

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0335b.2
(106 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
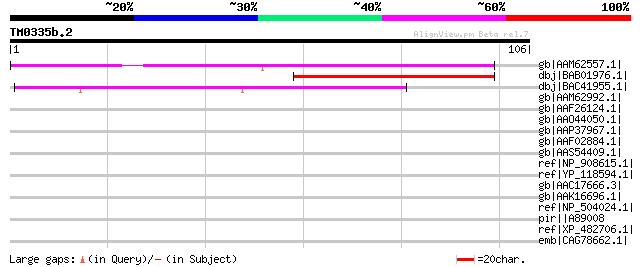
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM62557.1| unknown [Arabidopsis thaliana] gi|28394021|gb|AAO... 80 2e-14
dbj|BAB01976.1| unnamed protein product [Arabidopsis thaliana] 70 1e-11
dbj|BAC41955.1| unknown protein [Arabidopsis thaliana] gi|284168... 50 1e-05
gb|AAM62992.1| unknown [Arabidopsis thaliana] 40 0.018
gb|AAF26124.1| unknown protein [Arabidopsis thaliana] 37 0.12
gb|AAO44050.1| At3g03150 [Arabidopsis thaliana] gi|18396458|ref|... 37 0.12
gb|AAP37967.1| seed specific protein Bn15D1B [Brassica napus] 33 1.3
gb|AAF02884.1| Similar to late embryogenis abundant protein 5 [A... 33 2.2
gb|AAS54409.1| AGL081Wp [Ashbya gossypii ATCC 10895] gi|45201015... 32 2.9
ref|NP_908615.1| zinc-induced protein-like [Oryza sativa (japoni... 32 3.8
ref|YP_118594.1| putative dihydroxy-acid dehydratase [Nocardia f... 32 3.8
gb|AAC17666.3| Acetylcholine receptor protein 23 [Caenorhabditis... 32 3.8
gb|AAK16696.1| zinc-induced protein [Oryza sativa] 32 3.8
ref|NP_504024.1| acetylcholine receptor (acr-23) [Caenorhabditis... 32 3.8
pir||A89008 protein F59B1.9 [imported] - Caenorhabditis elegans 32 3.8
ref|XP_482706.1| hypothetical protein [Oryza sativa (japonica cu... 31 6.4
emb|CAG78662.1| unnamed protein product [Yarrowia lipolytica CLI... 31 6.4
>gb|AAM62557.1| unknown [Arabidopsis thaliana] gi|28394021|gb|AAO42418.1| unknown
protein [Arabidopsis thaliana]
gi|27754548|gb|AAO22721.1| unknown protein [Arabidopsis
thaliana] gi|18402280|ref|NP_566643.1| expressed
protein [Arabidopsis thaliana]
Length = 110
Score = 79.7 bits (195), Expect = 2e-14
Identities = 45/102 (44%), Positives = 58/102 (56%), Gaps = 7/102 (6%)
Query: 1 MARGGITKSTLLILRGARRAENRVMKFSGTTAKAAAAESSEEGVPKIISG---KTEDSSA 57
M R GI K+ L+LR ++ + R +G ++KAA + + IS K
Sbjct: 1 MGRTGIAKAPKLLLRSWKQFQGR----AGISSKAAKSNPMIVDYFEDISDHNLKFSGEEE 56
Query: 58 IWVPHPRTGIYYPKGHECVMEDVPEGAARFTQTYWFRNDDGV 99
WVPHPRTGI++P G E VMEDVP GAA F T+W RN DGV
Sbjct: 57 SWVPHPRTGIFFPPGQESVMEDVPNGAASFDMTFWLRNVDGV 98
>dbj|BAB01976.1| unnamed protein product [Arabidopsis thaliana]
Length = 75
Score = 70.5 bits (171), Expect = 1e-11
Identities = 29/41 (70%), Positives = 32/41 (77%)
Query: 59 WVPHPRTGIYYPKGHECVMEDVPEGAARFTQTYWFRNDDGV 99
WVPHPRTGI++P G E VMEDVP GAA F T+W RN DGV
Sbjct: 23 WVPHPRTGIFFPPGQESVMEDVPNGAASFDMTFWLRNVDGV 63
>dbj|BAC41955.1| unknown protein [Arabidopsis thaliana] gi|28416835|gb|AAO42948.1|
At1g73120 [Arabidopsis thaliana]
gi|15219378|ref|NP_177455.1| expressed protein
[Arabidopsis thaliana] gi|12324324|gb|AAG52133.1|
hypothetical protein; 69822-70342 [Arabidopsis
thaliana] gi|25373329|pir||A96757 hypothetical protein
T18K17.22 [imported] - Arabidopsis thaliana
Length = 109
Score = 50.4 bits (119), Expect = 1e-05
Identities = 31/86 (36%), Positives = 44/86 (51%), Gaps = 6/86 (6%)
Query: 2 ARGGITKSTLLI---LRGARRAENRVMKFSGTTAKAAAAESSEEGVPK---IISGKTEDS 55
+RG T +T+ + LR + ++ T K + S EE +G+T
Sbjct: 8 SRGAQTMNTMFVKPMLRKSIHKKSASHDIVRDTVKTEGSSSGEEVKTMRGFYGAGETSSP 67
Query: 56 SAIWVPHPRTGIYYPKGHECVMEDVP 81
++ WVPH TGIYYPKG E VM+DVP
Sbjct: 68 ASSWVPHEGTGIYYPKGQEKVMQDVP 93
>gb|AAM62992.1| unknown [Arabidopsis thaliana]
Length = 121
Score = 39.7 bits (91), Expect = 0.018
Identities = 17/43 (39%), Positives = 22/43 (50%), Gaps = 2/43 (4%)
Query: 54 DSSAIWVPHPRTGIYYPK--GHECVMEDVPEGAARFTQTYWFR 94
DS W PHP+TG++ P H ED + A +T WFR
Sbjct: 66 DSDKYWSPHPKTGVFGPSTTEHSATAEDAHQDTAVLEETAWFR 108
>gb|AAF26124.1| unknown protein [Arabidopsis thaliana]
Length = 86
Score = 37.0 bits (84), Expect = 0.12
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 2/43 (4%)
Query: 54 DSSAIWVPHPRTGIYYPK--GHECVMEDVPEGAARFTQTYWFR 94
DS W PHP+TG++ P H E + A +T WFR
Sbjct: 31 DSDKYWSPHPKTGVFGPSTTEHSATAEGAHQDTAVLEETAWFR 73
>gb|AAO44050.1| At3g03150 [Arabidopsis thaliana] gi|18396458|ref|NP_566194.1|
expressed protein [Arabidopsis thaliana]
Length = 121
Score = 37.0 bits (84), Expect = 0.12
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 2/43 (4%)
Query: 54 DSSAIWVPHPRTGIYYPK--GHECVMEDVPEGAARFTQTYWFR 94
DS W PHP+TG++ P H E + A +T WFR
Sbjct: 66 DSDKYWSPHPKTGVFGPSTTEHSATAEGAHQDTAVLEETAWFR 108
>gb|AAP37967.1| seed specific protein Bn15D1B [Brassica napus]
Length = 120
Score = 33.5 bits (75), Expect = 1.3
Identities = 15/43 (34%), Positives = 21/43 (47%), Gaps = 2/43 (4%)
Query: 54 DSSAIWVPHPRTGIYYPK--GHECVMEDVPEGAARFTQTYWFR 94
DS W PHP+TG++ P E + +A +T WFR
Sbjct: 65 DSDKYWSPHPQTGVFGPSTTDQTAAAEAARQDSAVLEETAWFR 107
>gb|AAF02884.1| Similar to late embryogenis abundant protein 5 [Arabidopsis
thaliana] gi|21618083|gb|AAM67133.1| late embryogenis
abundant protein, putative [Arabidopsis thaliana]
gi|15218614|ref|NP_171781.1| late embryogenesis
abundant 3 family protein / LEA3 family protein
[Arabidopsis thaliana] gi|25511624|pir||D86158
F22D16.18 protein - Arabidopsis thaliana
gi|51971691|dbj|BAD44510.1| unknown protein
[Arabidopsis thaliana]
Length = 91
Score = 32.7 bits (73), Expect = 2.2
Identities = 22/72 (30%), Positives = 30/72 (41%), Gaps = 4/72 (5%)
Query: 16 GARRAENRVMKFS-GTTAKAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYYPKGHE 74
G+ + N V + AK A S K +G+ A WVP P+TG Y P E
Sbjct: 15 GSEKLSNAVFRRGFAAAAKTALDGSVSTAEMKKRAGEASSEKAPWVPDPKTGYYRP---E 71
Query: 75 CVMEDVPEGAAR 86
V E++ R
Sbjct: 72 TVSEEIDPAELR 83
>gb|AAS54409.1| AGL081Wp [Ashbya gossypii ATCC 10895] gi|45201015|ref|NP_986585.1|
AGL081Wp [Eremothecium gossypii]
Length = 223
Score = 32.3 bits (72), Expect = 2.9
Identities = 11/25 (44%), Positives = 17/25 (68%)
Query: 41 EEGVPKIISGKTEDSSAIWVPHPRT 65
E+G+P + SGK + +WVPHP +
Sbjct: 168 EDGIPGVESGKAFGAYVVWVPHPES 192
>ref|NP_908615.1| zinc-induced protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 93
Score = 32.0 bits (71), Expect = 3.8
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Query: 20 AENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSSA----IWVPHPRTGIYYPKG 72
A+ R + AK A + E+ V ++ +T +A WVP P TG Y P G
Sbjct: 13 AQRRGYSVAAAVAKGAGRRADEKKVAAAVAKRTMAKAAEEKTAWVPDPVTGYYRPAG 69
>ref|YP_118594.1| putative dihydroxy-acid dehydratase [Nocardia farcinica IFM 10152]
gi|54015860|dbj|BAD57230.1| putative dihydroxy-acid
dehydratase [Nocardia farcinica IFM 10152]
Length = 629
Score = 32.0 bits (71), Expect = 3.8
Identities = 20/65 (30%), Positives = 32/65 (48%), Gaps = 9/65 (13%)
Query: 11 LLILRGARRAENRVMKFSGT-------TAKAAAAESSEEGVPKIISGKTEDSSAIWVPH- 62
L +LRG + V+K +G T +A AES EE V ++SG+ + + + +
Sbjct: 424 LAVLRGNLAVDGAVVKSAGVPADLHVFTGEAVVAESQEEAVTAVLSGRVRPGTVLVIRYE 483
Query: 63 -PRTG 66
PR G
Sbjct: 484 GPRGG 488
>gb|AAC17666.3| Acetylcholine receptor protein 23 [Caenorhabditis elegans]
gi|40763973|gb|AAR89634.1| acetylcholine receptor (62.5
kD) (acr-23) [Caenorhabditis elegans]
Length = 545
Score = 32.0 bits (71), Expect = 3.8
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 10/66 (15%)
Query: 33 KAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYY-PKGHECVMEDVPEGAARFTQTY 91
+A A + EE K +S K +++W T +Y+ + H +ME VP+GA +F Q
Sbjct: 379 QAYAKRAKEEKHRKRMSRK----NSMW-----TKVYHLARDHSKLMETVPDGAVKFNQIS 429
Query: 92 WFRNDD 97
F+N+D
Sbjct: 430 DFKNND 435
>gb|AAK16696.1| zinc-induced protein [Oryza sativa]
Length = 117
Score = 32.0 bits (71), Expect = 3.8
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Query: 20 AENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSSA----IWVPHPRTGIYYPKG 72
A+ R + AK A + E+ V ++ +T +A WVP P TG Y P G
Sbjct: 13 AQRRGYSVAAAVAKGAGRRADEKKVAAAVAKRTMAKAAEEKTAWVPDPVTGYYRPAG 69
>ref|NP_504024.1| acetylcholine receptor (acr-23) [Caenorhabditis elegans]
Length = 656
Score = 32.0 bits (71), Expect = 3.8
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 10/66 (15%)
Query: 33 KAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYY-PKGHECVMEDVPEGAARFTQTY 91
+A A + EE K +S K +++W T +Y+ + H +ME VP+GA +F Q
Sbjct: 490 QAYAKRAKEEKHRKRMSRK----NSMW-----TKVYHLARDHSKLMETVPDGAVKFNQIS 540
Query: 92 WFRNDD 97
F+N+D
Sbjct: 541 DFKNND 546
>pir||A89008 protein F59B1.9 [imported] - Caenorhabditis elegans
Length = 641
Score = 32.0 bits (71), Expect = 3.8
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 10/66 (15%)
Query: 33 KAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYY-PKGHECVMEDVPEGAARFTQTY 91
+A A + EE K +S K +++W T +Y+ + H +ME VP+GA +F Q
Sbjct: 475 QAYAKRAKEEKHRKRMSRK----NSMW-----TKVYHLARDHSKLMETVPDGAVKFNQIS 525
Query: 92 WFRNDD 97
F+N+D
Sbjct: 526 DFKNND 531
>ref|XP_482706.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|42407625|dbj|BAD08740.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 105
Score = 31.2 bits (69), Expect = 6.4
Identities = 18/56 (32%), Positives = 24/56 (42%), Gaps = 9/56 (16%)
Query: 18 RRAENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYYPKGH 73
R+ E +GTTAK AAE + G W+ PRTG + P+ H
Sbjct: 40 RQQEPATAAAAGTTAKLQAAEGAAAG---------SKEGFFWMREPRTGNWMPENH 86
>emb|CAG78662.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50556886|ref|XP_505851.1| hypothetical protein
[Yarrowia lipolytica]
Length = 239
Score = 31.2 bits (69), Expect = 6.4
Identities = 10/24 (41%), Positives = 17/24 (70%)
Query: 41 EEGVPKIISGKTEDSSAIWVPHPR 64
E+G+P ++SG+ ++ IWVP R
Sbjct: 182 EDGIPGVVSGRAAEAHVIWVPDQR 205
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.131 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 177,746,365
Number of Sequences: 2540612
Number of extensions: 6636418
Number of successful extensions: 14719
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 14712
Number of HSP's gapped (non-prelim): 17
length of query: 106
length of database: 863,360,394
effective HSP length: 82
effective length of query: 24
effective length of database: 655,030,210
effective search space: 15720725040
effective search space used: 15720725040
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0335b.2


