P ++L
Sbjct: 166 VLGFYRGLTPTLIGMAPYAGFSFFTFGTLKSLGLKHFPELLGRPSSDNPNVLVLKPQVNL 225
Query: 132 LAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYR 191
L G AG + +YPLD+AR ++ + + D K V + LT VY+
Sbjct: 226 LCGGMAGAVAQTISYPLDVARRRMQLGAV-----LPDSDKCV-------SLSKTLTYVYK 273
Query: 192 EGG-ARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
+ G +GLYRG+ +P + F TYE +K
Sbjct: 274 QYGIKKGLYRGLSLNYIRCVPSQAMAFTTYEFMK 307
Score = 38.5 bits (88), Expect = 0.29
Identities = 25/85 (29%), Positives = 39/85 (45%), Gaps = 6/85 (7%)
Query: 242 GALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGV 301
G +AG +T PLD +K +Q A + ++ LR + + +G+ L+ G
Sbjct: 24 GGVAGCCAKTTIAPLDRIKILLQ------AQNPHYKHLGVFATLRAVPQKEGFLGLYKGN 77
Query: 302 SINYIRIVPSAAISFTAYDTMKAWL 326
+RI P AI F A+D K L
Sbjct: 78 GAMMVRIFPYGAIQFMAFDNYKKLL 102
>ref|XP_624199.1| PREDICTED: similar to ENSANGP00000008222, partial [Apis mellifera]
Length = 315
Score = 210 bits (535), Expect = 4e-53
Identities = 130/309 (42%), Positives = 174/309 (56%), Gaps = 35/309 (11%)
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKGN 91
+K L+AGGVAG SKT VAPL+R+KIL Q + LGV L ++++ E F LYKGN
Sbjct: 15 LKSLIAGGVAGMCSKTTVAPLDRIKILLQAHNKYYKHLGVLSGLREVIQRERFFALYKGN 74
Query: 92 GASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFID-LLAGSAAGGTSVLCTYPLDL 150
A +RI PYAA F T+E YK ++ G ID LAGSAAG T+V TYPLD+
Sbjct: 75 FAQMIRIFPYAATQFTTFELYKKYLGG---LFGKHTHIDKFLAGSAAGVTAVTLTYPLDI 131
Query: 151 ARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGIL 210
R +LA+QV I GI H GI T EGG R LYRG PTI G++
Sbjct: 132 IRARLAFQVAGEH--IYIGI-------VHAGI----TIFKNEGGIRALYRGFWPTIFGMI 178
Query: 211 PYAGLKFYTYEKLKMHVPEEHQK---------------SILMRLSCGALAGLFGQTLTYP 255
PYAG FY++EKLK + +I RL CG +AG Q+ +YP
Sbjct: 179 PYAGFSFYSFEKLKYFCMKYASNYFCENCDRNTGGLVLTIPARLLCGGIAGAVAQSFSYP 238
Query: 256 LDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQG-WRQLFAGVSINYIRIVPSAAI 314
LDV +R MQ+G + +A H+ + + L ++ I + G + L+ G+SINY+R +P ++
Sbjct: 239 LDVTRRHMQLGIMHHANHKYS--SSMLQTIKMIYKENGIIKGLYRGMSINYLRAIPMVSV 296
Query: 315 SFTAYDTMK 323
SFT Y+ MK
Sbjct: 297 SFTTYEIMK 305
Score = 65.9 bits (159), Expect = 2e-09
Identities = 53/213 (24%), Positives = 92/213 (42%), Gaps = 26/213 (12%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-LGVCQSLNKLVKHEGFL 85
G ++ + +AG AG + T PL+ ++ + G H +G+ + + K+EG +
Sbjct: 104 GKHTHIDKFLAGSAAGVTAVTLTYPLDIIRARLAFQVAGEHIYIGIVHAGITIFKNEGGI 163
Query: 86 -GLYKGNGASAVRIVPYAALHFMTYERYK--------SWILNNCPALGSGPFID----LL 132
LY+G + ++PYA F ++E+ K ++ NC G + LL
Sbjct: 164 RALYRGFWPTIFGMIPYAGFSFYSFEKLKYFCMKYASNYFCENCDRNTGGLVLTIPARLL 223
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYRE 192
G AG + +YPLD+ TR ++ GI + + +Y+E
Sbjct: 224 CGGIAGAVAQSFSYPLDV-----------TRRHMQLGIMHHANHKYSSSMLQTIKMIYKE 272
Query: 193 GG-ARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
G +GLYRG+ +P + F TYE +K
Sbjct: 273 NGIIKGLYRGMSINYLRAIPMVSVSFTTYEIMK 305
Score = 50.8 bits (120), Expect = 6e-05
Identities = 30/99 (30%), Positives = 52/99 (52%), Gaps = 6/99 (6%)
Query: 229 EEHQKSILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKI 288
E+ + +L L G +AG+ +T PLD +K +Q A ++ ++ L GLR++
Sbjct: 8 EKDYEFLLKSLIAGGVAGMCSKTTVAPLDRIKILLQ------AHNKYYKHLGVLSGLREV 61
Query: 289 VRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAWLG 327
++ + + L+ G IRI P AA FT ++ K +LG
Sbjct: 62 IQRERFFALYKGNFAQMIRIFPYAATQFTTFELYKKYLG 100
>ref|XP_546134.1| PREDICTED: similar to solute carrier family 25, member 16 [Canis
familiaris]
Length = 515
Score = 205 bits (521), Expect = 2e-51
Identities = 120/303 (39%), Positives = 174/303 (56%), Gaps = 36/303 (11%)
Query: 38 GGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVR 97
G +AG +KT VAPL+RVK+L Q + LGV +L + + EG+LGLYKGNGA +R
Sbjct: 226 GCIAGCCAKTTVAPLDRVKVLLQAHNHHYKHLGVFSALRAVPQKEGYLGLYKGNGAMMIR 285
Query: 98 IVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAY 157
I PY A+ FM +E YK+ I SG L+AGS AG T+V+CTYPLD+ R +LA+
Sbjct: 286 IFPYGAIQFMAFEHYKTLITTKLGV--SGHVHRLMAGSMAGMTAVICTYPLDMVRVRLAF 343
Query: 158 QVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVY-REGGARGLYRGVGPTITGILPYAGLK 216
Q V+ + + GI ++Y +EGG G YRG+ PTI G+ PYAG+
Sbjct: 344 Q--------------VKGEHTYTGIIHAFKTIYAKEGGFLGFYRGLMPTILGMAPYAGVS 389
Query: 217 FYTYEKLK----------MHVPEEHQKSIL-----MRLSCGALAGLFGQTLTYPLDVVKR 261
F+T+ LK + P ++L + L CG +AG QT++YP DV +R
Sbjct: 390 FFTFGTLKSVGLSHAPTLLGRPSSDNPNVLVLKTHINLLCGGVAGAIAQTISYPFDVTRR 449
Query: 262 QMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQ-LFAGVSINYIRIVPSAAISFTAYD 320
+MQ+G+ A E + + ++ + + G R+ L+ G+S+NYIR VPS A++FT Y+
Sbjct: 450 RMQLGT---ALPEFEKCLTMWETMKYVYGHHGIRRGLYRGLSLNYIRCVPSQAVAFTTYE 506
Query: 321 TMK 323
MK
Sbjct: 507 LMK 509
Score = 74.3 bits (181), Expect = 5e-12
Identities = 62/216 (28%), Positives = 96/216 (43%), Gaps = 33/216 (15%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-LGVCQSLNKL-VKHEGF 84
GV +V L+AG +AG + PL+ V++ + G HT G+ + + K GF
Sbjct: 309 GVSGHVHRLMAGSMAGMTAVICTYPLDMVRVRLAFQVKGEHTYTGIIHAFKTIYAKEGGF 368
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGP------------FIDLL 132
LG Y+G + + + PYA + F T+ KS L++ P L P I+LL
Sbjct: 369 LGFYRGLMPTILGMAPYAGVSFFTFGTLKSVGLSHAPTLLGRPSSDNPNVLVLKTHINLL 428
Query: 133 AGSAAGGTSVLCTYPLDLARTKL----AYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTS 188
G AG + +YP D+ R ++ A + T+ + +K V H GI+
Sbjct: 429 CGGVAGAIAQTISYPFDVTRRRMQLGTALPEFEKCLTMWETMKYVY---GHHGIR----- 480
Query: 189 VYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
RGLYRG+ +P + F TYE +K
Sbjct: 481 -------RGLYRGLSLNYIRCVPSQAVAFTTYELMK 509
Score = 60.5 bits (145), Expect = 7e-08
Identities = 39/148 (26%), Positives = 72/148 (48%), Gaps = 7/148 (4%)
Query: 179 HIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKS-ILM 237
H+G+ S L +V ++ G GLY+G G + I PY ++F +E K + + S +
Sbjct: 256 HLGVFSALRAVPQKEGYLGLYKGNGAMMIRIFPYGAIQFMAFEHYKTLITTKLGVSGHVH 315
Query: 238 RLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQG-WRQ 296
RL G++AG+ TYPLD+V+ ++ + Y + + I +G +
Sbjct: 316 RLMAGSMAGMTAVICTYPLDMVRVRLAF-----QVKGEHTYTGIIHAFKTIYAKEGGFLG 370
Query: 297 LFAGVSINYIRIVPSAAISFTAYDTMKA 324
+ G+ + + P A +SF + T+K+
Sbjct: 371 FYRGLMPTILGMAPYAGVSFFTFGTLKS 398
Score = 39.7 bits (91), Expect = 0.13
Identities = 27/85 (31%), Positives = 43/85 (49%), Gaps = 4/85 (4%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTG---GFHTLGVCQSLNKLVKHEGFL-GLYKG 90
L+ GGVAGA+++T P + + Q T L + +++ + H G GLY+G
Sbjct: 427 LLCGGVAGAIAQTISYPFDVTRRRMQLGTALPEFEKCLTMWETMKYVYGHHGIRRGLYRG 486
Query: 91 NGASAVRIVPYAALHFMTYERYKSW 115
+ +R VP A+ F TYE K +
Sbjct: 487 LSLNYIRCVPSQAVAFTTYELMKQF 511
>sp|P16260|GDC_HUMAN Grave's disease carrier protein (GDC) (Grave's disease autoantigen)
(GDA) (Mitochondrial solute carrier protein homolog)
Length = 332
Score = 201 bits (512), Expect = 2e-50
Identities = 121/316 (38%), Positives = 178/316 (56%), Gaps = 48/316 (15%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKG 90
+++ +AG +AG +KT VAPL+RVK+L Q + LGV +L + + EGFLGLYKG
Sbjct: 36 WLRSFLAGSIAGCCAKTTVAPLDRVKVLLQAHNHHYKHLGVFSALRAVPQKEGFLGLYKG 95
Query: 91 NGASAVRIVPYAALHFMTYERYKSWILNNCPALG-SGPFIDLLAGSAAGGTSVLCTYPLD 149
NGA +RI PY A+ FM +E YK+ I LG SG L+AGS AG T+V+CT P+D
Sbjct: 96 NGAMMIRIFPYGAIQFMAFEHYKTLITTK---LGISGHVHRLMAGSMAGMTAVICTDPVD 152
Query: 150 LARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVY-REGGARGLYRGVGPTITG 208
+ R +LA+Q V+ + + GI ++Y +EGG G YRG+ PTI G
Sbjct: 153 MVRVRLAFQ--------------VKGEHRYTGIIHAFKTIYAKEGGFFGFYRGLMPTILG 198
Query: 209 ILPYAGLKFYTYEKLK----------MHVPEEHQKSIL-----MRLSCGALAGLFGQTLT 253
+ PYAG+ F+T+ LK + P ++L + L CG +A QT++
Sbjct: 199 MAPYAGVSFFTFGTLKSVGLSHAPTLLGSPSSDNPNVLVLKTHVNLLCGGVARAIAQTIS 258
Query: 254 YPLDVVKRQMQVGSLQNAAHEDTRYRNTL------DGLRKIVRNQGWRQLFAGVSINYIR 307
YP DV +R+MQ+G++ + R+T+ G+RK L+ G+S+NYIR
Sbjct: 259 YPFDVTRRRMQLGTVLPEFEKCLTMRDTMKYDYGHHGIRK--------GLYRGLSLNYIR 310
Query: 308 IVPSAAISFTAYDTMK 323
+PS A++FT Y+ MK
Sbjct: 311 CIPSQAVAFTTYELMK 326
Score = 66.6 bits (161), Expect = 1e-09
Identities = 54/216 (25%), Positives = 93/216 (43%), Gaps = 33/216 (15%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-LGVCQSLNKL-VKHEGF 84
G+ +V L+AG +AG + P++ V++ + G H G+ + + K GF
Sbjct: 126 GISGHVHRLMAGSMAGMTAVICTDPVDMVRVRLAFQVKGEHRYTGIIHAFKTIYAKEGGF 185
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGP------------FIDLL 132
G Y+G + + + PYA + F T+ KS L++ P L P ++LL
Sbjct: 186 FGFYRGLMPTILGMAPYAGVSFFTFGTLKSVGLSHAPTLLGSPSSDNPNVLVLKTHVNLL 245
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVI----DTRGTIKDGIKGVRPQPAHIGIKSVLTS 188
G A + +YP D+ R ++ + + T++D +K H GI+
Sbjct: 246 CGGVARAIAQTISYPFDVTRRRMQLGTVLPEFEKCLTMRDTMK---YDYGHHGIR----- 297
Query: 189 VYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
+GLYRG+ +P + F TYE +K
Sbjct: 298 -------KGLYRGLSLNYIRCIPSQAVAFTTYELMK 326
Score = 58.5 bits (140), Expect = 3e-07
Identities = 50/195 (25%), Positives = 87/195 (43%), Gaps = 22/195 (11%)
Query: 132 LAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYR 191
LAGS AG + PLD R K+ Q + H+G+ S L +V +
Sbjct: 41 LAGSIAGCCAKTTVAPLD--RVKVLLQAHNHHYK-------------HLGVFSALRAVPQ 85
Query: 192 EGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKS-ILMRLSCGALAGLFGQ 250
+ G GLY+G G + I PY ++F +E K + + S + RL G++AG+
Sbjct: 86 KEGFLGLYKGNGAMMIRIFPYGAIQFMAFEHYKTLITTKLGISGHVHRLMAGSMAGMTAV 145
Query: 251 TLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQG-WRQLFAGVSINYIRIV 309
T P+D+V+ ++ + RY + + I +G + + G+ + +
Sbjct: 146 ICTDPVDMVRVRLAF-----QVKGEHRYTGIIHAFKTIYAKEGGFFGFYRGLMPTILGMA 200
Query: 310 PSAAISFTAYDTMKA 324
P A +SF + T+K+
Sbjct: 201 PYAGVSFFTFGTLKS 215
Score = 35.0 bits (79), Expect = 3.2
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVK----HEGFL-GLYK 89
L+ GGVA A+++T P + + Q T C ++ +K H G GLY+
Sbjct: 244 LLCGGVARAIAQTISYPFDVTRRRMQLGTV-LPEFEKCLTMRDTMKYDYGHHGIRKGLYR 302
Query: 90 GNGASAVRIVPYAALHFMTYERYKSW 115
G + +R +P A+ F TYE K +
Sbjct: 303 GLSLNYIRCIPSQAVAFTTYELMKQF 328
>gb|EAA58310.1| hypothetical protein AN5801.2 [Aspergillus nidulans FGSC A4]
gi|67539262|ref|XP_663405.1| hypothetical protein
AN5801_2 [Aspergillus nidulans FGSC A4]
gi|49096956|ref|XP_409938.1| hypothetical protein
AN5801.2 [Aspergillus nidulans FGSC A4]
Length = 367
Score = 201 bits (510), Expect = 3e-50
Identities = 125/339 (36%), Positives = 191/339 (55%), Gaps = 22/339 (6%)
Query: 1 MDSSQGSTLAGLVDNASIQKD---QSCFDGVPVYVKELV-----AGGVAGALSKTAVAPL 52
+D S S ++ L S Q+ S G V V E V AGGVAGA+S+T V+PL
Sbjct: 17 VDKSALSKISQLEAPTSTQETYPRSSLLHGARVAVAEPVTAAFLAGGVAGAVSRTIVSPL 76
Query: 53 ERVKILWQTRTGGF--HTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYE 110
ER+KIL Q ++ G + L + Q+L K+ + EG+ G +GNG + +RI+PY+A+ F +Y
Sbjct: 77 ERLKILLQIQSVGREEYKLSIWQALKKIGREEGWRGFLRGNGTNCIRIIPYSAVQFGSYN 136
Query: 111 RYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGI 170
YK + P P L+ G AAG TSV+ TYPLDL RT+L+ Q +D
Sbjct: 137 FYKRF-AEPSPDADLTPIRRLICGGAAGITSVIVTYPLDLVRTRLSIQSASFAALKRDSA 195
Query: 171 KGVRPQPAHIGIKSVLTSVYR-EGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPE 229
P G+ + + VY+ EGG LYRG+ PT+ G+ PY GL F TYE ++ ++
Sbjct: 196 GEKLP-----GMFTTMVLVYKNEGGFLALYRGIIPTVAGVAPYVGLNFMTYESVRKYLTP 250
Query: 230 EHQK--SILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRK 287
E S L +L GA++G QT TYP DV++R+ Q+ ++ N ++ Y + D ++
Sbjct: 251 EGDSTPSALRKLLAGAISGAVAQTCTYPFDVLRRRFQINTMSNMGYQ---YASIFDAVKV 307
Query: 288 IVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAWL 326
IV +G R LF G++ N +++ PS A S+ +++ + +L
Sbjct: 308 IVAEEGVRGLFKGIAPNLLKVAPSMASSWLSFELTRDFL 346
Score = 48.5 bits (114), Expect = 3e-04
Identities = 23/96 (23%), Positives = 54/96 (55%), Gaps = 3/96 (3%)
Query: 26 DGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTG---GFHTLGVCQSLNKLVKHE 82
D P +++L+AG ++GA+++T P + ++ +Q T G+ + ++ +V E
Sbjct: 253 DSTPSALRKLLAGAISGAVAQTCTYPFDVLRRRFQINTMSNMGYQYASIFDAVKVIVAEE 312
Query: 83 GFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILN 118
G GL+KG + +++ P A ++++E + ++L+
Sbjct: 313 GVRGLFKGIAPNLLKVAPSMASSWLSFELTRDFLLS 348
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 579,600,509
Number of Sequences: 2540612
Number of extensions: 24238015
Number of successful extensions: 70407
Number of sequences better than 10.0: 2030
Number of HSP's better than 10.0 without gapping: 1378
Number of HSP's successfully gapped in prelim test: 654
Number of HSP's that attempted gapping in prelim test: 53340
Number of HSP's gapped (non-prelim): 5698
length of query: 340
length of database: 863,360,394
effective HSP length: 128
effective length of query: 212
effective length of database: 538,162,058
effective search space: 114090356296
effective search space used: 114090356296
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0349b.8


