

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.13
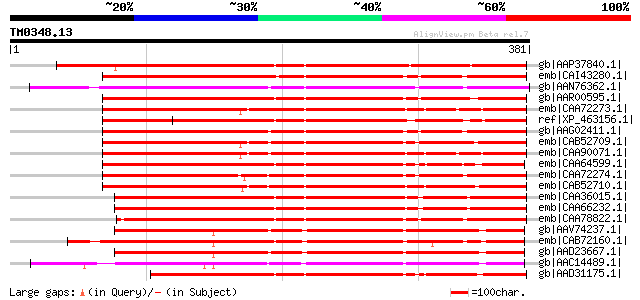
(381 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP37840.1| At1g47710 [Arabidopsis thaliana] gi|27311755|gb|A... 276 1e-72
emb|CAI43280.1| serpin [Cucumis sativus] 273 5e-72
gb|AAN76362.1| serpin-like protein [Citrus x paradisi] 271 3e-71
gb|AAR00595.1| putative serpin [Oryza sativa (japonica cultivar-... 262 1e-68
emb|CAA72273.1| serpin [Triticum aestivum] gi|7438221|pir||T0648... 262 1e-68
ref|XP_463156.1| putative serine protease inhibitor [Oryza sativ... 262 1e-68
gb|AAG02411.1| phloem serpin-1 [Cucurbita maxima] 255 2e-66
emb|CAB52709.1| serpin [Triticum aestivum] 254 2e-66
emb|CAA90071.1| serpin [Triticum aestivum] gi|2130110|pir||S6578... 253 9e-66
emb|CAA64599.1| serpin [Hordeum vulgare subsp. vulgare] gi|74382... 251 2e-65
emb|CAA72274.1| serpin [Triticum aestivum] gi|7438222|pir||T0659... 249 7e-65
emb|CAB52710.1| serpin [Triticum aestivum] 249 7e-65
emb|CAA36015.1| protein Z [Hordeum vulgare] gi|131091|sp|P06293|... 249 1e-64
emb|CAA66232.1| protein z-type serpin [Hordeum vulgare subsp. vu... 249 1e-64
emb|CAA78822.1| protein zx [Hordeum vulgare subsp. vulgare] gi|4... 246 6e-64
gb|AAV74237.1| At2g25240 [Arabidopsis thaliana] gi|42569306|ref|... 237 5e-61
emb|CAB72160.1| serpin-like protein [Arabidopsis thaliana] gi|15... 237 5e-61
gb|AAD23667.1| putative serpin [Arabidopsis thaliana] gi|2531086... 237 5e-61
gb|AAC14489.1| putative serpin [Arabidopsis thaliana] gi|1522529... 234 3e-60
gb|AAD31175.1| barley protein Z homolog [Avena fatua] 228 2e-58
>gb|AAP37840.1| At1g47710 [Arabidopsis thaliana] gi|27311755|gb|AAO00843.1| serpin,
putative [Arabidopsis thaliana]
gi|15220298|ref|NP_175202.1| serpin, putative / serine
protease inhibitor, putative [Arabidopsis thaliana]
gi|5668792|gb|AAD46018.1| Strong similarity to gb|Z15116
serpin (pazx) from Hordeum vulgare and is a member of
the PF|00079 Serpin family. ESTs gb|R65473, gb|N38150
and gb|AA712968 come from this gene. [Arabidopsis
thaliana] gi|25405891|pir||H96517 protein T2E6.22
[imported] - Arabidopsis thaliana
gi|9802595|gb|AAF99797.1| T2E6.22 [Arabidopsis thaliana]
Length = 391
Score = 276 bits (705), Expect = 1e-72
Identities = 159/349 (45%), Positives = 219/349 (62%), Gaps = 8/349 (2%)
Query: 35 AAHSTSFSPSSDLTPSTISTPSSLRGFPLCSPTKDAADADAH---HLSFTNGMFVDKSVS 91
AA S + L+ S+ L F + AD A+ LS NG ++DKS+S
Sbjct: 46 AAGSAGATKDQILSFLKFSSTDQLNSFSSEIVSAVLADGSANGGPKLSVANGAWIDKSLS 105
Query: 92 LSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHITQLLPPGTITNLTRL 151
F+ LL Y A+ DF+++ V+ +VNS E ++NG IT++LP G+ ++T+L
Sbjct: 106 FKPSFKQLLEDSYKAASNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKL 165
Query: 152 IFANALRFQGLWKHKFDGP-RYEFPFDLLNGTSVKVPFMTIKKKTHYIRAFDAFKILRLP 210
IFANAL F+G W KFD E F LL+G V PFMT KKK Y+ A+D FK+L LP
Sbjct: 166 IFANALYFKGTWNEKFDESLTQEGEFHLLDGNKVTAPFMTSKKK-QYVSAYDGFKVLGLP 224
Query: 211 YKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRKVRVDALQIPKFNIS 270
Y QG+D KR+FSM +LPDA++GLS L+ K+ S PGFL +PRR+V+V +IPKF S
Sbjct: 225 YLQGQD-KRQFSMYFYLPDANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFS 283
Query: 271 FTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIFHKAFIEVNEKGTKATA 330
F F+ASNVLK +G+ SPFS + T+M+E NLCV +IFHKA IEVNE+GT+A A
Sbjct: 284 FGFDASNVLKGLGLTSPFSGEEG-LTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAA 342
Query: 331 ASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQVLNPL 379
AS G + + I+F+A+HPFL ++ E+ TG +LF+GQV++PL
Sbjct: 343 ASAGVIKL-RGLLMEEDEIDFVADHPFLLVVTENITGVVLFIGQVVDPL 390
>emb|CAI43280.1| serpin [Cucumis sativus]
Length = 389
Score = 273 bits (699), Expect = 5e-72
Identities = 155/313 (49%), Positives = 212/313 (67%), Gaps = 10/313 (3%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ + LSF NG++VD+S+ L F+ ++ Y A L+ DFK + V +VNS
Sbjct: 83 DASPSGGPRLSFANGVWVDQSLPLKSSFKQVVDTLYKAKLSQADFKTKAAEVTSEVNSWA 142
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPR-YEFPFDLLNGTSVKVP 187
E Q+NG IT++LPPG++ +L++LI ANAL F+G W+ KFD + + F LL+G+SV+VP
Sbjct: 143 EKQTNGLITEVLPPGSVDSLSKLILANALYFKGEWEEKFDASKTKKQDFYLLDGSSVEVP 202
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FMT K K H I AFD FK+L L YKQG D R FSM IFLPD+ DGL +L+ +L S+ F
Sbjct: 203 FMTSKNKQH-IAAFDGFKVLGLSYKQGSD-PRHFSMYIFLPDSRDGLPSLIERLDSQSNF 260
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
+ +P K++V +IPKF ISF E SNVLK +G+V PFS + T+M+E + + N
Sbjct: 261 IDRHIPYEKLKVGEFKIPKFKISFGIEVSNVLKGLGLVLPFS--EGGLTEMVE-SQTAQN 317
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPG-PVINFIANHPFLFLIREDFT 366
L V IFHK+FIEVNE+GT+A AAS A V + P +I+F+A+ PFL+ IRED T
Sbjct: 318 LHVSKIFHKSFIEVNEEGTEAAAAS---AAVIKLRGLPSMDIIDFVADRPFLYAIREDKT 374
Query: 367 GTILFVGQVLNPL 379
G++LF+GQVLNPL
Sbjct: 375 GSLLFIGQVLNPL 387
>gb|AAN76362.1| serpin-like protein [Citrus x paradisi]
Length = 389
Score = 271 bits (692), Expect = 3e-71
Identities = 164/369 (44%), Positives = 220/369 (59%), Gaps = 15/369 (4%)
Query: 15 SSIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTISTPSSLRGFPLCSPTKDAADAD 74
SSIH LS + S P S S S D + S ++ D + +
Sbjct: 34 SSIHVLLSLISAGSKGPTLDQLLSFLKSKSDDQLNTFASELVAV-------VFADGSPSG 86
Query: 75 AHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNG 134
LS NG+++DKS+SL F+ ++ Y A+ +D + + V +VN E ++NG
Sbjct: 87 GPRLSVANGVWIDKSLSLKNTFKQVVDNVYKAASNQVDSQTKAAEVSREVNMWAEKETNG 146
Query: 135 HITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPR-YEFPFDLLNGTSVKVPFMTIKK 193
+ ++LPPG++ N TRLIFANAL F+G W FD + ++ F LLNG S+KVPFMT K
Sbjct: 147 LVKEVLPPGSVDNSTRLIFANALYFKGAWNETFDSSKTKDYDFHLLNGGSIKVPFMT-SK 205
Query: 194 KTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLP 253
K ++ AFD FK+L LPYKQG D KRRFSM FLPDA DGL L+ K+ SE FL LP
Sbjct: 206 KNQFVSAFDGFKVLGLPYKQGED-KRRFSMYFFLPDAKDGLPTLLEKMGSESKFLDHHLP 264
Query: 254 RRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSP-SNNLCVES 312
++V V +IP+F ISF E S VLK +G+V PFS +M ++SP NL V S
Sbjct: 265 SQRVEVGDFRIPRFKISFGIEVSKVLKGLGLVLPFSGEGGGLAEM--VDSPVGKNLYVSS 322
Query: 313 IFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFV 372
IF K+FIEVNE+GT+A AAS A V + I+F+A+HPF+F+IRED TG ++F+
Sbjct: 323 IFQKSFIEVNEEGTEAAAASA--ATVVLRSILLLDKIDFVADHPFVFMIREDMTGLVMFI 380
Query: 373 GQVLNPLDG 381
G VLNPL G
Sbjct: 381 GHVLNPLAG 389
>gb|AAR00595.1| putative serpin [Oryza sativa (japonica cultivar-group)]
Length = 396
Score = 262 bits (670), Expect = 1e-68
Identities = 145/313 (46%), Positives = 208/313 (66%), Gaps = 11/313 (3%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ A ++F +G+FVD S+SL F + +Y A S+DF+ + V VNS +
Sbjct: 90 DASGAGGPRVAFADGVFVDASLSLKKTFGDVAVGKYKAETHSVDFQTKAAEVASQVNSWV 149
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPR-YEFPFDLLNGTSVKVP 187
E ++G I ++LPPG++ + TRL+ NAL F+G W KFD + + F LL+G SV+ P
Sbjct: 150 EKVTSGLIKEILPPGSVDHTTRLVLGNALYFKGAWTEKFDASKTKDGEFHLLDGKSVQAP 209
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FM+ KK YI ++D K+L+LPY+QG D KR+FSM I LP+A DGL +L KL+SEP F
Sbjct: 210 FMSTSKK-QYILSYDNLKVLKLPYQQGGD-KRQFSMYILLPEAQDGLWSLAEKLNSEPEF 267
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
L+ +P R+V V ++PKF ISF FEAS++LK +G+ PFS +A T+M++ + N
Sbjct: 268 LEKHIPTRQVTVGQFKLPKFKISFGFEASDLLKSLGLHLPFS-SEADLTEMVD-SPEGKN 325
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVI-NFIANHPFLFLIREDFT 366
L V S+FHK+F+EVNE+GT+A AA+ + R P+ +F+A+HPFLFLI+ED T
Sbjct: 326 LFVSSVFHKSFVEVNEEGTEAAAATAAVITL-----RSAPIAEDFVADHPFLFLIQEDMT 380
Query: 367 GTILFVGQVLNPL 379
G +LFVG V+NPL
Sbjct: 381 GVVLFVGHVVNPL 393
>emb|CAA72273.1| serpin [Triticum aestivum] gi|7438221|pir||T06488 serpin WZS2 -
wheat
Length = 399
Score = 262 bits (670), Expect = 1e-68
Identities = 146/313 (46%), Positives = 209/313 (66%), Gaps = 8/313 (2%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ A ++F NG+FVD S+ L F+ L +Y A S+DF+ + V VNS +
Sbjct: 90 DASSAGGPRVAFANGVFVDASLLLKPSFQELAVCKYKAETQSVDFQTKAAEVTTQVNSWV 149
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFD--GPRYEFPFDLLNGTSVKV 186
E ++G I +LP G++ N T+L+ ANAL F+G W +FD G + ++ F LL+G+SV+
Sbjct: 150 EKVTSGRIKNILPSGSVDNTTKLVLANALYFKGAWTDQFDSYGTKNDY-FYLLDGSSVQT 208
Query: 187 PFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPG 246
PFM+ YI + D K+L+LPYKQG D R+FSM I LP+A GLS+L KLS+EP
Sbjct: 209 PFMSSMDDDQYISSSDGLKVLKLPYKQGGD-NRQFSMYILLPEAPGGLSSLAEKLSAEPD 267
Query: 247 FLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSN 306
FL+ +PR++V + ++PKF ISF EAS++LK +G+ PFS +A F++M++ P
Sbjct: 268 FLERHIPRQRVAIRQFKLPKFKISFGMEASDLLKCLGLQLPFSD-EADFSEMVDSPMP-Q 325
Query: 307 NLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFT 366
L V S+FH+AF+EVNE+GT+A AAS V Q AR P V++FIA+HPFLFL+RED +
Sbjct: 326 GLRVSSVFHQAFVEVNEQGTEA-AASTAIKMVPQQARPPS-VMDFIADHPFLFLLREDIS 383
Query: 367 GTILFVGQVLNPL 379
G +LF+G V+NPL
Sbjct: 384 GVVLFMGHVVNPL 396
>ref|XP_463156.1| putative serine protease inhibitor [Oryza sativa (japonica
cultivar-group)] gi|40539102|gb|AAR87358.1| putative
serine protease inhibitor [Oryza sativa (japonica
cultivar-group)]
Length = 653
Score = 262 bits (670), Expect = 1e-68
Identities = 145/313 (46%), Positives = 208/313 (66%), Gaps = 11/313 (3%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ A ++F +G+FVD S+SL F + +Y A S+DF+ + V VNS +
Sbjct: 90 DASGAGGPRVAFADGVFVDASLSLKKTFGDVAVGKYKAETHSVDFQTKAAEVASQVNSWV 149
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPR-YEFPFDLLNGTSVKVP 187
E ++G I ++LPPG++ + TRL+ NAL F+G W KFD + + F LL+G SV+ P
Sbjct: 150 EKVTSGLIKEILPPGSVDHTTRLVLGNALYFKGAWTEKFDASKTKDGEFHLLDGKSVQAP 209
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FM+ KK YI ++D K+L+LPY+QG D KR+FSM I LP+A DGL +L KL+SEP F
Sbjct: 210 FMSTSKK-QYILSYDNLKVLKLPYQQGGD-KRQFSMYILLPEAQDGLWSLAEKLNSEPEF 267
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
L+ +P R+V V ++PKF ISF FEAS++LK +G+ PFS +A T+M++ + N
Sbjct: 268 LEKHIPTRQVTVGQFKLPKFKISFGFEASDLLKSLGLHLPFS-SEADLTEMVD-SPEGKN 325
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVI-NFIANHPFLFLIREDFT 366
L V S+FHK+F+EVNE+GT+A AA+ + R P+ +F+A+HPFLFLI+ED T
Sbjct: 326 LFVSSVFHKSFVEVNEEGTEAAAATAAVITL-----RSAPIAEDFVADHPFLFLIQEDMT 380
Query: 367 GTILFVGQVLNPL 379
G +LFVG V+NPL
Sbjct: 381 GVVLFVGHVVNPL 393
Score = 218 bits (554), Expect = 3e-55
Identities = 128/264 (48%), Positives = 179/264 (67%), Gaps = 16/264 (6%)
Query: 120 VLHDVNSLIELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYEF-PFDL 178
VL VNS ++ ++G I + P +I + T+L+ ANAL F+G W KFD + E F L
Sbjct: 399 VLGQVNSWVDRVTSGLIKNIATPRSINHNTKLVLANALYFKGAWAEKFDVSKTEDGEFHL 458
Query: 179 LNGTSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALV 238
L+G SV+ PFM+ +KK Y+ ++D+ K+L+LPY QG D KR+FSM I LP+A DGL +L
Sbjct: 459 LDGESVQAPFMSTRKK-QYLSSYDSLKVLKLPYLQGGD-KRQFSMYILLPEAQDGLWSLA 516
Query: 239 HKLSSEPGFLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPF-SPMDACFTK 297
KL+SEP F++ +P R V V ++PKF ISF F AS +LK +G+ F S +D
Sbjct: 517 EKLNSEPEFMENHIPMRPVHVGQFKLPKFKISFGFGASGLLKGLGLPLLFGSEVD----- 571
Query: 298 MLE-INSP-SNNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANH 355
++E ++SP + NL V S+FHK+FIEVNE+GT+ATAA + V+ RP +NF+A+H
Sbjct: 572 LIEMVDSPGAQNLFVSSVFHKSFIEVNEEGTEATAAVM----VSMEHSRPRR-LNFVADH 626
Query: 356 PFLFLIREDFTGTILFVGQVLNPL 379
PF+FLIRED TG ILF+G V+NPL
Sbjct: 627 PFMFLIREDVTGVILFIGHVVNPL 650
>gb|AAG02411.1| phloem serpin-1 [Cucurbita maxima]
Length = 389
Score = 255 bits (651), Expect = 2e-66
Identities = 143/312 (45%), Positives = 202/312 (63%), Gaps = 9/312 (2%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ L+F NG+++D+S+SL F+ ++ Y A L +DF + + V+ +VNS +
Sbjct: 84 DASSCGGPRLAFVNGVWIDQSLSLKSSFQQVVDKYYKAELRQVDFLTKANEVISEVNSWV 143
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGP-RYEFPFDLLNGTSVKVP 187
E + G I ++LP G++ + T+L+ ANAL F+ W+ FD + F L++G+SVK P
Sbjct: 144 EKNTYGLIREILPAGSVGSSTQLVLANALYFKAAWQQAFDASITMKRDFYLIDGSSVKAP 203
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FM+ +K Y+ FD FK+L LPY QG D RRFSM FLPD DGL++L+ KL SEPGF
Sbjct: 204 FMS-GEKDQYVAVFDGFKVLALPYSQGPD-PRRFSMYFFLPDRKDGLASLIEKLDSEPGF 261
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
+ +P +K + IPKF ISF E S+VLK++G+V PF+ + M+E + + N
Sbjct: 262 IDRHIPCKKQELGGFLIPKFKISFGIEVSDVLKKLGLVLPFT--EGGLLGMVE-SPVAQN 318
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTG 367
L V +IFHKAFIEV+E+GTKA A+S A P I+FIAN PFL+LIRED +G
Sbjct: 319 LRVSNIFHKAFIEVDEEGTKAAASS---AVTVGIVSLPINRIDFIANRPFLYLIREDKSG 375
Query: 368 TILFVGQVLNPL 379
T+LF+GQVLNPL
Sbjct: 376 TLLFIGQVLNPL 387
>emb|CAB52709.1| serpin [Triticum aestivum]
Length = 398
Score = 254 bits (650), Expect = 2e-66
Identities = 143/314 (45%), Positives = 212/314 (66%), Gaps = 11/314 (3%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ A H++F NG+FVD S+ L F+ L +Y A S+DF+ + V VNS +
Sbjct: 90 DASSAGGPHVAFANGVFVDASLPLKPSFQELAVCKYKADTQSVDFQTKAAEVATQVNSWV 149
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFD--GPRYEFPFDLLNGTSVKV 186
E ++G I +LP G++ N T+L+ ANAL F+G W +FD G + ++ F L +G+SV+
Sbjct: 150 EKVTSGRIKDILPSGSVDNTTKLVLANALYFKGAWTDQFDSSGTKNDY-FYLPDGSSVQT 208
Query: 187 PFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPG 246
PFM+ Y+ + D K+L+LPYKQG D KR+FSM I LP+A GLS L KLS+EP
Sbjct: 209 PFMS-SMDDQYLSSSDGLKVLKLPYKQGGD-KRQFSMYILLPEAPGGLSNLAEKLSAEPD 266
Query: 247 FLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSP-S 305
FL+ +PR++V + ++PKF ISF EAS++LK +G+ PFS +A F++M ++SP +
Sbjct: 267 FLERHIPRQRVALRQFKLPKFKISFETEASDLLKCLGLQLPFS-NEADFSEM--VDSPMA 323
Query: 306 NNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDF 365
+ L V S+FH+AF+EVNE+GT+A A++ + Q+ RP V++FIA+HPFLFL+RED
Sbjct: 324 HGLRVSSVFHQAFVEVNEQGTEAAASTAIKMALLQA--RPPSVMDFIADHPFLFLLREDI 381
Query: 366 TGTILFVGQVLNPL 379
+G +LF+G V+NPL
Sbjct: 382 SGVVLFMGHVVNPL 395
>emb|CAA90071.1| serpin [Triticum aestivum] gi|2130110|pir||S65782 serpin - wheat
Length = 398
Score = 253 bits (645), Expect = 9e-66
Identities = 143/313 (45%), Positives = 205/313 (64%), Gaps = 9/313 (2%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ F NG+FVD S+ L F+ + +Y A S+DF+ + V VNS +
Sbjct: 90 DASSTGGSACRFANGVFVDASLLLKPSFQEIAVCKYKAETQSVDFQTKAAEVTTQVNSWV 149
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFD--GPRYEFPFDLLNGTSVKV 186
E ++G I +LPPG+I N T+L+ ANAL F+G W +FD G + ++ F LL+G+SV+
Sbjct: 150 EKVTSGRIKDILPPGSIDNTTKLVLANALYFKGAWTEQFDSYGTKNDY-FYLLDGSSVQT 208
Query: 187 PFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPG 246
PFM+ Y+ + D K+L+LPYKQG D R+F M I LP+A GLS+L KLS+EP
Sbjct: 209 PFMS-SMDDQYLLSSDGLKVLKLPYKQGGD-NRQFFMYILLPEAPGGLSSLAEKLSAEPD 266
Query: 247 FLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSN 306
FL+ +PR++V + ++PKF ISF EAS++LK +G+ PF +A F++M++ P
Sbjct: 267 FLERHIPRQRVALRQFKLPKFKISFGIEASDLLKCLGLQLPFGD-EADFSEMVDSLMP-Q 324
Query: 307 NLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFT 366
L V S+FH+AF+EVNE+GT+A AAS V Q AR P V++FIA+HPFLFL+RED +
Sbjct: 325 GLRVSSVFHQAFVEVNEQGTEA-AASTAIKMVLQQARPPS-VMDFIADHPFLFLVREDIS 382
Query: 367 GTILFVGQVLNPL 379
G +LF+G V+NPL
Sbjct: 383 GVVLFMGHVVNPL 395
>emb|CAA64599.1| serpin [Hordeum vulgare subsp. vulgare] gi|7438218|pir||T06183
serpin - barley
Length = 397
Score = 251 bits (642), Expect = 2e-65
Identities = 142/312 (45%), Positives = 202/312 (64%), Gaps = 13/312 (4%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ ++F NG+FVD S+SL F+ L Y + + S+DFK + VNS +
Sbjct: 93 DASINSGPRIAFANGVFVDASLSLKPSFQELAVCNYKSEVQSVDFKTKAPEAASQVNSWV 152
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVP 187
+ + G I ++LP G+I N TRL+ NAL F+GLW KFD + ++ F LLNG++V+ P
Sbjct: 153 KNVTAGLIEEILPAGSIDNTTRLVLGNALYFKGLWTKKFDESKTKYDDFHLLNGSTVQTP 212
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FM+ K Y+ + D K+L+LPY+ G D R+FSM I LP+A DGLS L KLS+EP F
Sbjct: 213 FMSSTNK-QYLSSSDGLKVLKLPYQHGGD-NRQFSMYILLPEAHDGLSRLAQKLSTEPDF 270
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
L+ ++P +V V +PKF ISF FEA+ +LK +G+ PFS ++A ++M +NSP
Sbjct: 271 LENRIPTEEVEVGQFMLPKFKISFGFEANKLLKTLGLQLPFS-LEANLSEM--VNSPM-G 326
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPV-INFIANHPFLFLIREDFT 366
L + S+FHK F+EV+E+GTKA AA+ G V +S P+ ++F+ANHPFLFLIRED
Sbjct: 327 LYISSVFHKTFVEVDEEGTKAGAAT-GDVIVDRSL----PIRMDFVANHPFLFLIREDIA 381
Query: 367 GTILFVGQVLNP 378
G +LF+G V NP
Sbjct: 382 GVVLFIGHVANP 393
>emb|CAA72274.1| serpin [Triticum aestivum] gi|7438222|pir||T06597 serpin homolog
WZS3 - wheat
Length = 398
Score = 249 bits (637), Expect = 7e-65
Identities = 143/314 (45%), Positives = 202/314 (63%), Gaps = 11/314 (3%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA++ ++F NG+FVD S+ L F+ L +Y A S+DF+ + V VNS +
Sbjct: 90 DASNIGGPRVAFANGVFVDASLQLKPSFQELAVCKYKAEAQSVDFQTKAAEVTAQVNSWV 149
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPR--YEFPFDLLNGTSVKV 186
E + G I +LP G+I N TRL+ NAL F+G W +FD PR F LL+G+S++
Sbjct: 150 EKVTTGLIKDILPAGSIDNTTRLVLGNALYFKGAWTDQFD-PRATQSDDFYLLDGSSIQT 208
Query: 187 PFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPG 246
PFM + YI + D K+L+LPYKQG D KR+FSM I LP+A GL +L KLS+EP
Sbjct: 209 PFM-YSSEEQYISSSDGLKVLKLPYKQGGD-KRQFSMYILLPEALSGLWSLAEKLSAEPE 266
Query: 247 FLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSP-S 305
FL+ +PR+KV + ++PKF IS EAS++LK +G++ PF +A ++M ++SP +
Sbjct: 267 FLEQHIPRQKVALRQFKLPKFKISLGIEASDLLKGLGLLLPFG-AEADLSEM--VDSPMA 323
Query: 306 NNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDF 365
NL + SIFHKAF+EVNE GT+A A ++ A V P V++FI +HPFLFLIRED
Sbjct: 324 QNLYISSIFHKAFVEVNETGTEAAATTI--AKVVLRQAPPPSVLDFIVDHPFLFLIREDT 381
Query: 366 TGTILFVGQVLNPL 379
+G +LF+G V+NPL
Sbjct: 382 SGVVLFIGHVVNPL 395
>emb|CAB52710.1| serpin [Triticum aestivum]
Length = 398
Score = 249 bits (637), Expect = 7e-65
Identities = 139/313 (44%), Positives = 205/313 (65%), Gaps = 9/313 (2%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
DA+ AD+ ++F NG+FVD S+ L F+ L +Y A S+DF+ + V VNS +
Sbjct: 90 DASYADSPRVTFANGVFVDASLPLKPSFQELAVCKYKAEAQSVDFQTKAAEVTAQVNSWV 149
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDG--PRYEFPFDLLNGTSVKV 186
E + G I +LP G+I+N TRL+ NAL F+G W +FD + ++ F LL+G+S++
Sbjct: 150 EKVTTGLIKDILPAGSISNTTRLVLGNALYFKGAWTDQFDSRVTKSDY-FYLLDGSSIQT 208
Query: 187 PFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPG 246
PFM + YI + D K+L+LPYKQG D KR+FSM I LP+A G+ +L KLS+EP
Sbjct: 209 PFM-YSSEEQYISSSDGLKVLKLPYKQGGD-KRQFSMYILLPEAPSGIWSLAEKLSAEPE 266
Query: 247 FLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSN 306
L+ +PR+KV + ++PKF ISF EAS++LK +G+ PFS +A ++M++ P
Sbjct: 267 LLERHIPRQKVALRQFKLPKFKISFGIEASDLLKHLGLQLPFSD-EADLSEMVDSPMP-Q 324
Query: 307 NLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFT 366
L + S+FHK F+EVNE GT+A AA++ A + ++ P ++FIA+HPFLFLIRED +
Sbjct: 325 GLRISSVFHKTFVEVNETGTEAAAATIAKAVLLSAS--PPSDMDFIADHPFLFLIREDTS 382
Query: 367 GTILFVGQVLNPL 379
G +LF+G V+NPL
Sbjct: 383 GVVLFIGHVVNPL 395
>emb|CAA36015.1| protein Z [Hordeum vulgare] gi|131091|sp|P06293|PRTZ_HORVU Protein
Z (Z4) (Major endosperm albumin)
Length = 399
Score = 249 bits (636), Expect = 1e-64
Identities = 137/303 (45%), Positives = 197/303 (64%), Gaps = 8/303 (2%)
Query: 78 LSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHIT 137
++F NG+FVD S+SL F L QY A S+DF+++ + VNS +E + G I
Sbjct: 101 IAFANGIFVDASLSLKPSFEELAVCQYKAKTQSVDFQHKTLEAVGQVNSWVEQVTTGLIK 160
Query: 138 QLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVPFMTIKKKTH 196
Q+LPPG++ N T+LI NAL F+G W KFD + F LL+G+S++ FM+ KK
Sbjct: 161 QILPPGSVDNTTKLILGNALYFKGAWDQKFDESNTKCDSFHLLDGSSIQTQFMSSTKK-Q 219
Query: 197 YIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRK 256
YI + D K+L+LPY +G D KR+FSM I LP A DGL +L +LS+EP F++ +P++
Sbjct: 220 YISSSDNLKVLKLPYAKGHD-KRQFSMYILLPGAQDGLWSLAKRLSTEPEFIENHIPKQT 278
Query: 257 VRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIFHK 316
V V Q+PKF IS+ FEAS++L+ +G+ PFS +A ++M++ S L + +FHK
Sbjct: 279 VEVGRFQLPKFKISYQFEASSLLRALGLQLPFSE-EADLSEMVD---SSQGLEISHVFHK 334
Query: 317 AFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQVL 376
+F+EVNE+GT+A AA+V V S +++F+ANHPFLFLIRED G ++FVG V
Sbjct: 335 SFVEVNEEGTEAGAATVAMG-VAMSMPLKVDLVDFVANHPFLFLIREDIAGVVVFVGHVT 393
Query: 377 NPL 379
NPL
Sbjct: 394 NPL 396
>emb|CAA66232.1| protein z-type serpin [Hordeum vulgare subsp. vulgare]
Length = 400
Score = 249 bits (635), Expect = 1e-64
Identities = 136/303 (44%), Positives = 197/303 (64%), Gaps = 8/303 (2%)
Query: 78 LSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHIT 137
++F NG+FVD S+SL F L QY A S+DF+++ + VNS +E + G I
Sbjct: 102 IAFANGIFVDASLSLKPSFEELAVCQYKAKTQSVDFQHKTLEAVGQVNSWVEQVTTGLIK 161
Query: 138 QLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVPFMTIKKKTH 196
Q+LPPG++ N T+L+ NAL F+G W KFD + F LL+G+S++ FM+ KK
Sbjct: 162 QILPPGSVDNTTKLVLGNALYFKGAWDQKFDESNTKCDSFHLLDGSSIQTQFMSSTKK-Q 220
Query: 197 YIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRK 256
YI + D K+L+LPY +G D KR+FSM I LP A DGL +L +LS+EP F++ +P++
Sbjct: 221 YISSSDNLKVLKLPYAKGHD-KRQFSMYILLPGAQDGLWSLAKRLSTEPEFIENHIPKQT 279
Query: 257 VRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIFHK 316
V V Q+PKF IS+ FEAS++L+ +G+ PFS +A ++M++ S L + +FHK
Sbjct: 280 VEVGRFQLPKFKISYQFEASSLLRALGLQLPFSE-EADLSEMVD---SSQGLEISHVFHK 335
Query: 317 AFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQVL 376
+F+EVNE+GT+A AA+V V S +++F+ANHPFLFLIRED G ++FVG V
Sbjct: 336 SFVEVNEEGTEAGAATVAMG-VAMSMPLKVDLVDFVANHPFLFLIREDIAGVVVFVGHVT 394
Query: 377 NPL 379
NPL
Sbjct: 395 NPL 397
>emb|CAA78822.1| protein zx [Hordeum vulgare subsp. vulgare] gi|421989|pir||S29819
serpin - barley gi|444778|prf||1908213A protein Zx
Length = 398
Score = 246 bits (629), Expect = 6e-64
Identities = 140/303 (46%), Positives = 203/303 (66%), Gaps = 9/303 (2%)
Query: 79 SFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHITQ 138
SF N +FVD S+ L F+ L+ +Y S+DF+ + V VNS +E + G I +
Sbjct: 100 SFAN-VFVDSSLKLKPSFKDLVVGKYKGETQSVDFQTKAPEVAGQVNSWVEKITTGLIKE 158
Query: 139 LLPPGTITNLTRLIFANALRFQGLWKHKFDGPRY-EFPFDLLNGTSVKVPFMTIKKKTHY 197
+LP G++ + TRL+ NAL F+G W KFD + + F LL+G+SV+ PFM+ KK Y
Sbjct: 159 ILPAGSVDSTTRLVLGNALYFKGSWTEKFDASKTKDEKFHLLDGSSVQTPFMSSTKK-QY 217
Query: 198 IRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRKV 257
I ++D+ K+L+LPY+QG D KR+FSM I LP+A DGL L +KLS+EP F++ +P +KV
Sbjct: 218 ISSYDSLKVLKLPYQQGGD-KRQFSMYILLPEAQDGLWNLANKLSTEPEFMEKHMPMQKV 276
Query: 258 RVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIFHKA 317
V ++PKF ISF FEAS++LK +G+ PFS +A ++M++ + + +L V S+FHK+
Sbjct: 277 PVGQFKLPKFKISFGFEASDMLKGLGLQLPFS-SEADLSEMVD-SPAARSLYVSSVFHKS 334
Query: 318 FIEVNEKGTKATAASVGFAPVTQSARRPGPV-INFIANHPFLFLIREDFTGTILFVGQVL 376
F+EVNE+GT+A A + VT + PV ++F+A+HPFLFLIRED TG +LFVG V
Sbjct: 335 FVEVNEEGTEAAARTA--RVVTLRSLPVEPVKVDFVADHPFLFLIREDLTGVVLFVGHVF 392
Query: 377 NPL 379
NPL
Sbjct: 393 NPL 395
>gb|AAV74237.1| At2g25240 [Arabidopsis thaliana] gi|42569306|ref|NP_180096.2|
serpin, putative / serine protease inhibitor, putative
[Arabidopsis thaliana] gi|63003890|gb|AAY25474.1|
At2g25240 [Arabidopsis thaliana]
Length = 324
Score = 237 bits (604), Expect = 5e-61
Identities = 134/304 (44%), Positives = 193/304 (63%), Gaps = 12/304 (3%)
Query: 78 LSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHIT 137
LS NG+++DK SL F+ LL Y A+ + +DF ++ V+ +VN+ E+ +NG I
Sbjct: 27 LSIANGVWIDKFFSLKLSFKDLLENSYKATCSQVDFASKPSEVIDEVNTWAEVHTNGLIK 86
Query: 138 QLLPPGTITNL--TRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVPFMTIKKK 194
Q+L +I + + L+ ANA+ F+G W KFD + F LL+GTSVKVPFMT +
Sbjct: 87 QILSRDSIDTIRSSTLVLANAVYFKGAWSSKFDANMTKKNDFHLLDGTSVKVPFMT-NYE 145
Query: 195 THYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPR 254
Y+R++D FK+LRLPY + + R+FSM I+LP+ +GL+ L+ K+ SEP F +P
Sbjct: 146 DQYLRSYDGFKVLRLPYIEDQ---RQFSMYIYLPNDKEGLAPLLEKIGSEPSFFDNHIPL 202
Query: 255 RKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIF 314
+ V A +IPKF SF F AS VLK++G+ SPF+ T+M++ S ++L V SI
Sbjct: 203 HCISVGAFRIPKFKFSFEFNASEVLKDMGLTSPFN-NGGGLTEMVDSPSNGDDLYVSSIL 261
Query: 315 HKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQ 374
HKA IEV+E+GT+A A SVG T R P +F+A+ PFLF +RED +G ILF+GQ
Sbjct: 262 HKACIEVDEEGTEAAAVSVGVVSCTSFRRNP----DFVADRPFLFTVREDKSGVILFMGQ 317
Query: 375 VLNP 378
VL+P
Sbjct: 318 VLDP 321
>emb|CAB72160.1| serpin-like protein [Arabidopsis thaliana]
gi|15230638|ref|NP_190108.1| serpin, putative / serine
protease inhibitor, putative [Arabidopsis thaliana]
gi|11279069|pir||T47462 serpin-like protein -
Arabidopsis thaliana
Length = 393
Score = 237 bits (604), Expect = 5e-61
Identities = 142/345 (41%), Positives = 212/345 (61%), Gaps = 27/345 (7%)
Query: 43 PSSDLTPSTISTPSSLRGFPLCSPTKDAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAY 102
PSSD + ++ S+ D + HLS G+++DKS+S F+ LL
Sbjct: 64 PSSDYLNAVLAKTVSVA-------LNDGMERSDLHLSTAYGVWIDKSLSFKPSFKDLLEN 116
Query: 103 QYNASLASLDFKNRGDRVLHDVNSLIELQSNGHITQLLPPGTITNL--TRLIFANALRFQ 160
YNA+ +DF + V+++VN+ E+ +NG I ++L +I + + LI ANA+ F+
Sbjct: 117 SYNATCNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFK 176
Query: 161 GLWKHKFDGPRYE-FPFDLLNGTSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKR 219
G W KFD + + F LL+GT VKVPFMT KK Y+ +D FK+LRLPY + + R
Sbjct: 177 GAWSKKFDAKLTKSYDFHLLDGTMVKVPFMTNYKK-QYLEYYDGFKVLRLPYVEDQ---R 232
Query: 220 RFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRKVRVDALQIPKFNISFTFEASNVL 279
+F+M I+LP+ DGL L+ ++SS+P FL +PR+++ +A +IPKF SF F+AS+VL
Sbjct: 233 QFAMYIYLPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVL 292
Query: 280 KEVGVVSPFSPMDACFTKMLEINSPSNNLC------VESIFHKAFIEVNEKGTKATAASV 333
KE+G+ PF+ T+M+E S NLC V ++FHKA IEV+E+GT+A A SV
Sbjct: 293 KEMGLTLPFT--HGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVSV 350
Query: 334 GFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQVLNP 378
A +T+ G +F+A+HPFLF +RE+ +G ILF+GQVL+P
Sbjct: 351 --ASMTKDMLLMG---DFVADHPFLFTVREEKSGVILFMGQVLDP 390
>gb|AAD23667.1| putative serpin [Arabidopsis thaliana] gi|25310862|pir||A84646
probable serpin [imported] - Arabidopsis thaliana
Length = 385
Score = 237 bits (604), Expect = 5e-61
Identities = 134/304 (44%), Positives = 193/304 (63%), Gaps = 12/304 (3%)
Query: 78 LSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHIT 137
LS NG+++DK SL F+ LL Y A+ + +DF ++ V+ +VN+ E+ +NG I
Sbjct: 88 LSIANGVWIDKFFSLKLSFKDLLENSYKATCSQVDFASKPSEVIDEVNTWAEVHTNGLIK 147
Query: 138 QLLPPGTITNL--TRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVPFMTIKKK 194
Q+L +I + + L+ ANA+ F+G W KFD + F LL+GTSVKVPFMT +
Sbjct: 148 QILSRDSIDTIRSSTLVLANAVYFKGAWSSKFDANMTKKNDFHLLDGTSVKVPFMT-NYE 206
Query: 195 THYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPR 254
Y+R++D FK+LRLPY + + R+FSM I+LP+ +GL+ L+ K+ SEP F +P
Sbjct: 207 DQYLRSYDGFKVLRLPYIEDQ---RQFSMYIYLPNDKEGLAPLLEKIGSEPSFFDNHIPL 263
Query: 255 RKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIF 314
+ V A +IPKF SF F AS VLK++G+ SPF+ T+M++ S ++L V SI
Sbjct: 264 HCISVGAFRIPKFKFSFEFNASEVLKDMGLTSPFN-NGGGLTEMVDSPSNGDDLYVSSIL 322
Query: 315 HKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQ 374
HKA IEV+E+GT+A A SVG T R P +F+A+ PFLF +RED +G ILF+GQ
Sbjct: 323 HKACIEVDEEGTEAAAVSVGVVSCTSFRRNP----DFVADRPFLFTVREDKSGVILFMGQ 378
Query: 375 VLNP 378
VL+P
Sbjct: 379 VLDP 382
>gb|AAC14489.1| putative serpin [Arabidopsis thaliana] gi|15225297|ref|NP_180207.1|
serpin, putative / serine protease inhibitor, putative
[Arabidopsis thaliana] gi|7438225|pir||T00972 probable
serpin [imported] - Arabidopsis thaliana
Length = 389
Score = 234 bits (597), Expect = 3e-60
Identities = 150/371 (40%), Positives = 213/371 (56%), Gaps = 30/371 (8%)
Query: 16 SIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTIS--TPSSLRGFPLCSPTKDAADA 73
SI+ LS + S P ++ S SPS+D + ++ LC
Sbjct: 38 SINVLLSLIAAGSNPVTKEEILSFLMSPSTDHLNAVLAKIADGGTERSDLC--------- 88
Query: 74 DAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSN 133
LS +G+++DKS L F+ LL Y AS + +DF + V+ +VN ++ +N
Sbjct: 89 ----LSTAHGVWIDKSSYLKPSFKELLENSYKASCSQVDFATKPVEVIDEVNIWADVHTN 144
Query: 134 GHITQLLP---PGTITNL--TRLIFANALRFQGLWKHKFDGP-RYEFPFDLLNGTSVKVP 187
G I Q+L TI + + LI ANA+ F+ W KFD + F LL+G +VKVP
Sbjct: 145 GLIKQILSRDCTDTIKEIRNSTLILANAVYFKAAWSRKFDAKLTKDNDFHLLDGNTVKVP 204
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FM + K Y+R +D F++LRLPY + KR FSM I+LP+ DGL+AL+ K+S+EPGF
Sbjct: 205 FM-MSYKDQYLRGYDGFQVLRLPYVED---KRHFSMYIYLPNDKDGLAALLEKISTEPGF 260
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
L +P + VDAL+IPK N SF F+AS VLK++G+ SPF+ T+M++ S +
Sbjct: 261 LDSHIPLHRTPVDALRIPKLNFSFEFKASEVLKDMGLTSPFT-SKGNLTEMVDSPSNGDK 319
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTG 367
L V SI HKA IEV+E+GT+A A SV R P +F+A+HPFLF +RED +G
Sbjct: 320 LHVSSIIHKACIEVDEEGTEAAAVSVAIMMPQCLMRNP----DFVADHPFLFTVREDNSG 375
Query: 368 TILFVGQVLNP 378
ILF+GQVL+P
Sbjct: 376 VILFIGQVLDP 386
>gb|AAD31175.1| barley protein Z homolog [Avena fatua]
Length = 280
Score = 228 bits (581), Expect = 2e-58
Identities = 130/277 (46%), Positives = 182/277 (64%), Gaps = 11/277 (3%)
Query: 104 YNASLASLDFKNRGDRVLHDVNSLIELQSNGHITQLLPPGTITNLTRLIFANALRFQGLW 163
Y A + S DF+ + + V S +E + G I +LP G++ N T L+ ANAL F+G W
Sbjct: 11 YKADVQSADFQTKPAEAVAQVISWVEKATAGLIEDVLPQGSVDNSTALVLANALYFKGSW 70
Query: 164 KHKFDGPRY-EFPFDLLNGTSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFS 222
KFD + E F LL+G+SV+ PFM+ KK YI + D KIL+LPY QG D KR+FS
Sbjct: 71 HEKFDSSKTRENMFHLLDGSSVQTPFMSTTKK-QYISSLDNLKILKLPYHQGGD-KRKFS 128
Query: 223 MCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEV 282
M I LP+A DGL +L +LS+EP F++ +P+ KV V ++PKF ISF FEASN+L+ +
Sbjct: 129 MYILLPEALDGLWSLAKRLSTEPEFIENHIPKEKVEVGQFKLPKFKISFGFEASNLLQGL 188
Query: 283 GVVSPFSPMDACFTKMLEINSPSNNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSA 342
G+ PFS +A T+M ++SP NL + ++ HK+F+EVNE+GT+A AA+ T
Sbjct: 189 GLQLPFS-TEADLTEM--VDSP-ENLHISAVQHKSFVEVNEEGTEAVAATATTIMQTSMP 244
Query: 343 RRPGPVINFIANHPFLFLIREDFTGTILFVGQVLNPL 379
R I+F+A+HPFLFLIRED +G +LFVG V+NP+
Sbjct: 245 R----TIDFVADHPFLFLIREDVSGVVLFVGHVVNPI 277
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 640,242,920
Number of Sequences: 2540612
Number of extensions: 26221054
Number of successful extensions: 63421
Number of sequences better than 10.0: 1200
Number of HSP's better than 10.0 without gapping: 671
Number of HSP's successfully gapped in prelim test: 529
Number of HSP's that attempted gapping in prelim test: 60104
Number of HSP's gapped (non-prelim): 1274
length of query: 381
length of database: 863,360,394
effective HSP length: 130
effective length of query: 251
effective length of database: 533,080,834
effective search space: 133803289334
effective search space used: 133803289334
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0348.13


