

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0342a.5
(1069 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
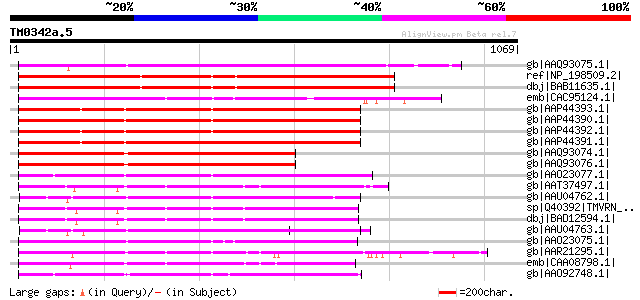
gb|AAQ93075.1| TIR-NBS-LRR type R protein 7 [Malus baccata] 669 0.0
ref|NP_198509.2| disease resistance protein (TIR-NBS-LRR class),... 613 e-173
dbj|BAB11635.1| TMV resistance protein N [Arabidopsis thaliana] 611 e-173
emb|CAC95124.1| TIR/NBS/LRR protein [Populus deltoides] 546 e-153
gb|AAP44393.1| nematode resistance-like protein [Solanum tuberosum] 519 e-145
gb|AAP44390.1| nematode resistance protein [Solanum tuberosum] 517 e-145
gb|AAP44392.1| nematode resistance-like protein [Solanum tuberosum] 515 e-144
gb|AAP44391.1| nematode resistance-like protein [Solanum tuberosum] 514 e-144
gb|AAQ93074.1| putative TIR-NBS type R protein 4 [Malus baccata] 505 e-141
gb|AAQ93076.1| putative TIR-NBS type R protein 4 [Malus baccata] 503 e-140
gb|AAO23077.1| R 8 protein [Glycine max] 475 e-132
gb|AAT37497.1| N-like protein [Nicotiana tabacum] 475 e-132
gb|AAU04762.1| MRGH21 [Cucumis melo] 473 e-131
sp|Q40392|TMVRN_NICGU TMV resistance protein N gi|558887|gb|AAA5... 471 e-131
dbj|BAD12594.1| N protein [Nicotiana tabacum] 471 e-131
gb|AAU04763.1| MRGH8 [Cucumis melo] 466 e-129
gb|AAO23075.1| R 5 protein [Glycine max] 464 e-129
gb|AAR21295.1| bacterial spot disease resistance protein 4 [Lyco... 464 e-129
emb|CAA08798.1| NL27 [Solanum tuberosum] 462 e-128
gb|AAO92748.1| candidate disease-resistance protein SR1 [Glycine... 461 e-128
>gb|AAQ93075.1| TIR-NBS-LRR type R protein 7 [Malus baccata]
Length = 1095
Score = 669 bits (1726), Expect = 0.0
Identities = 401/960 (41%), Positives = 559/960 (57%), Gaps = 41/960 (4%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR+ F HL+AAL + G ++D + L +G E+K L RAIEGSRIS+ V S YA
Sbjct: 31 GEDTRNGFTGHLHAALKDRGYQAYMDQDDLNRGEEIKEELFRAIEGSRISIIVFSKRYAD 90
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLL-------------GKQ 124
S WCLDELVKI+ECR G+ V+P+FYHVDPSHVR Q G + GK+
Sbjct: 91 SSWCLDELVKIMECRSKLGRHVLPIFYHVDPSHVRKQDGDLAEAFLKHEEGIGEGTDGKK 150
Query: 125 LQVEDTQRKT-RRGGHEVASFSGYNLSPDRSEAEAI---NEIVKNVFRN--LDIRLMSIT 178
+ + + K ++ E A+ SG++L + EA EIV N+ + + +
Sbjct: 151 REAKQERVKQWKKALTEAANLSGHDLRITDNGREANLCPREIVDNIITKWLMSTNKLRVA 210
Query: 179 DFPVGLESHLQEVIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIEN 238
VG+ S +Q++I + S S V MVGIWGMGGLGKTT AKAI+NQIH +F SF+ +
Sbjct: 211 KHQVGINSRIQDIISRLSSGGSNVIMVGIWGMGGLGKTTAAKAIYNQIHHEFQFKSFLPD 270
Query: 239 IREVCENDSRGHMHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSE 298
+ G ++LQ++L+ D+LKTK KI S+ G +I+ +R LVI+D++
Sbjct: 271 VGNAASK--HGLVYLQKELIYDILKTKSKISSVDEGIGLIEDQFRHRRVLVIMDNIDEVG 328
Query: 299 QLRALCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEAS 358
QL A+ GN +WF GS II+TTRD LL + D Y +++DE E+LELFSWHAFG
Sbjct: 329 QLDAIVGNPDWFGPGSRIIITTRDEHLLKQV--DKTYVAQKLDEREALELFSWHAFGNNW 386
Query: 359 PREDFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRIS 418
P E++ ELS +VV+YCGGLPLALEVLGS+L +R EW+S L KL+R P ++ + LRIS
Sbjct: 387 PNEEYLELSEKVVSYCGGLPLALEVLGSFLFKRPIAEWKSQLEKLKRTPEGKIIKSLRIS 446
Query: 419 YDGLKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKL 478
++GL D+ +K IFLDI CFF+G+D+ YV +L+GCG YA IGI++L ER L+ VE +NKL
Sbjct: 447 FEGL-DDAQKAIFLDISCFFIGEDKDYVAKVLDGCGFYATIGISVLRERCLVTVE-HNKL 504
Query: 479 GMHDLVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRV 538
MHDL+R+M + I+ E S D GK SRLW +V +VLT +GTE VEGL L
Sbjct: 505 NMHDLLREMAKVIISEKSPGDPGKWSRLWDKREVINVLTNKSGTEEVEGLALPWGYRHDT 564
Query: 539 CFSTNAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFY-LGNLVVL 597
FST AF ++KLRLLQL V+L G+Y+HL EL W++W IPD F+ LVVL
Sbjct: 565 AFSTEAFANLKKLRLLQLCRVELNGEYKHLPKELIWLHWFECPLKSIPDDFFNQDKLVVL 624
Query: 598 ELKYSSVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHT 657
E+++S + QVW+ +K L LK LDLS S L+ +PDFS++PNLE+LIL +C L E+H +
Sbjct: 625 EMQWSKLVQVWEGSKSLHNLKTLDLSESRSLQKSPDFSQVPNLEELILYNCKELSEIHPS 684
Query: 658 IGDLSNLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIA 717
IG L L L+NLE C L +LPG Y+ KSV+ L+++GC + +L EDI +M SL TL A
Sbjct: 685 IGHLKRLSLVNLEWCDKLISLPGDFYKSKSVEALLLNGCLILRELHEDIGEMISLRTLEA 744
Query: 718 KDTAIKEVPYSILGLKSIGYISLCGYEGL----SRDVFPSLLRSWISPTMNPLSHIPLFA 773
+ T I+EVP SI+ LK++ +SL E + S SL +S IP
Sbjct: 745 EYTDIREVPPSIVRLKNLTRLSLSSVESIHLPHSLHGLNSLRELNLSSFELADDEIPKDL 804
Query: 774 AMSLPLVSMDIQNYSSGNSSSWLSSLSKLRSVWIQCHSEIQLAQESRRILDDQYDVNSMD 833
+ L +++Q S LS LSKL ++ + +++ + L
Sbjct: 805 GSLISLQDLNLQRNDFHTLPS-LSGLSKLETLRLHHCEQLRTITDLPTNLKFLLANGCPA 863
Query: 834 LEASSSSSYASQISDFSLKSLLIRLGSCYTVIDTLASSISQGSTTNASSNFFLPGGDDPY 893
LE + S S I + + S + L +I QG T+ FL P
Sbjct: 864 LETMPNFSEMSNIRELKVSD------SPNNLSTHLRKNILQGWTSCGFGGIFLHANYVPD 917
Query: 894 WLAYKGEGPSVHFQVPEDRDCCLKGIVLCVVYSSTPENMATECLTSVMINNHTKFTIQIY 953
W + EG V F +P +G+ L +Y S + L ++INN + ++ Y
Sbjct: 918 WFEFVNEGTKVTFDIPPSDGRNFEGLTLFCMYHS----YRSRQLAIIVINNTQRTELRAY 973
>ref|NP_198509.2| disease resistance protein (TIR-NBS-LRR class), putative
[Arabidopsis thaliana]
Length = 1188
Score = 613 bits (1580), Expect = e-173
Identities = 350/802 (43%), Positives = 502/802 (61%), Gaps = 15/802 (1%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
G D R +F+SHLY +L GI+ F+DD +L++G + P LL AIE S+I + VL+ +YAS
Sbjct: 22 GADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPELLNAIETSKILIVVLTKDYAS 81
Query: 78 SRWCLDELVKILECRRTY-GQLVIPLFYHVDPSHVRNQKGPV---LLLGKQLQVEDTQRK 133
S WCLDELV I++ + +V P+F +VDPS +R Q+G K + +
Sbjct: 82 SAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQGSYAKSFSKHKNSHPLNKLKD 141
Query: 134 TRRGGHEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEVIK 193
R +VA+ SG+++ +R+EAE I +I + + + L + + + + VGL S LQ +
Sbjct: 142 WREALTKVANISGWDIK-NRNEAECIADITREILKRLPCQYLHVPSYAVGLRSRLQHISS 200
Query: 194 LIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHMHL 253
L+ S V ++ I+GMGG+GKTT AK FN+ F +SF+EN RE + G HL
Sbjct: 201 LLSIGSDGVRVIVIYGMGGIGKTTLAKVAFNEFSHLFEGSSFLENFREYSKKPE-GRTHL 259
Query: 254 QEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFASG 313
Q QLLSD+L+ + I G A +K+ KR L+++DDV QL + +R+ F G
Sbjct: 260 QHQLLSDILRRND-IEFKGLDHA-VKERFRSKRVLLVVDDVDDVHQLNSAAIDRDCFGHG 317
Query: 314 SVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVVAY 373
S II+TTR++ LL L+A+ Y+ KE+D DESLELFSWHAF + P ++F + S VV Y
Sbjct: 318 SRIIITTRNMHLLKQLRAEGSYSPKELDGDESLELFSWHAFRTSEPPKEFLQHSEEVVTY 377
Query: 374 CGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIFLD 433
C GLPLA+EVLG++L ER EWES L L+RIPND +Q KL+IS++ L E +KD+FLD
Sbjct: 378 CAGLPLAVEVLGAFLIERSIREWESTLKLLKRIPNDNIQAKLQISFNALTIE-QKDVFLD 436
Query: 434 ICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGRAIVR 493
I CFF+G D YV IL+GC LY DI ++LL+ER L+ + NN + MHDL+RDMGR IVR
Sbjct: 437 IACFFIGVDSYYVACILDGCNLYPDIVLSLLMERCLITISGNN-IMMHDLLRDMGRQIVR 495
Query: 494 ESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMRKLRL 553
E S K G+RSRLW H DV VL K +GT +EGL LK F AF +M++LRL
Sbjct: 496 EISPKKCGERSRLWSHNDVVGVLKKKSGTNAIEGLSLKADVMDFQYFEVEAFAKMQELRL 555
Query: 554 LQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWK---E 610
L+L VDL G Y H +L+W+ W GF+ P + L +L L+L+YS++K+ WK
Sbjct: 556 LELRYVDLNGSYEHFPKDLRWLCWHGFSLECFPINLSLESLAALDLQYSNLKRFWKAQSP 615
Query: 611 TKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDL-SNLLLINL 669
+ +K LDLSHS YL TPDFS PN+EKLIL +C SL +H +IG L L+L+NL
Sbjct: 616 PQPANMVKYLDLSHSVYLRETPDFSYFPNVEKLILINCKSLVLVHKSIGILDKKLVLLNL 675
Query: 670 EGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSI 729
C L+ LP + Y+LKS+++L +S CSK+++L++ + ++ESLTTL+A TA++E+P +I
Sbjct: 676 SSCIELDVLPEEIYKLKSLESLFLSNCSKLERLDDALGELESLTTLLADFTALREIPSTI 735
Query: 730 LGLKSIGYISLCGYEGLSRDVFPSLLRSWISPTMNPLSHIPLFAAMSLPLVSMDIQNYSS 789
LK + +SL G +GL D +L S S +++ L + L + ++S+ N S
Sbjct: 736 NQLKKLKRLSLNGCKGLLSDDIDNLY-SEKSHSVSLLRPVSLSGLTYMRILSLGYCNLSD 794
Query: 790 GNSSSWLSSLSKLRSVWIQCHS 811
+ SLS LR + ++ +S
Sbjct: 795 ELIPEDIGSLSFLRDLDLRGNS 816
Score = 40.0 bits (92), Expect = 0.42
Identities = 29/75 (38%), Positives = 36/75 (47%), Gaps = 1/75 (1%)
Query: 598 ELKYSSVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHT 657
EL S ++ L L LD+ L+ TPD SK L KL L DC SLFE+
Sbjct: 832 ELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRTPDISKCSALFKLQLNDCISLFEI-PG 890
Query: 658 IGDLSNLLLINLEGC 672
I + L I L+GC
Sbjct: 891 IHNHEYLSFIVLDGC 905
Score = 39.7 bits (91), Expect = 0.55
Identities = 48/201 (23%), Positives = 86/201 (41%), Gaps = 28/201 (13%)
Query: 523 ETVEGLILKLQSTSRVCFSTNAFKEMRKL--RLLQLECVDLTGDYRHLSHELKWVYWKGF 580
E ++ + +L+S + + A +E+ +L +L+ + L G LS ++ +Y +
Sbjct: 706 ERLDDALGELESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLYSEKS 765
Query: 581 TSTYIPDHFYLGNLV---VLELKYSSVKQ--VWKETKLLEKLKILDLSHSWYLENTPDFS 635
S + L L +L L Y ++ + ++ L L+ LDL + + DF+
Sbjct: 766 HSVSLLRPVSLSGLTYMRILSLGYCNLSDELIPEDIGSLSFLRDLDLRGNSFCNLPTDFA 825
Query: 636 KLPNLEKLILKDCSSL------------------FELHHT--IGDLSNLLLINLEGCTSL 675
LPNL +L+L DCS L L T I S L + L C SL
Sbjct: 826 TLPNLGELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRTPDISKCSALFKLQLNDCISL 885
Query: 676 NNLPGKTYQLKSVKTLIISGC 696
+PG + + + +++ GC
Sbjct: 886 FEIPG-IHNHEYLSFIVLDGC 905
>dbj|BAB11635.1| TMV resistance protein N [Arabidopsis thaliana]
Length = 1130
Score = 611 bits (1575), Expect = e-173
Identities = 350/806 (43%), Positives = 501/806 (61%), Gaps = 18/806 (2%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
G D R +F+SHLY +L GI+ F+DD +L++G + P LL AIE S+I + VL+ +YAS
Sbjct: 22 GADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPELLNAIETSKILIVVLTKDYAS 81
Query: 78 SRWCLDELVKILECRRTY-GQLVIPLFYHVDPSHVRNQKGPV---LLLGKQLQVEDTQRK 133
S WCLDELV I++ + +V P+F +VDPS +R Q+G K + +
Sbjct: 82 SAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQGSYAKSFSKHKNSHPLNKLKD 141
Query: 134 TRRGGHEVASFSGYNLS----PDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQ 189
R +VA+ SG+++ R+EAE I +I + + + L + + + + VGL S LQ
Sbjct: 142 WREALTKVANISGWDIKNRIYDSRNEAECIADITREILKRLPCQYLHVPSYAVGLRSRLQ 201
Query: 190 EVIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRG 249
+ L+ S V ++ I+GMGG+GKTT AK FN+ F +SF+EN RE + G
Sbjct: 202 HISSLLSIGSDGVRVIVIYGMGGIGKTTLAKVAFNEFSHLFEGSSFLENFREYSKKPE-G 260
Query: 250 HMHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREW 309
HLQ QLLSD+L+ + I G A +K+ KR L+++DDV QL + +R+
Sbjct: 261 RTHLQHQLLSDILRRND-IEFKGLDHA-VKERFRSKRVLLVVDDVDDVHQLNSAAIDRDC 318
Query: 310 FASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRR 369
F GS II+TTR++ LL L+A+ Y+ KE+D DESLELFSWHAF + P ++F + S
Sbjct: 319 FGHGSRIIITTRNMHLLKQLRAEGSYSPKELDGDESLELFSWHAFRTSEPPKEFLQHSEE 378
Query: 370 VVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKD 429
VV YC GLPLA+EVLG++L ER EWES L L+RIPND +Q KL+IS++ L E +KD
Sbjct: 379 VVTYCAGLPLAVEVLGAFLIERSIREWESTLKLLKRIPNDNIQAKLQISFNALTIE-QKD 437
Query: 430 IFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGR 489
+FLDI CFF+G D YV IL+GC LY DI ++LL+ER L+ + NN + MHDL+RDMGR
Sbjct: 438 VFLDIACFFIGVDSYYVACILDGCNLYPDIVLSLLMERCLITISGNN-IMMHDLLRDMGR 496
Query: 490 AIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMR 549
IVRE S K G+RSRLW H DV VL K +GT +EGL LK F AF +M+
Sbjct: 497 QIVREISPKKCGERSRLWSHNDVVGVLKKKSGTNAIEGLSLKADVMDFQYFEVEAFAKMQ 556
Query: 550 KLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWK 609
+LRLL+L VDL G Y H +L+W+ W GF+ P + L +L L+L+YS++K+ WK
Sbjct: 557 ELRLLELRYVDLNGSYEHFPKDLRWLCWHGFSLECFPINLSLESLAALDLQYSNLKRFWK 616
Query: 610 ---ETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDL-SNLL 665
+ +K LDLSHS YL TPDFS PN+EKLIL +C SL +H +IG L L+
Sbjct: 617 AQSPPQPANMVKYLDLSHSVYLRETPDFSYFPNVEKLILINCKSLVLVHKSIGILDKKLV 676
Query: 666 LINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEV 725
L+NL C L+ LP + Y+LKS+++L +S CSK+++L++ + ++ESLTTL+A TA++E+
Sbjct: 677 LLNLSSCIELDVLPEEIYKLKSLESLFLSNCSKLERLDDALGELESLTTLLADFTALREI 736
Query: 726 PYSILGLKSIGYISLCGYEGLSRDVFPSLLRSWISPTMNPLSHIPLFAAMSLPLVSMDIQ 785
P +I LK + +SL G +GL D +L S S +++ L + L + ++S+
Sbjct: 737 PSTINQLKKLKRLSLNGCKGLLSDDIDNLY-SEKSHSVSLLRPVSLSGLTYMRILSLGYC 795
Query: 786 NYSSGNSSSWLSSLSKLRSVWIQCHS 811
N S + SLS LR + ++ +S
Sbjct: 796 NLSDELIPEDIGSLSFLRDLDLRGNS 821
Score = 40.0 bits (92), Expect = 0.42
Identities = 29/75 (38%), Positives = 36/75 (47%), Gaps = 1/75 (1%)
Query: 598 ELKYSSVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHT 657
EL S ++ L L LD+ L+ TPD SK L KL L DC SLFE+
Sbjct: 837 ELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRTPDISKCSALFKLQLNDCISLFEI-PG 895
Query: 658 IGDLSNLLLINLEGC 672
I + L I L+GC
Sbjct: 896 IHNHEYLSFIVLDGC 910
Score = 39.7 bits (91), Expect = 0.55
Identities = 48/201 (23%), Positives = 86/201 (41%), Gaps = 28/201 (13%)
Query: 523 ETVEGLILKLQSTSRVCFSTNAFKEMRKL--RLLQLECVDLTGDYRHLSHELKWVYWKGF 580
E ++ + +L+S + + A +E+ +L +L+ + L G LS ++ +Y +
Sbjct: 711 ERLDDALGELESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLYSEKS 770
Query: 581 TSTYIPDHFYLGNLV---VLELKYSSVKQ--VWKETKLLEKLKILDLSHSWYLENTPDFS 635
S + L L +L L Y ++ + ++ L L+ LDL + + DF+
Sbjct: 771 HSVSLLRPVSLSGLTYMRILSLGYCNLSDELIPEDIGSLSFLRDLDLRGNSFCNLPTDFA 830
Query: 636 KLPNLEKLILKDCSSL------------------FELHHT--IGDLSNLLLINLEGCTSL 675
LPNL +L+L DCS L L T I S L + L C SL
Sbjct: 831 TLPNLGELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRTPDISKCSALFKLQLNDCISL 890
Query: 676 NNLPGKTYQLKSVKTLIISGC 696
+PG + + + +++ GC
Sbjct: 891 FEIPG-IHNHEYLSFIVLDGC 910
>emb|CAC95124.1| TIR/NBS/LRR protein [Populus deltoides]
Length = 1147
Score = 546 bits (1406), Expect = e-153
Identities = 359/947 (37%), Positives = 524/947 (54%), Gaps = 73/947 (7%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR +F HLY AL AGI+ F DD++L +G E+ LRAI+ S+IS+AV S YAS
Sbjct: 47 GEDTRKTFTDHLYTALVQAGIHTFRDDDELPRGEEISDHFLRAIQESKISIAVFSKGYAS 106
Query: 78 SRWCLDELVKILECR-RTYGQLVIPLFYHVDPSHVRNQKGP---VLLLGKQLQVEDTQRK 133
SRWCL+ELV+IL+C+ R GQ+V+P+FY +DPS VR Q G + ++ E ++
Sbjct: 107 SRWCLNELVEILKCKKRKTGQIVLPIFYDIDPSDVRKQNGSFAEAFVKHEERFEEKLVKE 166
Query: 134 TRRGGHEVASFSGYNLSP--DRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEV 191
R+ E + SG+NL+ + EA+ I EI+K V L+ + + + + VG++ + +
Sbjct: 167 WRKALEEAGNLSGWNLNDMANGHEAKFIKEIIKVVLNKLEPKYLYVPEHLVGMDQLARNI 226
Query: 192 IKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHM 251
+ + + V +VGI GM G+GKTT A+A+FNQ+ F + F+ +I E + + G +
Sbjct: 227 FDFLSAATDDVRIVGIHGMPGIGKTTIAQAVFNQLCYGFEGSCFLSSINERSKQVN-GLV 285
Query: 252 HLQEQLLSDVLKTK-EKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWF 310
LQ+QL D+LK RG +IK+ L KR LV+ DDV EQL AL G+R WF
Sbjct: 286 PLQKQLHHDILKQDVANFDCADRGKVLIKERLRRKRVLVVADDVAHLEQLNALMGDRSWF 345
Query: 311 ASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRV 370
GS +I+TTRD LL +AD +Y ++E+ DESL+LFS HAF ++ P +D+ ELS++
Sbjct: 346 GPGSRVIITTRDSNLLR--EADQIYQIEELKPDESLQLFSRHAFKDSKPAQDYIELSKKA 403
Query: 371 VAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDI 430
V YCGGLPLALEV+G+ L + S + L RIPN +Q KL ISY L EL++
Sbjct: 404 VGYCGGLPLALEVIGALLYRKNRGRCVSEIDNLSRIPNQDIQGKLLISYHALDGELQR-A 462
Query: 431 FLDICCFFVGKDRAYVTDILNG-CGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGR 489
FLDI CFF+G +R YVT +L C ++ + L ERSL+QV + MHDL+RDMGR
Sbjct: 463 FLDIACFFIGIEREYVTKVLGARCRPNPEVVLETLSERSLIQV-FGETVSMHDLLRDMGR 521
Query: 490 AIVRESSTKDLGKRSRLWFHEDVHDVL--TKNTGTETVEGLILKLQSTSRVCFSTNAFKE 547
+V ++S K GKR+R+W ED +VL K GT+ V+GL L ++++ S +F E
Sbjct: 522 EVVCKASPKQPGKRTRIWNQEDAWNVLEQQKVRGTDVVKGLALDVRASEAKSLSAGSFAE 581
Query: 548 MRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQV 607
M+ L LLQ+ V LTG + S EL W+ W Y+P F L NL VL+++YS++K++
Sbjct: 582 MKCLNLLQINGVHLTGSLKLFSKELMWICWHECPLKYLPFDFTLDNLAVLDMQYSNLKEL 641
Query: 608 WKETK---LLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNL 664
WK K +L+ K L Y+ LEKL LK CSSL E+H +IG+L++L
Sbjct: 642 WKGKKVRNMLQSPKFLQYVIYIYI-----------LEKLNLKGCSSLVEVHQSIGNLTSL 690
Query: 665 LLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKE 724
+NLEGC L NLP +KS++TL ISGCS+++KL E + MESL L+A ++
Sbjct: 691 DFLNLEGCWRLKNLPESIGNVKSLETLNISGCSQLEKLPESMGDMESLIELLADGIENEQ 750
Query: 725 VPYSILGLKSIGYISLCGYEG-------LSRDV------FPSLLRSWISPTMNPLSHIPL 771
SI LK + +SL GY +S V P+ WIS L H L
Sbjct: 751 FLSSIGQLKHVRRLSLRGYSSTPPSSSLISAGVLNLKRWLPTSFIQWISVKRLELPHGGL 810
Query: 772 ---------FAAMSLPLVSMDIQNYSSGNSSSWLSSLSKLRSVWIQ-CHSEIQLAQ--ES 819
F+ +S L +D+ + S + LSKL+ + ++ C + + S
Sbjct: 811 SDRAAKCVDFSGLS-ALEVLDLIGNKFSSLPSGIGFLSKLKFLSVKACKYLVSIPDLPSS 869
Query: 820 RRILDDQYDVN--------------SMDLEASSSSSYASQISDFSLKSLLIRLGSCYTVI 865
LD Y + ++L S S I S + + +
Sbjct: 870 LDCLDASYCKSLERVRIPIEPKKELDINLYKSHSLEEIQGIEGLSNNIWSLEVDTSRHSP 929
Query: 866 DTLASSISQGSTTNASSNFF---LPGGDDPYWLAYKGEGPSVHFQVP 909
+ L S+ + + N + +PGG+ P W++Y GEG S+ F +P
Sbjct: 930 NKLQKSVVE-AICNGRHRYCIHGIPGGNMPNWMSYSGEGCSLSFHIP 975
>gb|AAP44393.1| nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 519 bits (1336), Expect = e-145
Identities = 302/731 (41%), Positives = 442/731 (60%), Gaps = 14/731 (1%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GED R +FV HLY AL IN F DDEKL+KG + P L+ +IE SRI+L + S NYA+
Sbjct: 26 GEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELVSSIEESRIALIIFSKNYAN 85
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQK---GPVLLLGKQLQVEDTQRKT 134
S WCLDEL KI+EC+ GQ+V+P+FY VDPS VR QK G + ED +K
Sbjct: 86 STWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEARFQEDKVQKW 145
Query: 135 RRGGHEVASFSGYNLSPDRS---EAEAINEIVKNVFRNLDI-RLMSITDFPVGLESHLQE 190
R E A+ SG++L P+ S EA + +I +++ L R S VG+ESH+ +
Sbjct: 146 RAALEEAANISGWDL-PNTSNGHEARVMEKIAEDIMARLGSQRHASNARNLVGMESHMHQ 204
Query: 191 VIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGH 250
V K++ S V +GI GM G+GKTT A+ I++ I +F F+ +R+ + +G
Sbjct: 205 VYKMLGIGSGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACFLHEVRD--RSAKQGL 262
Query: 251 MHLQEQLLSDVLKTKE-KIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREW 309
LQE LLS++L K+ +I+ G M K+ L K+ L++LDDV +QL AL G REW
Sbjct: 263 ERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREW 322
Query: 310 FASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRR 369
F GS II+TT+D LL + + +Y MK ++ ESL+LF HAF + P ++F +LS +
Sbjct: 323 FGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKKNRPTKEFEDLSAQ 382
Query: 370 VVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKD 429
V+ + GLPLAL+VLGS+L R EW S + +L++IP +++ +KL S+ GL + E+
Sbjct: 383 VIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFTGLHNT-EQK 441
Query: 430 IFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGR 489
IFLDI CFF GK + VT IL IGI +L+E+ L+ + ++ +H L++DMG
Sbjct: 442 IFLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLITTLQG-RITIHQLIQDMGW 500
Query: 490 AIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMR 549
IVR +T D SRLW ED+ VL +N GT+ +EG+ L L + V F AF +M
Sbjct: 501 HIVRREATDDPRMCSRLWKREDICPVLERNLGTDKIEGMSLHLTNEEEVNFGGKAFMQMT 560
Query: 550 KLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWK 609
+LR L+ + + L EL+W+ W G+ S +P+ F LV L+LK S + Q+WK
Sbjct: 561 RLRFLKFQNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVSLKLKKSRIIQLWK 620
Query: 610 ETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINL 669
+K L KLK ++LSHS L PDFS PNLE+L+L++C+SL E++ +I +L L+L+NL
Sbjct: 621 TSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLERLVLEECTSLVEINFSIENLGKLVLLNL 680
Query: 670 EGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSI 729
+ C +L LP K +L+ ++ L+++GCSK+ E +M L L T++ E+P S+
Sbjct: 681 KNCRNLKTLP-KRIRLEKLEILVLTGCSKLRTFPEIEEKMNCLAELYLDATSLSELPASV 739
Query: 730 LGLKSIGYISL 740
L +G I+L
Sbjct: 740 ENLSGVGVINL 750
>gb|AAP44390.1| nematode resistance protein [Solanum tuberosum]
Length = 1136
Score = 517 bits (1332), Expect = e-145
Identities = 300/730 (41%), Positives = 441/730 (60%), Gaps = 12/730 (1%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GED R +FV HLY AL IN F DDEKL+KG + P L+ +IE SRI+L + S NYA+
Sbjct: 26 GEDVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELVSSIEESRIALIIFSKNYAN 85
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQK---GPVLLLGKQLQVEDTQRKT 134
S WCLDEL KI+EC+ GQ+V+P+FY VDPS VR QK G + ED +K
Sbjct: 86 STWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEARFQEDKVQKW 145
Query: 135 RRGGHEVASFSGYNL--SPDRSEAEAINEIVKNVFRNLDI-RLMSITDFPVGLESHLQEV 191
R E A+ SG++L + + EA + +I +++ L R S VG+ESH+ +V
Sbjct: 146 RAALEEAANISGWDLPNTANGHEARVMEKIAEDIMARLGSQRHASNARNLVGMESHMHKV 205
Query: 192 IKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHM 251
K++ S V +GI GM G+GKTT A+ I++ I +F F+ +R+ + +G
Sbjct: 206 YKMLGIGSGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACFLHEVRD--RSAKQGLE 263
Query: 252 HLQEQLLSDVLKTKE-KIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWF 310
LQE LLS++L K+ +I+ G M K+ L K+ L++LDDV +QL AL G REWF
Sbjct: 264 RLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREWF 323
Query: 311 ASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRV 370
GS II+TT+D LL + + +Y MK ++ ESL+LF HAF + P ++F +LS +V
Sbjct: 324 GDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKKNRPTKEFEDLSAQV 383
Query: 371 VAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDI 430
+ + GLPLAL+VLGS+L R EW S + +L++IP +++ +KL S+ GL + E+ I
Sbjct: 384 IKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFTGLHNT-EQKI 442
Query: 431 FLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGRA 490
FLDI CFF GK + VT IL IGI +L+E+ L+ + + ++ +H L++DMG
Sbjct: 443 FLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLITILQG-RITIHQLIQDMGWH 501
Query: 491 IVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMRK 550
IVR +T D SR+W ED+ VL +N GT+ EG+ L L + V F AF +M +
Sbjct: 502 IVRREATDDPRMCSRMWKREDICPVLERNLGTDKNEGMSLHLTNEEEVNFGGKAFMQMTR 561
Query: 551 LRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWKE 610
LR L+ + L EL+W+ W G+ S +P+ F LV L+LK S + Q+WK
Sbjct: 562 LRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVGLKLKKSRIIQLWKT 621
Query: 611 TKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINLE 670
+K L KLK ++LSHS L TPDFS PNLE+L+L++C+SL E++ +I +L L+L+NL+
Sbjct: 622 SKDLGKLKYMNLSHSQKLIRTPDFSVTPNLERLVLEECTSLVEINFSIENLGKLVLLNLK 681
Query: 671 GCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSIL 730
C +L LP K +L+ ++ L+++GCSK+ E +M L L T++ E+P S+
Sbjct: 682 NCRNLKTLP-KRIRLEKLEILVLTGCSKLRTFPEIEEKMNCLAELYLGATSLSELPASVE 740
Query: 731 GLKSIGYISL 740
L +G I+L
Sbjct: 741 NLSGVGVINL 750
>gb|AAP44392.1| nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 515 bits (1327), Expect = e-144
Identities = 302/731 (41%), Positives = 442/731 (60%), Gaps = 14/731 (1%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GED R +FV HLY AL IN F DDEKL+KG + P L+ +IE SRI+L + S NYA+
Sbjct: 26 GEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELMSSIEESRIALIIFSKNYAN 85
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQK---GPVLLLGKQLQVEDTQRKT 134
S WCLDEL KI+EC+ GQ+V+P+FY VDPS VR QK G + ED +K
Sbjct: 86 STWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEARFQEDKVQKW 145
Query: 135 RRGGHEVASFSGYNLSPDRS---EAEAINEIVKNVFRNLDI-RLMSITDFPVGLESHLQE 190
R E A+ SG++L P+ S EA + +I +++ L R S VG+ESH+ +
Sbjct: 146 RAALEEAANISGWDL-PNTSNGHEARVMEKIAEDIMARLGSQRHASNARNLVGMESHMLK 204
Query: 191 VIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGH 250
V K++ S V +GI GM G+GKTT A+ I++ I +F F+ +R+ + +G
Sbjct: 205 VYKMLGIGSGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACFLHEVRD--RSAKQGL 262
Query: 251 MHLQEQLLSDVLKTKE-KIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREW 309
LQE LLS++L K+ +I++ G M K+ L K+ L++LDDV +QL AL G REW
Sbjct: 263 ERLQEILLSEILVVKKLRINNSFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREW 322
Query: 310 FASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRR 369
F GS II+TT+D LL + + +Y MK ++ ESL+LF HAF + P ++F +LS +
Sbjct: 323 FGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKKNRPTKEFEDLSAQ 382
Query: 370 VVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKD 429
V+ + GLPLAL+VLGS+L R EW S + +L++IP +++ +KL S+ GL + E+
Sbjct: 383 VIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFTGLHNT-EQK 441
Query: 430 IFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGR 489
IFLDI CFF GK + VT IL IGI +L+E+ L+ + + ++ +H L++DMG
Sbjct: 442 IFLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLITILQG-RITIHQLIQDMGW 500
Query: 490 AIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMR 549
IVR +T D SRLW ED+ VL +N GT+ EG+ L L + V F AF +M
Sbjct: 501 HIVRREATDDPRMCSRLWKREDICPVLERNLGTDKNEGMSLHLTNEEEVNFGGKAFMQMT 560
Query: 550 KLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWK 609
+LR L+ + L EL+W+ W G+ S +P+ F LV L+LK S + Q+WK
Sbjct: 561 RLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVGLKLKKSRIIQLWK 620
Query: 610 ETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINL 669
+K L KLK ++LSHS L TPDFS PNLE+L+L++C+SL E++ +I +L L+L+NL
Sbjct: 621 TSKDLGKLKYMNLSHSQKLIRTPDFSVTPNLERLVLEECTSLVEINFSIENLGKLVLLNL 680
Query: 670 EGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSI 729
+ C +L LP K +L+ ++ L+++GCSK+ E +M L L T++ +P S+
Sbjct: 681 KNCRNLKTLP-KRIRLEKLEILVLTGCSKLRTFPEIEEKMNCLAELYLGATSLSGLPASV 739
Query: 730 LGLKSIGYISL 740
L +G I+L
Sbjct: 740 ENLSGVGVINL 750
>gb|AAP44391.1| nematode resistance-like protein [Solanum tuberosum]
Length = 1121
Score = 514 bits (1323), Expect = e-144
Identities = 300/731 (41%), Positives = 441/731 (60%), Gaps = 14/731 (1%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GE+ R +FV HLY AL IN F DDEKL+KG + P L+ +IE SRI+L + S NYA+
Sbjct: 26 GENVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELMSSIEESRIALIIFSKNYAN 85
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQK---GPVLLLGKQLQVEDTQRKT 134
S WCLDEL KI+EC+ GQ+V+P+FY VDPS VR QK G + ED +K
Sbjct: 86 STWCLDELTKIIECKNVKGQIVVPVFYDVDPSTVRRQKNIFGEAFSKHEARFEEDKVKKW 145
Query: 135 RRGGHEVASFSGYNLSPDRS---EAEAINEIVKNVFRNLDI-RLMSITDFPVGLESHLQE 190
R E A+ SG++L P+ S EA I +I +++ L R S VG+ESH+ +
Sbjct: 146 RAALEEAANISGWDL-PNTSNGHEARVIEKITEDIMVRLGSQRHASNARNVVGMESHMHQ 204
Query: 191 VIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGH 250
V K++ S V +GI GM G+GKTT A+ I++ I +F F+ +R+ + +G
Sbjct: 205 VYKMLGIGSGGVRFLGILGMSGVGKTTLARVIYDNIQSQFEGACFLHEVRD--RSAKQGL 262
Query: 251 MHLQEQLLSDVLKTKE-KIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREW 309
HLQE LLS++L K+ +I+ G M K+ L K+ L++LDDV +QL AL G REW
Sbjct: 263 EHLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREW 322
Query: 310 FASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRR 369
F GS II+TT+D LL + + +Y M +D+ ESL+LF HAF + ++F +LS +
Sbjct: 323 FGDGSRIIITTKDKHLLVKYETEKIYRMGTLDKYESLQLFKQHAFKKNHSTKEFEDLSAQ 382
Query: 370 VVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKD 429
V+ + GGLPLAL+VLGS+L R EW S + +L++IP +++ +KL S+ GL + +E+
Sbjct: 383 VIEHTGGLPLALKVLGSFLYGRGLDEWISEVERLKQIPQNEILKKLEPSFTGLNN-IEQK 441
Query: 430 IFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGR 489
IFLDI CFF GK + VT IL IGI +L+E+ L+ + K ++ +H L+++MG
Sbjct: 442 IFLDIACFFSGKKKDSVTRILESFHFSPVIGIKVLMEKCLITILKG-RITIHQLIQEMGW 500
Query: 490 AIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMR 549
IVR ++ + SRLW ED+ VL +N T+ +EG+ L L + V F A +M
Sbjct: 501 HIVRREASYNPRICSRLWKREDICPVLEQNLCTDKIEGMSLHLTNEEEVNFGGKALMQMT 560
Query: 550 KLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWK 609
LR L+ + L EL+W+ W G+ S +P+ F LV L+LK S + Q+WK
Sbjct: 561 SLRFLKFRNAYVYQGPEFLPDELRWLDWHGYPSKNLPNSFKGDQLVSLKLKKSRIIQLWK 620
Query: 610 ETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINL 669
+K L KLK ++LSHS L PDFS PNLE+L+L++C+SL E++ +IGDL L+L+NL
Sbjct: 621 TSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLERLVLEECTSLVEINFSIGDLGKLVLLNL 680
Query: 670 EGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSI 729
+ C +L +P K +L+ ++ L++SGCSK+ E +M L L T++ E+P S+
Sbjct: 681 KNCRNLKTIP-KRIRLEKLEVLVLSGCSKLRTFPEIEEKMNRLAELYLGATSLSELPASV 739
Query: 730 LGLKSIGYISL 740
+G I+L
Sbjct: 740 ENFSGVGVINL 750
>gb|AAQ93074.1| putative TIR-NBS type R protein 4 [Malus baccata]
Length = 726
Score = 505 bits (1301), Expect = e-141
Identities = 269/587 (45%), Positives = 375/587 (63%), Gaps = 9/587 (1%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR +F HL+ AL+ AGIN F+DDE+L++G ++ L++AI+GSRIS+ V S YA
Sbjct: 133 GEDTRKNFTGHLHEALTKAGINAFIDDEELRRGEDITTELVQAIQGSRISIIVFSRRYAD 192
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLLGKQLQVEDTQRKTRRG 137
S WCL+ELVKI+ECRRT GQLV+P+FY VDPS+VR G + E + R
Sbjct: 193 SSWCLEELVKIMECRRTLGQLVLPIFYDVDPSNVRKLTGSFAQSFLKHTDEKKVERWRAA 252
Query: 138 GHEVASFSGYNLSP--DRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEVIKLI 195
E ++ SG++L DR EA+ I I V L+ R ++ + VG+++ + + +
Sbjct: 253 LTEASNLSGWDLKNTLDRHEAKFIRMITNQVTVKLNNRYFNVAPYQVGIDTRVLNISNYL 312
Query: 196 E-SQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHMHLQ 254
S V ++GI GMGG+GKTT KAI+N+ + +F SF+E +RE + + LQ
Sbjct: 313 GIGDSDDVRVIGISGMGGIGKTTIVKAIYNEFYERFEGKSFLEKVRE------KKLVKLQ 366
Query: 255 EQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFASGS 314
+QLL D+L+TK K+ S+ GTA++ + R LVI+DDV +QLR L GN F GS
Sbjct: 367 KQLLFDILQTKTKVSSVAVGTALVGERFRRLRVLVIVDDVDDVKQLRELVGNCHSFGPGS 426
Query: 315 VIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVVAYC 374
II+TTR+ R+L D +Y MD++E+LEL SWHAF + + L+R VV YC
Sbjct: 427 RIIITTRNERVLKEFAVDEIYRENGMDQEEALELLSWHAFKSSWCPSQYLVLTREVVNYC 486
Query: 375 GGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIFLDI 434
GGLPLALEVLGS + +R EW S+L +L+ IP ++Q +L+ISYDGL D ++ IFLDI
Sbjct: 487 GGLPLALEVLGSTIFKRSVNEWRSILDELKMIPRGEIQAQLKISYDGLNDHYKRQIFLDI 546
Query: 435 CCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGRAIVRE 494
FF+G D+ V IL+GCG YA GI +L++R L+ + + NK+ MHDL+RDMGR IV
Sbjct: 547 AFFFIGMDKNDVMQILDGCGFYATTGIEVLLDRCLVTIGRKNKIMMHDLLRDMGRDIVHA 606
Query: 495 SSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMRKLRLL 554
+ +RSRLW +DVHDVL +GTE +EGL L L S FST+AF+ M++LRLL
Sbjct: 607 ENPGFPRERSRLWHPKDVHDVLIDKSGTEKIEGLALNLPSLEETSFSTDAFRNMKRLRLL 666
Query: 555 QLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKY 601
QL V LTG YR LS +L+W+ W GF +IP N+V ++++Y
Sbjct: 667 QLNYVRLTGGYRCLSKKLRWLCWHGFPLEFIPIELCQPNIVAIDMQY 713
>gb|AAQ93076.1| putative TIR-NBS type R protein 4 [Malus baccata]
Length = 726
Score = 503 bits (1295), Expect = e-140
Identities = 268/587 (45%), Positives = 374/587 (63%), Gaps = 9/587 (1%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR +F HL+ AL+ AGIN F+DDE+L++G ++ L++AI+GSRIS+ V S YA
Sbjct: 133 GEDTRKNFTGHLHEALTKAGINAFIDDEELRRGEDITTELVQAIQGSRISIIVFSRRYAD 192
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLLGKQLQVEDTQRKTRRG 137
S WCL+ELVKI+ECRRT GQLV+P+FY VDPS+VR G + E + R
Sbjct: 193 SSWCLEELVKIMECRRTLGQLVLPIFYDVDPSNVRKLTGSFAQSFLKHTDEKKVERWRAA 252
Query: 138 GHEVASFSGYNLSP--DRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEVIKLI 195
E ++ SG++L DR EA+ I I V L+ R ++ + VG+++ + + +
Sbjct: 253 LTEASNLSGWDLKNTLDRHEAKFIRMITNQVTVKLNNRYFNVAPYQVGIDTRVLNISNYL 312
Query: 196 E-SQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHMHLQ 254
S V ++GI G GG+GKTT KAI+N+ + +F SF+E +RE + + LQ
Sbjct: 313 GIGDSDDVRVIGISGSGGIGKTTIVKAIYNEFYERFEGKSFLEKVRE------KKLVKLQ 366
Query: 255 EQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFASGS 314
+QLL D+L+TK K+ S+ GTA++ + R LVI+DDV +QLR L GN F GS
Sbjct: 367 KQLLFDILQTKTKVSSVAVGTALVGERFRRLRVLVIVDDVDDVKQLRELVGNCHSFGPGS 426
Query: 315 VIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVVAYC 374
II+TTR+ R+L D +Y MD++E+LEL SWHAF + + L+R VV YC
Sbjct: 427 RIIITTRNERVLKEFAVDEIYRENGMDQEEALELLSWHAFKSSWCPSQYLVLTREVVNYC 486
Query: 375 GGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIFLDI 434
GGLPLALEVLGS + +R EW S+L +L+ IP ++Q +L+ISYDGL D ++ IFLDI
Sbjct: 487 GGLPLALEVLGSTIFKRSVNEWRSILDELKMIPRGEIQAQLKISYDGLNDHYKRQIFLDI 546
Query: 435 CCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGRAIVRE 494
FF+G D+ V IL+GCG YA GI +L++R L+ + + NK+ MHDL+RDMGR IV
Sbjct: 547 AFFFIGMDKNDVMQILDGCGFYATTGIEVLLDRCLVTIGRKNKIMMHDLLRDMGRDIVHA 606
Query: 495 SSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMRKLRLL 554
+ +RSRLW +DVHDVL +GTE +EGL L L S FST+AF+ M++LRLL
Sbjct: 607 ENPGFPRERSRLWHPKDVHDVLIDKSGTEKIEGLALNLPSLEETSFSTDAFRNMKRLRLL 666
Query: 555 QLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKY 601
QL V LTG YR LS +L+W+ W GF +IP N+V ++++Y
Sbjct: 667 QLNYVRLTGGYRCLSKKLRWLCWHGFPLEFIPIELCQPNIVAIDMQY 713
>gb|AAO23077.1| R 8 protein [Glycine max]
Length = 892
Score = 475 bits (1222), Expect = e-132
Identities = 293/765 (38%), Positives = 445/765 (57%), Gaps = 24/765 (3%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
G+DTR F +LY AL + GI F+DD++L++G E+KPAL AI+ SRI++ VLS NYAS
Sbjct: 20 GQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAITVLSQNYAS 79
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGP----VLLLGKQLQV-EDTQR 132
S +CLDELV IL C+ + G LVIP+FY VDPSHVR+QKG + K+ + ++ +
Sbjct: 80 SSFCLDELVTILHCK-SQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFKANKEKLQ 138
Query: 133 KTRRGGHEVASFSGYNLSP-DRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEV 191
K R H+VA SGY+ D E E I IV+ + R + + D+PVGLES + EV
Sbjct: 139 KWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRKFSRASLHVADYPVGLESEVTEV 198
Query: 192 IKLIESQSSKVC-MVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGH 250
+KL++ S V ++GI GMGGLGKTT A A+ N I F + F++N+RE E++ G
Sbjct: 199 MKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALHFDESCFLQNVRE--ESNKHGL 256
Query: 251 MHLQEQLLSDVLKTKE-KIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREW 309
HLQ LLS +L K+ + S G +MI+ L K+ L+ILDDV +QL+A+ G +W
Sbjct: 257 KHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAIVGRPDW 316
Query: 310 FASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRR 369
F GS +I+TTRD LL + + Y +K +++ +L+L +W+AF + ++ R
Sbjct: 317 FGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLTWNAFKREKIDPSYEDVLNR 376
Query: 370 VVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKD 429
VV Y GLPLALEV+GS L E+ EWES + +RIP+D++Q+ L++S+D L +E +K+
Sbjct: 377 VVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDEIQEILKVSFDALGEE-QKN 435
Query: 430 IFLDICCFFVGKDRAYVTDILNGC-GLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMG 488
+FLDI C F G + V +IL G I +L+E+SL++V + + MHD+++DMG
Sbjct: 436 VFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLVKVSCCDTVEMHDMIQDMG 495
Query: 489 RAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSR---VCFSTNAF 545
R I R+ S ++ GK RL +D+ VL NTGT +E + L + + V ++ NAF
Sbjct: 496 REIERQRSPEEPGKCKRLLLPKDIIQVLKDNTGTSKIEIICLDFSISDKEETVEWNENAF 555
Query: 546 KEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVK 605
+M+ L++L + + + L+ + W + S +P +F NLV+ +L SS+
Sbjct: 556 MKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSNCLPSNFDPINLVICKLPDSSIT 615
Query: 606 --QVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSN 663
+ +K L L +L+ +L PD S LPNL++L C SL + +IG L+
Sbjct: 616 SFEFHGSSKKLGHLTVLNFDRCEFLTKIPDVSDLPNLKELSFNWCESLVAVDDSIGFLNK 675
Query: 664 LLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIK 723
L ++ GC L + P L S++TL + GCS ++ E + +M+++T L D IK
Sbjct: 676 LKTLSAYGCRKLTSFP--PLNLTSLETLNLGGCSSLEYFPEILGEMKNITVLALHDLPIK 733
Query: 724 EVPYSILGLKSIGYISL--CGYEGL--SRDVFPSLLRSWISPTMN 764
E+P+S L + ++ L CG L S P L I+ + N
Sbjct: 734 ELPFSFQNLIGLLFLWLDSCGIVQLRCSLATMPKLCEFCITDSCN 778
>gb|AAT37497.1| N-like protein [Nicotiana tabacum]
Length = 941
Score = 475 bits (1222), Expect = e-132
Identities = 296/804 (36%), Positives = 459/804 (56%), Gaps = 46/804 (5%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR +F SHLY L + GI F D+++L+ G+ + L +AIE S+ ++ V S NYA+
Sbjct: 20 GEDTRKTFTSHLYEVLKDRGIKTFQDEKRLEYGATIPEELCKAIEESQFAIVVFSENYAT 79
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLLGKQLQVEDTQRKT--- 134
SRWCL+ELVKI+EC+ + Q +IP+FY VDPSHVRNQK K + +T+ K
Sbjct: 80 SRWCLNELVKIMECKTQFRQTIIPIFYDVDPSHVRNQKES---FAKAFEEHETKYKDDVE 136
Query: 135 -----RRGGHEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQ 189
R + A+ G + D+++A+ I +IV + L +S VG+++HL+
Sbjct: 137 GIQRWRTALNAAANLKGSCDNRDKTDADCIRQIVDQISSKLSKISLSYLQNIVGIDTHLE 196
Query: 190 EVIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQI------HRKFMCTSFIENIREVC 243
E+ L+ + V +VGIWGMGG+GKTT A+A+F+ + +F F+++I+E
Sbjct: 197 EIESLLGIGINDVRIVGIWGMGGVGKTTIARAMFDTLLGRRDSSYQFDGACFLKDIKE-- 254
Query: 244 ENDSRGHMHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQ-LRA 302
+ RG LQ LL ++L+ ++ G + L K+ L++LDD+ + L
Sbjct: 255 --NKRGMHSLQNTLLFELLRENANYNNEDDGKHQMASRLRSKKVLIVLDDIDDKDHYLEY 312
Query: 303 LCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPRED 362
L G+ +WF +GS IIVTTRD L+ K D +Y + + + E+++LF HAF + P E
Sbjct: 313 LAGDLDWFGNGSRIIVTTRDKHLIG--KNDIIYEVTALPDHEAIQLFYQHAFKKEVPDEC 370
Query: 363 FNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGL 422
F ELS VV + GLPLAL+V GS L++R+ W+S + +++ PN ++ +KL+ISYDGL
Sbjct: 371 FKELSLEVVNHAKGLPLALKVWGSSLHKRDITVWKSAIEQMKINPNSKIVEKLKISYDGL 430
Query: 423 KDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHD 482
+ +++++FLDI CFF G+ + Y+ +L C A+ G+ +LIE+SL+ + + N++ MHD
Sbjct: 431 -ESMQQEMFLDIACFFRGRQKDYIMQVLKSCHFGAEYGLDVLIEKSLVFISEYNQVEMHD 489
Query: 483 LVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFST 542
L++DMG+ IV + KD G+RSRLW EDV +V+ N GT +VE ++ + + FS
Sbjct: 490 LIQDMGKYIV--NFKKDPGERSRLWLAEDVEEVMNNNAGTMSVE--VIWVHYDFGLYFSN 545
Query: 543 NAFKEMRKLRLLQ----LECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLE 598
+A K M++LR+L L G +L L+W + +P F L LV LE
Sbjct: 546 DAMKNMKRLRILHIKGYLSSTSHDGSIEYLPSNLRWFVLDDYPWESLPSTFDLKMLVHLE 605
Query: 599 LKYSSVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTI 658
L SS+ +W ETK L L+ +DLS S L TPDF+ +PNLE L + C +L E+HH++
Sbjct: 606 LSRSSLHYLWTETKHLPSLRRIDLSSSRRLRRTPDFTGMPNLEYLNMLYCRNLEEVHHSL 665
Query: 659 GDLSNLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAK 718
S L+ +NL C SL P ++S++ L + CS ++K E +M+ + +
Sbjct: 666 RCCSKLIRLNLNNCKSLKRFP--CVNVESLEYLSLEYCSSLEKFPEIHGRMKPEIQIHMQ 723
Query: 719 DTAIKEVPYSILGLKS-IGYISLCGYEGLSRDVFPS---LLRSWISPTMNPLSHIPLFAA 774
+ I+E+P SI ++ I + L G E L PS L+S +S LS F
Sbjct: 724 GSGIRELPSSITQYQTHITKLDLRGMEKLV--ALPSSICRLKSLVS-----LSVSGCFKL 776
Query: 775 MSLPLVSMDIQNYSSGNSSSWLSS 798
SLP D++N ++S L S
Sbjct: 777 ESLPEEVGDLENLEELDASCTLIS 800
Score = 39.7 bits (91), Expect = 0.55
Identities = 26/80 (32%), Positives = 41/80 (50%)
Query: 591 LGNLVVLELKYSSVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSS 650
L +L L L ++ + + + L L+IL+L + L P+F+ + NLE L L+ CS
Sbjct: 860 LSSLKKLYLSGNNFEHLPRSIAQLGALRILELRNCKRLTQLPEFTGMLNLEYLDLEGCSY 919
Query: 651 LFELHHTIGDLSNLLLINLE 670
L E+HH G L + E
Sbjct: 920 LEEVHHFPGVLQKTHSVKFE 939
Score = 37.4 bits (85), Expect = 2.7
Identities = 33/118 (27%), Positives = 55/118 (45%), Gaps = 5/118 (4%)
Query: 591 LGNLVVLELKYSSVKQVWKETKLLEKLKILDLSHSW---YLENTPDFSKLPNLEKLILKD 647
L NL L+ + + + L KLKI D S + E P +LE L L++
Sbjct: 786 LENLEELDASCTLISRPPSSIIRLSKLKIFDFGSSKDRVHFELPPVVEGFRSLETLSLRN 845
Query: 648 CSSLFE-LHHTIGDLSNLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEE 704
C+ + L +G LS+L + L G + +LP QL +++ L + C ++ +L E
Sbjct: 846 CNLIDGGLPEDMGSLSSLKKLYLSG-NNFEHLPRSIAQLGALRILELRNCKRLTQLPE 902
>gb|AAU04762.1| MRGH21 [Cucumis melo]
Length = 1020
Score = 473 bits (1216), Expect = e-131
Identities = 269/729 (36%), Positives = 429/729 (57%), Gaps = 16/729 (2%)
Query: 20 DTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYASSR 79
DT SF+S L+ AL++ GI VF+D E + G + ++A++ SR S+ V S NY S
Sbjct: 50 DTGRSFISDLHEALTSQGIVVFIDKEDEEDGGKPLTEKMKAVDESRSSIVVFSENYGS-- 107
Query: 80 W-CLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLL-------LGKQLQVEDTQ 131
W C+ E+ KI C+++ QLV+P+FY VDP VR Q+G L+ + +E+
Sbjct: 108 WVCMKEIRKIRMCQKSRDQLVLPIFYKVDPGDVRKQEGESLVKFFNEHEANPNISIEEV- 166
Query: 132 RKTRRGGHEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEV 191
+K R+ ++V + SG++L + E I E+V ++F L L D VG+ L E+
Sbjct: 167 KKWRKSMNKVGNLSGWHLQDSQFEEGIIKEVVDHIFNKLRPDLFRYDDKLVGISRRLHEI 226
Query: 192 IKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGHM 251
KL+ V +GIWGM G+GKTT A+ I+ + F F++N++E + + G
Sbjct: 227 NKLMGIGLDDVRFIGIWGMSGIGKTTIARIIYKSVSHLFDGCYFLDNVKEALKKE--GIA 284
Query: 252 HLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFA 311
LQ++LL+ L + G +IK+ +S +AL+ILDDV QLR L G+ +WF
Sbjct: 285 SLQQKLLTGALMKRNIDIPNADGATLIKRRISNIKALIILDDVDNVSQLRQLAGSLDWFG 344
Query: 312 SGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVV 371
SGS +IVTT+ +L S + Y ++ + DE ++LFS AFGE P+E + +L +VV
Sbjct: 345 SGSRVIVTTKHEDILVSHGIERRYNVEVLKIDEGIQLFSQKAFGEDYPKEGYFDLCSQVV 404
Query: 372 AYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIF 431
Y GGLPLA+EVLGS L + +W + KL + + ++ +KL+ISY L+++ +++IF
Sbjct: 405 DYAGGLPLAIEVLGSSLRNKPMEDWIDAVKKLWEVRDKEINEKLKISYYMLEND-DREIF 463
Query: 432 LDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDMGRAI 491
LDI CFF K + +IL G A +G+ +L E+SL+ + K+ MHDL+++MG+ I
Sbjct: 464 LDIACFFKRKSKRRAIEILESFGFPAVLGLDILKEKSLITTP-HEKIQMHDLIQEMGQKI 522
Query: 492 VRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKEMRKL 551
V E + KRSRLW ED++ L+++ GTE +EG+++ L + +F M L
Sbjct: 523 VNEEFPDEPEKRSRLWLREDINRALSRDQGTEEIEGIMMDLDEEGESHLNAKSFSSMTNL 582
Query: 552 RLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWKET 611
R+L+L V L + +LS +L+++ W G+ +P +F NL+ LEL SS+ +W +
Sbjct: 583 RVLKLNNVHLCEEIEYLSDQLRFLNWHGYPLKTLPSNFNPTNLLELELPNSSIHLLWTTS 642
Query: 612 KLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNLLLINLEG 671
K +E LK+++LS S +L TPDFS +PNLE+L+L C L +LHH++G+L +L+ ++L
Sbjct: 643 KSMETLKVINLSDSQFLSKTPDFSVVPNLERLVLSGCVELHQLHHSLGNLKHLIQLDLRN 702
Query: 672 CTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSILG 731
C L N+P L+S+K L++SGCS + + + M L L ++T+IK + SI
Sbjct: 703 CKKLTNIPFNIC-LESLKILVLSGCSSLTHFPKISSNMNYLLELHLEETSIKVLHSSIGH 761
Query: 732 LKSIGYISL 740
L S+ ++L
Sbjct: 762 LTSLVVLNL 770
>sp|Q40392|TMVRN_NICGU TMV resistance protein N gi|558887|gb|AAA50763.1| N
Length = 1144
Score = 471 bits (1211), Expect = e-131
Identities = 273/732 (37%), Positives = 434/732 (58%), Gaps = 30/732 (4%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR +F SHLY L++ GI F DD++L+ G+ + L +AIE S+ ++ V S NYA+
Sbjct: 20 GEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCKAIEESQFAIVVFSENYAT 79
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLLGKQLQVEDTQRKTRRG 137
SRWCL+ELVKI+EC+ + Q VIP+FY VDPSHVRNQK K + +T+ K
Sbjct: 80 SRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKES---FAKAFEEHETKYKDDVE 136
Query: 138 G--------HEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQ 189
G +E A+ G + D+++A+ I +IV + L +S VG+++HL+
Sbjct: 137 GIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLCKISLSYLQNIVGIDTHLE 196
Query: 190 EVIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQI------HRKFMCTSFIENIREVC 243
++ L+E + V ++GIWGMGG+GKTT A+AIF+ + +F F+++I+E
Sbjct: 197 KIESLLEIGINGVRIMGIWGMGGVGKTTIARAIFDTLLGRMDSSYQFDGACFLKDIKE-- 254
Query: 244 ENDSRGHMHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQ-LRA 302
+ RG LQ LLS++L+ K ++ G + L K+ L++LDD+ + L
Sbjct: 255 --NKRGMHSLQNALLSELLREKANYNNEEDGKHQMASRLRSKKVLIVLDDIDNKDHYLEY 312
Query: 303 LCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPRED 362
L G+ +WF +GS II+TTRD L+ K D +Y + + + ES++LF HAFG+ P E+
Sbjct: 313 LAGDLDWFGNGSRIIITTRDKHLIE--KNDIIYEVTALPDHESIQLFKQHAFGKEVPNEN 370
Query: 363 FNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGL 422
F +LS VV Y GLPLAL+V GS L+ EW+S + ++ + KL+ISYDGL
Sbjct: 371 FEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMKNNSYSGIIDKLKISYDGL 430
Query: 423 KDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHD 482
+ + ++++FLDI CF G+++ Y+ IL C + A+ G+ +LI++SL+ + + N++ MHD
Sbjct: 431 EPK-QQEMFLDIACFLRGEEKDYILQILESCHIGAEYGLRILIDKSLVFISEYNQVQMHD 489
Query: 483 LVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFST 542
L++DMG+ IV + KD G+RSRLW ++V +V++ NTGT +E + + S S + FS
Sbjct: 490 LIQDMGKYIV--NFQKDPGERSRLWLAKEVEEVMSNNTGTMAMEAIWVSSYS-STLRFSN 546
Query: 543 NAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYS 602
A K M++LR+ + +L + L+ + P F L LV L+L+++
Sbjct: 547 QAVKNMKRLRVFNMGRSSTHYAIDYLPNNLRCFVCTNYPWESFPSTFELKMLVHLQLRHN 606
Query: 603 SVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLS 662
S++ +W ETK L L+ +DLS S L TPDF+ +PNLE + L CS+L E+HH++G S
Sbjct: 607 SLRHLWTETKHLPSLRRIDLSWSKRLTRTPDFTGMPNLEYVNLYQCSNLEEVHHSLGCCS 666
Query: 663 NLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAI 722
++ + L C SL P ++S++ L + C ++KL E +M+ + + + I
Sbjct: 667 KVIGLYLNDCKSLKRFP--CVNVESLEYLGLRSCDSLEKLPEIYGRMKPEIQIHMQGSGI 724
Query: 723 KEVPYSILGLKS 734
+E+P SI K+
Sbjct: 725 RELPSSIFQYKT 736
>dbj|BAD12594.1| N protein [Nicotiana tabacum]
Length = 1128
Score = 471 bits (1211), Expect = e-131
Identities = 273/732 (37%), Positives = 434/732 (58%), Gaps = 30/732 (4%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR +F SHLY L++ GI F DD++L+ G+ + L +AIE S+ ++ V S NYA+
Sbjct: 12 GEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCKAIEESQFAIVVFSENYAT 71
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLLGKQLQVEDTQRKTRRG 137
SRWCL+ELVKI+EC+ + Q VIP+FY VDPSHVRNQK K + +T+ K
Sbjct: 72 SRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKES---FAKAFEEHETKYKDDVE 128
Query: 138 G--------HEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQ 189
G +E A+ G + D+++A+ I +IV + L +S VG+++HL+
Sbjct: 129 GIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLCKISLSYLQNIVGIDTHLE 188
Query: 190 EVIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQI------HRKFMCTSFIENIREVC 243
++ L+E + V ++GIWGMGG+GKTT A+AIF+ + +F F+++I+E
Sbjct: 189 KIESLLEIGINGVRIMGIWGMGGVGKTTIARAIFDTLLGRMDSSYQFDGACFLKDIKE-- 246
Query: 244 ENDSRGHMHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQ-LRA 302
+ RG LQ LLS++L+ K ++ G + L K+ L++LDD+ + L
Sbjct: 247 --NKRGMHSLQNALLSELLREKANYNNEEDGKHQMASRLRSKKVLIVLDDIDNKDHYLEY 304
Query: 303 LCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPRED 362
L G+ +WF +GS II+TTRD L+ K D +Y + + + ES++LF HAFG+ P E+
Sbjct: 305 LAGDLDWFGNGSRIIITTRDKHLIE--KNDIIYEVTALPDHESIQLFKQHAFGKEVPNEN 362
Query: 363 FNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGL 422
F +LS VV Y GLPLAL+V GS L+ EW+S + ++ + KL+ISYDGL
Sbjct: 363 FEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMKNNSYSGIIDKLKISYDGL 422
Query: 423 KDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHD 482
+ + ++++FLDI CF G+++ Y+ IL C + A+ G+ +LI++SL+ + + N++ MHD
Sbjct: 423 EPK-QQEMFLDIACFLRGEEKDYILQILESCHIGAEYGLRILIDKSLVFISEYNQVQMHD 481
Query: 483 LVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFST 542
L++DMG+ IV + KD G+RSRLW ++V +V++ NTGT +E + + S S + FS
Sbjct: 482 LIQDMGKYIV--NFQKDPGERSRLWLAKEVEEVMSNNTGTMAMEAIWVSSYS-STLRFSN 538
Query: 543 NAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYS 602
A K M++LR+ + +L + L+ + P F L LV L+L+++
Sbjct: 539 QAVKNMKRLRVFNMGRSSTHYAIDYLPNNLRCFVCTNYPWESFPSTFELKMLVHLQLRHN 598
Query: 603 SVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLS 662
S++ +W ETK L L+ +DLS S L TPDF+ +PNLE + L CS+L E+HH++G S
Sbjct: 599 SLRHLWTETKHLPSLRRIDLSWSKRLTRTPDFTGMPNLEYVNLYQCSNLEEVHHSLGCCS 658
Query: 663 NLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAI 722
++ + L C SL P ++S++ L + C ++KL E +M+ + + + I
Sbjct: 659 KVIGLYLNDCKSLKRFP--CVNVESLEYLGLRSCDSLEKLPEIYGRMKPEIQIHMQGSGI 716
Query: 723 KEVPYSILGLKS 734
+E+P SI K+
Sbjct: 717 RELPSSIFQYKT 728
>gb|AAU04763.1| MRGH8 [Cucumis melo]
Length = 1058
Score = 466 bits (1198), Expect = e-129
Identities = 271/739 (36%), Positives = 431/739 (57%), Gaps = 26/739 (3%)
Query: 20 DTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYASSR 79
DT SF+S L+ AL++ GI VF+D E + G + ++A++ SR S+ V S NY S
Sbjct: 49 DTGRSFISDLHEALTSQGIVVFIDKEDEEDGGKPLTEKMKAVDESRSSIVVFSENYGS-- 106
Query: 80 W-CLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLL-------LGKQLQVEDTQ 131
W C+ E+ KI C++ QLV+P+FY VDP VR Q+G L+ + +E+
Sbjct: 107 WVCMKEIRKIRMCQKLRDQLVLPIFYKVDPGDVRKQEGESLVKFFNEHEANPNISIEEV- 165
Query: 132 RKTRRGGHEVASFSGYNLSPDR----------SEAEAINEIVKNVFRNLDIRLMSITDFP 181
+K R+ ++V + SG++L + SE AI EIV +VF L L D
Sbjct: 166 KKWRKSMNKVGNLSGWHLQDSQLNITFKQFCSSEEGAIKEIVNHVFNKLRPDLFRYDDKL 225
Query: 182 VGLESHLQEVIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIRE 241
VG+ L ++ L+ + VGIWGMGG+GKTT A+ I+ + F F++N++E
Sbjct: 226 VGISQRLHQINMLLGIGLDDIRFVGIWGMGGIGKTTLARIIYRSVSHLFDGCYFLDNVKE 285
Query: 242 VCENDSRGHMHLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLR 301
+ +G LQE+LL+ L + G +IK+ +S +AL+ILDDV QL+
Sbjct: 286 ALKK--QGIASLQEKLLTGALMKRNIDIPNADGATLIKRRISNIKALIILDDVDHLSQLQ 343
Query: 302 ALCGNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPRE 361
L G+ +WF SGS IIVTTR+ LL S + Y ++ ++ +E+L+LFS AFG P++
Sbjct: 344 QLAGSSDWFGSGSRIIVTTRNEHLLVSHGIEKRYKVEGLNVEEALQLFSQKAFGTNYPKK 403
Query: 362 DFNELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDG 421
D+ +LS +VV Y G LPLA+EVLGS L ++ W++ + KL+ I + ++ + LR+SYD
Sbjct: 404 DYFDLSIQVVEYSGDLPLAIEVLGSSLRDKSREVWKNAVEKLKEIRDKKILEILRVSYD- 462
Query: 422 LKDELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMH 481
L D+ EK+IFLD+ CFF K + ++L G A IG+ +L ERSL+ + K+ MH
Sbjct: 463 LLDKSEKEIFLDLACFFKKKSKKQAIEVLQSFGFQAIIGLEILEERSLITTP-HEKIQMH 521
Query: 482 DLVRDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFS 541
DL+++MG+ +VR + KR+RLW EDV+ L+ + G E +EG+++ +
Sbjct: 522 DLIQEMGQEVVRRMFPNNPEKRTRLWLREDVNLALSHDQGAEAIEGIVMDSSEEGESHLN 581
Query: 542 TNAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKY 601
F M LR+L++ V L G+ +LS +L+++ W G+ S Y+P +F+ +++ LEL
Sbjct: 582 AKVFSTMTNLRILKINNVSLCGELDYLSDQLRFLSWHGYPSKYLPPNFHPKSILELELPN 641
Query: 602 SSVKQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDL 661
S + +WK +K L++LK ++LS S ++ TPDFS +PNLE+LIL C L +LH ++G L
Sbjct: 642 SFIHYLWKGSKRLDRLKTVNLSDSQFISKTPDFSGVPNLERLILSGCVRLTKLHQSLGSL 701
Query: 662 SNLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTA 721
L+ ++L+ C +L +P + L+S+ L +S CS + + M++LT L T+
Sbjct: 702 KRLIQLDLKNCKALKAIP-FSISLESLIVLSLSNCSSLKNFPNIVGNMKNLTELHLDGTS 760
Query: 722 IKEVPYSILGLKSIGYISL 740
I+E+ SI L + ++L
Sbjct: 761 IQELHPSIGHLTGLVLLNL 779
Score = 89.7 bits (221), Expect = 5e-16
Identities = 62/171 (36%), Positives = 92/171 (53%), Gaps = 5/171 (2%)
Query: 591 LGNLVVLELKYSSVKQVWKETKLLEKLKILDLSHSWYLENTPDF-SKLPNLEKLILKDCS 649
L L+ L+LK + + LE L +L LS+ L+N P+ + NL +L L D +
Sbjct: 701 LKRLIQLDLKNCKALKAIPFSISLESLIVLSLSNCSSLKNFPNIVGNMKNLTELHL-DGT 759
Query: 650 SLFELHHTIGDLSNLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQM 709
S+ ELH +IG L+ L+L+NLE CT+L LP L +KTL + GCSK+ ++ E + +
Sbjct: 760 SIQELHPSIGHLTGLVLLNLENCTNLLELPNTIGSLICLKTLTLHGCSKLTRIPESLGFI 819
Query: 710 ESLTTLIAKDTAIKEVPYSILGLKSIGYISLCGYEGLSRDVFPSLLRSWIS 760
SL L +T I + P L L+ + + + GLSR SL SW S
Sbjct: 820 ASLEKLDVTNTCINQAP---LSLQLLTNLEILDCRGLSRKFIHSLFPSWNS 867
>gb|AAO23075.1| R 5 protein [Glycine max]
Length = 907
Score = 464 bits (1195), Expect = e-129
Identities = 274/728 (37%), Positives = 425/728 (57%), Gaps = 21/728 (2%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR+ F +LY AL + GI+ F D++KL G E+ PALL+AI+ SRI++ VLS ++AS
Sbjct: 20 GEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAITVLSEDFAS 79
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPV--LLLGKQLQVEDTQRKTR 135
S +CLDEL IL C + G +VIP+FY V P VR+QKG L + + D +K
Sbjct: 80 SSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRFPDKLQKWE 139
Query: 136 RGGHEVASFSGYNLSP-DRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEVIKL 194
R +VA+ SG + D E + I IV +V ++ + + D PVGLES +QEV KL
Sbjct: 140 RALRQVANLSGLHFKDRDEYEYKFIGRIVASVSEKINPASLHVADLPVGLESKVQEVRKL 199
Query: 195 IE-SQSSKVCMVGIWGMGGLGKTTTAKAIFNQ--IHRKFMCTSFIENIREVCENDSRGHM 251
++ VCM+GI GMGG+GK+T A+A++N I F F+EN+RE N G
Sbjct: 200 LDVGNHDGVCMIGIHGMGGIGKSTLARAVYNDLIITENFDGLCFLENVRESSNN--HGLQ 257
Query: 252 HLQEQLLSDVLKTKEKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREWFA 311
HLQ LLS++L K+ S +G + I+ +L GK+ L+ILDDV +QL+ + G R+WF
Sbjct: 258 HLQSILLSEILGEDIKVRSKQQGISKIQSMLKGKKVLLILDDVDKPQQLQTIAGRRDWFG 317
Query: 312 SGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRRVV 371
GS+II+TTRD +LL Y ++ ++++ +L+L +W+AF + ++ RVV
Sbjct: 318 PGSIIIITTRDKQLLAPHGVKKRYEVEVLNQNAALQLLTWNAFKREKIDPSYEDVLNRVV 377
Query: 372 AYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKDIF 431
Y GLPLALEV+GS + + EW+S + +RIPND++ + L++S+D L +E +K++F
Sbjct: 378 TYASGLPLALEVIGSNMFGKRVAEWKSAVEHYKRIPNDEILEILKVSFDALGEE-QKNVF 436
Query: 432 LDICCFFVGKDRAYVTDILNGCGLYADI---GITLLIERSLLQVEKNNKLGMHDLVRDMG 488
LDI C F G V +L GLY + I +L+++SL++V ++ + MHDL++ +G
Sbjct: 437 LDIACCFKGCKLTEVEHMLR--GLYNNCMKHHIDVLVDKSLIKV-RHGTVNMHDLIQVVG 493
Query: 489 RAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSR---VCFSTNAF 545
R I R+ S ++ GK RLW +D+ VL NTGT +E + L + + V ++ NAF
Sbjct: 494 REIERQISPEEPGKCKRLWLPKDIIQVLKHNTGTSKIEIICLDFSISDKEQTVEWNQNAF 553
Query: 546 KEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSVK 605
+M L++L + + + L+ + W + S +P +F+ NL++ +L SS+
Sbjct: 554 MKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSKCLPSNFHPNNLLICKLPDSSMA 613
Query: 606 QV-WKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNL 664
+ + L +L + +L PD S LPNL +L K C SL + +IG L+ L
Sbjct: 614 SFEFHGSSKFGHLTVLKFDNCKFLTQIPDVSDLPNLRELSFKGCESLVAVDDSIGFLNKL 673
Query: 665 LLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKE 724
+N GC L + P L S++TL +SGCS ++ E + +ME++ L+ +D IKE
Sbjct: 674 KKLNAYGCRKLTSFP--PLNLTSLETLQLSGCSSLEYFPEILGEMENIKQLVLRDLPIKE 731
Query: 725 VPYSILGL 732
+P+S L
Sbjct: 732 LPFSFQNL 739
>gb|AAR21295.1| bacterial spot disease resistance protein 4 [Lycopersicon esculentum]
Length = 1146
Score = 464 bits (1193), Expect = e-129
Identities = 344/1112 (30%), Positives = 568/1112 (50%), Gaps = 151/1112 (13%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
GEDTR +F HLY L N GIN F DD++L+ G + LLRAIE S+++L + S NYA+
Sbjct: 28 GEDTRKTFTGHLYEGLRNRGINTFQDDKRLEHGDSIPKELLRAIEDSQVALIIFSKNYAT 87
Query: 78 SRWCLDELVKILECR-RTYGQLVIPLFYHVDPSHVRNQK---GPVLLLGKQLQVEDTQ-- 131
SRWCL+ELVKI+EC+ GQ VIP+FY+VDPSHVR Q G + +D +
Sbjct: 88 SRWCLNELVKIMECKEEENGQTVIPIFYNVDPSHVRYQTESFGAAFAKHESKYKDDVEGM 147
Query: 132 ---RKTRRGGHEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFP--VGLES 186
++ R A+ GY++ + E+E I +IV + S++ VG+ +
Sbjct: 148 QKVQRWRTALTAAANLKGYDIR-NGIESENIQQIVDCISSKFCTNAYSLSFLQDIVGINA 206
Query: 187 HLQEVIKLIESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCEND 246
HL+++ ++ + + V ++GIWG+GG+GKT AKAIF+ + +F + F+ +++E +
Sbjct: 207 HLEKLKSKLQIEINDVRILGIWGIGGVGKTRIAKAIFDTLSYQFEASCFLADVKEFAK-- 264
Query: 247 SRGHMH-LQEQLLSDVLKTK-EKIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALC 304
+ +H LQ LLS++L+ K + +++ G MI L + L++LDD+ +Q+ L
Sbjct: 265 -KNKLHSLQNILLSELLRKKNDYVYNKYDGKCMIPNRLCSLKVLIVLDDIDHGDQMEYLA 323
Query: 305 GNREWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFN 364
G+ WF +GS +IVTTR+ L+ K D +Y + + + E+++LF+ HAF + P EDF
Sbjct: 324 GDICWFGNGSRVIVTTRNKHLIE--KDDAIYEVSTLPDHEAMQLFNMHAFKKEVPNEDFK 381
Query: 365 ELSRRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKD 424
EL+ +V + GLPLAL+V G L+++ W+ + ++++ N ++ ++L+ISYDGL+
Sbjct: 382 ELALEIVNHAKGLPLALKVWGCLLHKKNLSLWKITVEQIKKDSNSEIVEQLKISYDGLES 441
Query: 425 ELEKDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLV 484
E E++IFLDI CFF G+ R V IL C A+ G+ +LI +SL+ + +N+++ MHDL+
Sbjct: 442 E-EQEIFLDIACFFRGEKRKEVMQILKSCDFGAEYGLDVLINKSLVFISENDRIEMHDLI 500
Query: 485 RDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNA 544
RDMGR +V+ + KRSR+W ED +V+ TGT TVE + V F+ A
Sbjct: 501 RDMGRYVVKMQKLQK--KRSRIWDVEDFKEVMIDYTGTMTVEA--IWFSCFEEVRFNKEA 556
Query: 545 FKEMRKLRLLQL-----------------------ECVDLTGDY-----RHLSHELKWVY 576
K+M++LR+L + + DL D+ +LS+ L+W+
Sbjct: 557 MKKMKRLRILHIFDGFVKFFSSPPSSNSNDSEEEDDSYDLVVDHHDDSIEYLSNNLRWLV 616
Query: 577 WKGFTSTYIPDHFYLGNLVVLELKYSSVKQVWKETKLLEKLKILDLSHSWYLENTPDFSK 636
W ++ +P++F LV LEL++SS+ +WK+T+ L L+ LDLS S L TPDF+
Sbjct: 617 WNHYSWKSLPENFKPEKLVHLELRWSSLHYLWKKTEHLPSLRKLDLSLSKSLVQTPDFTG 676
Query: 637 LPNLEKLILKDCSSLFELHHTIGDLSNLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGC 696
+PNLE L L+ CS L E+H+++ L+ +NL CT L P ++S+++L + C
Sbjct: 677 MPNLEYLNLEYCSKLEEVHYSLAYCEKLIELNLSWCTKLRRFP--YINMESLESLDLQYC 734
Query: 697 SKIDKLEEDIAQMESLTTLIAKDTAIKEVPYSILGLKSIGYISLCGYEGLSRDVFPSLL- 755
I E I M+ +++ +T I E+P S+ + + L G E L + PS +
Sbjct: 735 YGIMVFPEIIGTMKPELMILSANTMITELPSSLQYPTHLTELDLSGMENL--EALPSSIV 792
Query: 756 -----------------------------------RSWISP------TMNPLSHIPL--- 771
R+ IS +N L + L
Sbjct: 793 KLKDLVKLNVSYCLTLKSLPEEIGDLENLEELDASRTLISQPPSSIVRLNKLKSLKLMKR 852
Query: 772 ----------FAAMSLPLVSMDI-----QNYSSGNSSSWLSSLSKLRSVWIQCHSEIQLA 816
F ++ L+S++I N+ G + LS L+ + ++ + L
Sbjct: 853 NTLTDDVCFVFPPVNNGLLSLEILELGSSNFEDGRIPEDIGCLSSLKELRLEGDNFNHLP 912
Query: 817 QESRRI-------LDDQYDVNSM-DLEASSSSSYASQISDFSLKSLLIRLGSCYTVIDTL 868
Q ++ + D + S+ + + +A +D KSL + + S I +
Sbjct: 913 QSIAQLGALRFLYIKDCRSLTSLPEFPPQLDTIFADWSNDLICKSLFLNISSFQHNI-SA 971
Query: 869 ASSISQGSTTNASSNFFLPGGDDPYWLAYKGEGPSVHFQVPED---RDCCLKGIVLCVVY 925
+ S+S T+ S+ P W ++G SV +PE+ D L G +C Y
Sbjct: 972 SDSLSLRVFTSLGSSI-------PIWFHHQGTDTSVSVNLPENWYVSDNFL-GFAVC-YY 1022
Query: 926 SSTPENMA--------TECLT-SVMINNHTKFTIQIYKQGTVMSFNDEDWESVKSN-LDP 975
+ EN A C+T ++++NH++ T I ++ F W++ +N P
Sbjct: 1023 GNLTENTAELIMSSAGMPCITWKLLLSNHSECT-YIRIHFFLVPFAGL-WDTSNANGKTP 1080
Query: 976 GDDTEIVVSFGRGLTVKETAVYLIYAQSIIME 1007
D I++SF + L KE V L Y ++E
Sbjct: 1081 NDYKHIMLSFPQEL--KECGVRLFYEDESVLE 1110
>emb|CAA08798.1| NL27 [Solanum tuberosum]
Length = 821
Score = 462 bits (1190), Expect = e-128
Identities = 274/725 (37%), Positives = 427/725 (58%), Gaps = 31/725 (4%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
G DTR +F SHLY L N GI F DD++L+ G + LL+AIE S+++L + S NYA+
Sbjct: 28 GVDTRRTFTSHLYEGLKNRGIFTFQDDKRLENGDSIPEELLKAIEESQVALIIFSKNYAT 87
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGPVLLLGKQLQ------VEDTQ 131
SRWCL+ELVKI+EC+ GQ+VIP+FY VDPS VR Q + + +E Q
Sbjct: 88 SRWCLNELVKIMECKEEKGQIVIPIFYDVDPSEVRKQTKSFAEAFTEHESKYANDIEGMQ 147
Query: 132 RKT--RRGGHEVASFSGYNLSPDRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQ 189
+ R + A GY++S +R E++ I IV ++ L I + VG+++H +
Sbjct: 148 KVKGWRTALSDAADLKGYDIS-NRIESDYIQHIVDHISVLCKGSLSYIKNL-VGIDTHFK 205
Query: 190 EVIKLI-ESQSSKVCMVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSR 248
+ L+ E Q S V +VGIWGM G+GKTT A+AIF+++ +F F+ +I+E +
Sbjct: 206 NIRSLLAELQMSGVLIVGIWGMPGVGKTTIARAIFDRLSYQFEAVCFLADIKE----NKC 261
Query: 249 GHMHLQEQLLSDVLKTKEK-IHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNR 307
G LQ LLS++LK K+ +++ G +++ L K+ LV+LDD+ +QL L GN
Sbjct: 262 GMHSLQNILLSELLKEKDNCVNNKEDGRSLLAHRLRFKKVLVVLDDIDHIDQLDYLAGNL 321
Query: 308 EWFASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELS 367
+WF +GS II TTRD L+ + VY + + + ++++LF +AF E + F EL+
Sbjct: 322 DWFGNGSRIIATTRDKHLIGK---NVVYELPTLHDHDAIKLFERYAFKEQVSDKCFKELT 378
Query: 368 RRVVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELE 427
VV++ GLPLAL+V G + +ER+ EW S + +++ PN ++ +KL+ISYDGL + ++
Sbjct: 379 LEVVSHAKGLPLALKVFGCFFHERDITEWRSAIKQIKNNPNSEIVEKLKISYDGL-ETIQ 437
Query: 428 KDIFLDICCFFVGKDRAYVTDILNGCGLYADIGITLLIERSLLQVEKNNKLGMHDLVRDM 487
+ IFLDI CF G+ + YV IL C ADIG+++LI++SL+ + NN + MHDL++DM
Sbjct: 438 QSIFLDIACFLRGRRKDYVMQILESCDFGADIGLSVLIDKSLVSISGNNTIEMHDLIQDM 497
Query: 488 GRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSRVCFSTNAFKE 547
G+ +V++ KD G+RSRLW +D +V+ NTGT+ VE + + + +R FS A
Sbjct: 498 GKYVVKKQ--KDPGERSRLWLTKDFEEVMINNTGTKAVEA--IWVPNFNRPRFSKEAMTI 553
Query: 548 MRKLRLLQL---ECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKYSSV 604
M++LR+L + C+D G +L + L+W W + +P++F LV L+L SS+
Sbjct: 554 MQRLRILCIHDSNCLD--GSIEYLPNSLRWFVWNNYPCESLPENFEPQKLVHLDLSLSSL 611
Query: 605 KQVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIGDLSNL 664
+W K L L+ LDL S L TPDF+ +PNL+ L L C +L E+HH++G L
Sbjct: 612 HHLWTGKKHLPFLQKLDLRDSRSLMQTPDFTWMPNLKYLDLSYCRNLSEVHHSLGYSREL 671
Query: 665 LLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKDTAIKE 724
+ +NL C L P ++S+ + + CS ++K M+ + + IKE
Sbjct: 672 IELNLYNCGRLKRFP--CVNVESLDYMDLEFCSSLEKFPIIFGTMKPELKIKMGLSGIKE 729
Query: 725 VPYSI 729
+P S+
Sbjct: 730 LPSSV 734
>gb|AAO92748.1| candidate disease-resistance protein SR1 [Glycine max]
Length = 1137
Score = 461 bits (1187), Expect = e-128
Identities = 288/745 (38%), Positives = 441/745 (58%), Gaps = 33/745 (4%)
Query: 18 GEDTRHSFVSHLYAALSNAGINVFLDDEKLKKGSELKPALLRAIEGSRISLAVLSSNYAS 77
G+DTRH F +LY AL + GI F+DD++L +G E+KPAL AI+GSRI++ VLS NYA
Sbjct: 20 GQDTRHGFTGYLYKALDDRGIYTFIDDQELPRGDEIKPALSDAIQGSRIAITVLSQNYAF 79
Query: 78 SRWCLDELVKILECRRTYGQLVIPLFYHVDPSHVRNQKGP----VLLLGKQLQV-EDTQR 132
S +CLDELV IL C+ + G LVIP+FY VDPSHVR+QKG + K+ + ++ +
Sbjct: 80 STFCLDELVTILHCK-SEGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFKANKEKLQ 138
Query: 133 KTRRGGHEVASFSGYNLSP-DRSEAEAINEIVKNVFRNLDIRLMSITDFPVGLESHLQEV 191
K R +VA SGY+ D E + I IV+ V R ++ + + D+PVGL S + EV
Sbjct: 139 KWRMALQQVADLSGYHFKDGDAYEYKFIQSIVEQVSREINRAPLHVADYPVGLGSQVIEV 198
Query: 192 IKLIESQSSKVC-MVGIWGMGGLGKTTTAKAIFNQIHRKFMCTSFIENIREVCENDSRGH 250
KL++ S V ++GI GMGGLGKTT A A++N I F + F++N+RE E++ +
Sbjct: 199 RKLLDVGSDDVVHIIGIHGMGGLGKTTLAVAVYNLIAPHFDESCFLQNVRE--ESNLK-- 254
Query: 251 MHLQEQLLSDVLKTKE-KIHSIGRGTAMIKKILSGKRALVILDDVTTSEQLRALCGNREW 309
HLQ LLS +L K+ + S G +MI+ L K+ L+ILDDV EQL+A+ G +W
Sbjct: 255 -HLQSSLLSKLLGEKDITLTSWQEGASMIQHRLRRKKVLLILDDVDKREQLKAIVGKPDW 313
Query: 310 FASGSVIIVTTRDVRLLNSLKADYVYTMKEMDEDESLELFSWHAFGEASPREDFNELSRR 369
F GS +I+TTRD LL + + Y +K ++ + +L L +W+AF ++++ R
Sbjct: 314 FGPGSRVIITTRDKHLLKYHEVERTYEVKVLNHNAALHLLTWNAFKREKIDPIYDDVLNR 373
Query: 370 VVAYCGGLPLALEVLGSYLNEREEIEWESVLSKLERIPNDQVQQKLRISYDGLKDELEKD 429
VV Y GLPLALEV+GS L + EWES L +RIP++++ + L++S+D L++E +++
Sbjct: 374 VVTYASGLPLALEVIGSNLYGKTVAEWESALETYKRIPSNEILKILQVSFDALEEE-QQN 432
Query: 430 IFLDICCFFVGKDRAYVTDI---LNGCGLYADIGITLLIERSLLQVEKNNK--LGMHDLV 484
+FLDI C F G + V DI L G G IG+ L+E+SL++ +NN+ + MH+L+
Sbjct: 433 VFLDIACCFKGHEWTEVDDIFRALYGNGKKYHIGV--LVEKSLIKYNRNNRGTVQMHNLI 490
Query: 485 RDMGRAIVRESSTKDLGKRSRLWFHEDVHDVLTKNTGTETVEGLILKLQSTSR---VCFS 541
+DMGR I R+ S ++ GKR RLW +D+ VL NTGT +E + L + + V ++
Sbjct: 491 QDMGREIERQRSPEEPGKRKRLWSPKDIIQVLKHNTGTSKIEIICLDSSISDKEETVEWN 550
Query: 542 TNAFKEMRKLRLLQLECVDLTGDYRHLSHELKWVYWKGFTSTYIPDHFYLGNLVVLELKY 601
NAF +M L++L + + ++ L+ + W + S +P +F NLV+ +L
Sbjct: 551 ENAFMKMENLKILIIRNGKFSIGPNYIPEGLRVLEWHRYPSNCLPSNFDPINLVICKLPD 610
Query: 602 SSVK--QVWKETKLLEKLKILDLSHSWYLENTPDFSKLPNLEKLILKDCSSLFELHHTIG 659
SS+ + +K L L +L+ +L PD S LPNL++L + C SL + ++G
Sbjct: 611 SSITSFEFHGSSKKLGHLTVLNFDKCKFLTQIPDVSDLPNLKELSFRKCESLVAVDDSVG 670
Query: 660 DLSNLLLINLEGCTSLNNLPGKTYQLKSVKTLIISGCSKIDKLEEDIAQMESLTTLIAKD 719
L+ L ++ GC L + P L S++ L ISGCS ++ E + +M + L D
Sbjct: 671 FLNKLKKLSAYGCRKLTSFP--PLNLTSLRRLQISGCSSLEYFPEILGEMVKIRVLELHD 728
Query: 720 TAIKEVPYS---ILGLKSIGYISLC 741
IKE+P+S ++GL + Y+ C
Sbjct: 729 LPIKELPFSFQNLIGLSRL-YLRRC 752
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,769,015,671
Number of Sequences: 2540612
Number of extensions: 74262811
Number of successful extensions: 247836
Number of sequences better than 10.0: 3086
Number of HSP's better than 10.0 without gapping: 1388
Number of HSP's successfully gapped in prelim test: 1728
Number of HSP's that attempted gapping in prelim test: 235792
Number of HSP's gapped (non-prelim): 6697
length of query: 1069
length of database: 863,360,394
effective HSP length: 139
effective length of query: 930
effective length of database: 510,215,326
effective search space: 474500253180
effective search space used: 474500253180
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 81 (35.8 bits)
Lotus: description of TM0342a.5


