

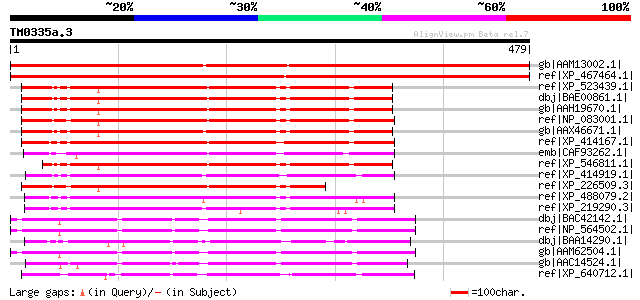
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0335a.3
(479 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM13002.1| putative steroid dehydrogenase [Arabidopsis thali... 684 0.0
ref|XP_467464.1| putative NAD(P)-dependent cholesterol dehydroge... 662 0.0
ref|XP_523439.1| PREDICTED: similar to NAD(P) dependent steroid ... 281 4e-74
dbj|BAE00861.1| unnamed protein product [Macaca fascicularis] 278 3e-73
gb|AAH19670.1| NAD(P) dependent steroid dehydrogenase-like [Homo... 276 8e-73
ref|NP_083001.1| RIKEN cDNA 4632417N05 [Mus musculus] gi|2441656... 269 1e-70
gb|AAX46671.1| NAD(P) dependent steroid dehydrogenase-like [Bos ... 269 2e-70
ref|XP_414167.1| PREDICTED: similar to NAD(P) dependent steroid ... 265 3e-69
emb|CAF93262.1| unnamed protein product [Tetraodon nigroviridis] 263 1e-68
ref|XP_546811.1| PREDICTED: similar to NAD(P) dependent steroid ... 256 1e-66
ref|XP_414919.1| PREDICTED: similar to NAD(P) dependent steroid ... 243 7e-63
ref|XP_226509.3| PREDICTED: similar to RIKEN cDNA 4632417N05 [Ra... 233 8e-60
ref|XP_488079.2| PREDICTED: similar to NAD(P) dependent steroid ... 213 1e-53
ref|XP_219290.3| PREDICTED: similar to NAD(P) dependent steroid ... 210 9e-53
dbj|BAC42142.1| putative 3-beta-hydroxysteroid dehydrogenase [Ar... 190 7e-47
ref|NP_564502.1| 3-beta hydroxysteroid dehydrogenase/isomerase f... 190 7e-47
dbj|BAA14290.1| NAD(P)-dependent cholesterol dehydrogenase [Noca... 189 1e-46
gb|AAM62504.1| 3-beta-hydroxysteroid dehydrogenase, putative [Ar... 188 3e-46
gb|AAC14524.1| 3-beta-hydroxysteroid dehydrogenase [Arabidopsis ... 186 1e-45
ref|XP_640712.1| hypothetical protein DDB0205620 [Dictyostelium ... 186 1e-45
>gb|AAM13002.1| putative steroid dehydrogenase [Arabidopsis thaliana]
gi|2459443|gb|AAB80678.1| putative steroid dehydrogenase
[Arabidopsis thaliana] gi|16226673|gb|AAL16229.1|
At2g33630/F4P9.40 [Arabidopsis thaliana]
gi|25408295|pir||H84747 probable steroid dehydrogenase
[imported] - Arabidopsis thaliana
gi|15226138|ref|NP_180921.1| 3-beta hydroxysteroid
dehydrogenase/isomerase family protein [Arabidopsis
thaliana]
Length = 480
Score = 684 bits (1765), Expect = 0.0
Identities = 329/479 (68%), Positives = 398/479 (82%), Gaps = 2/479 (0%)
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVR 60
MHLSENEG+EG T VVTGGLGFVG+ALCLELVRRGA +VR+FDLR SSPW LK++GVR
Sbjct: 1 MHLSENEGVEGNTFVVTGGLGFVGAALCLELVRRGARQVRSFDLRHSSPWSDDLKNSGVR 60
Query: 61 CIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG 120
CIQGD+ +K+DV++AL GADCV HLA++GMSGKEML+ R D+VNI GTC+VL+A
Sbjct: 61 CIQGDVTKKQDVDNALDGADCVLHLASYGMSGKEMLRFGRCDEVNINGTCNVLEAAFKHE 120
Query: 121 IKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKND 180
I R+VY STYNVVF G+ I+NGNE LPYFP+D HVD Y R+KSIAEQLVLK+N RPFKN
Sbjct: 121 ITRIVYVSTYNVVFGGKEILNGNEGLPYFPLDDHVDAYSRTKSIAEQLVLKSNGRPFKN- 179
Query: 181 AGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALI 240
G +YTCA+RPAAIYGPGEDRHLPRI+T KLGL LF IG+ +VKSDW++VENLVLA+I
Sbjct: 180 GGKRMYTCAIRPAAIYGPGEDRHLPRIVTLTKLGLALFKIGEPSVKSDWIYVENLVLAII 239
Query: 241 LASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHA 300
LASMGLLDD G+ Q P+AAGQ YF+SDG PVNTFEFL+PLL+SL+Y+LPK +++V A
Sbjct: 240 LASMGLLDDIPGREGQ-PVAAGQPYFVSDGYPVNTFEFLRPLLKSLDYDLPKCTISVPFA 298
Query: 301 LVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSR 360
L LG+I QG YT+LYPWL + WLPQP +LP+EV+KVGVTHYFSYLKAKEE+GYVP SS+
Sbjct: 299 LSLGKIFQGFYTVLYPWLSKSWLPQPLVLPAEVYKVGVTHYFSYLKAKEELGYVPFKSSK 358
Query: 361 EGMASTISYWQQRKMITLDGPTIYTWLFCVIGMISLFCAAFLPDVGIVFLLRATSLFVFR 420
EGMA+TISYWQ+RK +LDGPT++TW+ IGM +LF A +LPD+G V LRA LF FR
Sbjct: 359 EGMAATISYWQERKRRSLDGPTMFTWIAVTIGMSALFAAGWLPDIGPVPFLRAIHLFFFR 418
Query: 421 SMWMIRLVFILATAAHVFEAIYAWYLAKRVDHANARGWFWQTFALGYFSLRFLLKRARK 479
++ +++ VFI+A HV E IYAW+LAKRVD NA GWF QT ALG+FS+RFLLKRA++
Sbjct: 419 TITIVKAVFIVAVVLHVAEGIYAWFLAKRVDPGNAMGWFLQTSALGFFSMRFLLKRAKE 477
>ref|XP_467464.1| putative NAD(P)-dependent cholesterol dehydrogenase [Oryza sativa
(japonica cultivar-group)] gi|42408030|dbj|BAD09166.1|
putative NAD(P)-dependent cholesterol dehydrogenase
[Oryza sativa (japonica cultivar-group)]
Length = 478
Score = 662 bits (1708), Expect = 0.0
Identities = 325/479 (67%), Positives = 383/479 (79%), Gaps = 1/479 (0%)
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVR 60
MHLSENEGIEG VTGG GFVGSALCLEL+RRGA EVR+ DLR SSPW L GVR
Sbjct: 1 MHLSENEGIEGVRFAVTGGQGFVGSALCLELLRRGAREVRSLDLRASSPWSDQLLGAGVR 60
Query: 61 CIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG 120
QGD+ +KEDV ALRG DCVFHLA++GMSGKEM+Q R D+VNI GTC+VLDAC G
Sbjct: 61 FFQGDVRKKEDVGKALRGVDCVFHLASYGMSGKEMVQAGRADEVNINGTCNVLDACHEHG 120
Query: 121 IKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKND 180
++RLVY STYNVVF G+ IVNGNE+LPYFP++ HVD Y RSKSIAEQLVLK+N R K+D
Sbjct: 121 VRRLVYVSTYNVVFGGEPIVNGNEALPYFPVEDHVDAYARSKSIAEQLVLKSNGRQTKSD 180
Query: 181 AGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALI 240
+ LYTC++RPAAIYGPGE+RHLPRI++ AKLGL F IGD VKSDWV+V+NLVLALI
Sbjct: 181 KSSRLYTCSIRPAAIYGPGEERHLPRILSLAKLGLAFFKIGDPNVKSDWVYVDNLVLALI 240
Query: 241 LASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHA 300
LASMGLLDD + K P+AAGQAYFI DGSPVNTFEFL PL +SL+Y +P+ + A
Sbjct: 241 LASMGLLDDIPDR-KGIPVAAGQAYFICDGSPVNTFEFLSPLFQSLDYTVPRVRMDTSVA 299
Query: 301 LVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSR 360
L + R +YT+LYPWLD W+PQP +LP+EV+KVGVTHYFSYLKA+EEIGYVPMVS R
Sbjct: 300 LAISRFFVFMYTLLYPWLDSKWIPQPLLLPAEVYKVGVTHYFSYLKAREEIGYVPMVSPR 359
Query: 361 EGMASTISYWQQRKMITLDGPTIYTWLFCVIGMISLFCAAFLPDVGIVFLLRATSLFVFR 420
EG+A+TISYWQ+RK LDGPTI+ WLF IGM++LF AA+LP VG + + LFVFR
Sbjct: 360 EGLAATISYWQERKRKELDGPTIFPWLFVTIGMLALFSAAYLPPVGPLKWVLDLHLFVFR 419
Query: 421 SMWMIRLVFILATAAHVFEAIYAWYLAKRVDHANARGWFWQTFALGYFSLRFLLKRARK 479
S +IRLVF++ATA HV EA+YAW+LAK+ D NA GWFWQTF LG+FSLR+LLKR R+
Sbjct: 420 SKLVIRLVFVIATALHVGEAVYAWFLAKKYDPRNATGWFWQTFMLGFFSLRYLLKRMRE 478
>ref|XP_523439.1| PREDICTED: similar to NAD(P) dependent steroid dehydrogenase-like
[Pan troglodytes]
Length = 630
Score = 281 bits (718), Expect = 4e-74
Identities = 165/344 (47%), Positives = 214/344 (61%), Gaps = 18/344 (5%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+T+++TGG G+ G L L ++G H + FD+ SSP I G++ IQGDI D
Sbjct: 246 ETVLITGGSGYFGFRLGCALNQKGVHVI-LFDI--SSPAQTI--PEGIKFIQGDIRHLSD 300
Query: 72 VESALRGAD--CVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCST 129
VE A + AD CVFH+A++GMSG+E L +++VN+ GT ++L C + RLVY ST
Sbjct: 301 VEKAFQDADVTCVFHIASYGMSGREQLNRNLIEEVNVRGTDNILQVCQRRSVPRLVYTST 360
Query: 130 YNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCA 189
+NV+F GQ I NG+ESLPY P+ H D Y R+KSIAEQ VL+ NA P G L TCA
Sbjct: 361 FNVIFGGQVIRNGDESLPYLPLHLHPDHYSRTKSIAEQKVLEANATPLDRGDG-VLRTCA 419
Query: 190 VRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDD 249
+RPA IYGPGE RHLPRI++ + GL F GD ++V V+NLV A ILAS L
Sbjct: 420 LRPAGIYGPGEQRHLPRIVSYIEKGLFKFVYGDPRSLVEFVHVDNLVQAHILASEAL--- 476
Query: 250 SAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQG 309
A KG IA+GQ YFISDG PVN FEF +PL+ L Y P T L + + +
Sbjct: 477 RADKGH---IASGQPYFISDGRPVNNFEFFRPLVEGLGYTFPSTRLPLTLVYCFAFLTEM 533
Query: 310 VYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGY 353
V+ I L R + QPF+ +EV+K GVTHYFS KAK+E+GY
Sbjct: 534 VHFI----LGRLYNFQPFLTRTEVYKTGVTHYFSLEKAKKELGY 573
>dbj|BAE00861.1| unnamed protein product [Macaca fascicularis]
Length = 393
Score = 278 bits (711), Expect = 3e-73
Identities = 164/344 (47%), Positives = 214/344 (61%), Gaps = 18/344 (5%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+T+++TGG G+ G L L ++G H + FD+ SSP I G++ IQGDI D
Sbjct: 9 ETVLITGGGGYFGFRLGCALNQKGVHVI-LFDI--SSPAETI--PEGIKFIQGDICHLSD 63
Query: 72 VESALRGAD--CVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCST 129
+E A + AD CVFH+A++GMSG+E L +++VNI GT ++L AC + RLVY ST
Sbjct: 64 IEKAFQDADITCVFHIASYGMSGREQLNRNLIEEVNIGGTDNILQACQRRRVPRLVYTST 123
Query: 130 YNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCA 189
+NV+F GQ I NG+ESLPY P+ H D Y R+KSIAE+ VL+ N P G L TCA
Sbjct: 124 FNVIFGGQVIRNGDESLPYLPLHLHPDHYSRTKSIAEKKVLEANGTPLDRGDG-VLRTCA 182
Query: 190 VRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDD 249
+RPA IYGPGE RHLPRI++ + GL F GD ++V V+NLV A ILAS L
Sbjct: 183 LRPAGIYGPGEQRHLPRIVSYIEKGLFKFVYGDPRSLVEFVHVDNLVQAHILASEAL--- 239
Query: 250 SAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQG 309
A KG IA+GQ YFISDG PVN FEF +PL+ L Y P T L + + +
Sbjct: 240 RADKGH---IASGQPYFISDGRPVNNFEFFRPLVEGLGYTFPSTRLPLTLVYCFAFLTEM 296
Query: 310 VYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGY 353
V+ I L R + QPF+ +EV+K GVTHYFS KAK+E+GY
Sbjct: 297 VHFI----LGRLYNFQPFLTRTEVYKTGVTHYFSLEKAKKELGY 336
>gb|AAH19670.1| NAD(P) dependent steroid dehydrogenase-like [Homo sapiens]
gi|21553311|ref|NP_660151.1| NAD(P) dependent steroid
dehydrogenase-like [Homo sapiens]
Length = 383
Score = 276 bits (707), Expect = 8e-73
Identities = 164/344 (47%), Positives = 212/344 (60%), Gaps = 18/344 (5%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+++++TGG G+ G L L + G H + FD+ SSP I G++ IQGDI D
Sbjct: 9 ESVLITGGSGYFGFRLGCALNQNGVHVI-LFDI--SSPAQTI--PEGIKFIQGDIRHLSD 63
Query: 72 VESALRGAD--CVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCST 129
VE A + AD CVFH+A++GMSG+E L + +VN+ GT ++L C + RLVY ST
Sbjct: 64 VEKAFQDADVTCVFHIASYGMSGREQLNRNLIKEVNVRGTDNILQVCQRRRVPRLVYTST 123
Query: 130 YNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCA 189
+NV+F GQ I NG+ESLPY P+ H D Y R+KSIAEQ VL+ NA P G L TCA
Sbjct: 124 FNVIFGGQVIRNGDESLPYLPLHLHPDHYSRTKSIAEQKVLEANATPLDRGDG-VLRTCA 182
Query: 190 VRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDD 249
+RPA IYGPGE RHLPRI++ + GL F GD ++V V+NLV A ILAS L
Sbjct: 183 LRPAGIYGPGEQRHLPRIVSYIEKGLFKFVYGDPRSLVEFVHVDNLVQAHILASEAL--- 239
Query: 250 SAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQG 309
A KG IA+GQ YFISDG PVN FEF +PL+ L Y P T L + + +
Sbjct: 240 RADKGH---IASGQPYFISDGRPVNNFEFFRPLVEGLGYTFPSTRLPLTLVYCFAFLTEM 296
Query: 310 VYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGY 353
V+ I L R + QPF+ +EV+K GVTHYFS KAK+E+GY
Sbjct: 297 VHFI----LGRLYNFQPFLTRTEVYKTGVTHYFSLEKAKKELGY 336
>ref|NP_083001.1| RIKEN cDNA 4632417N05 [Mus musculus] gi|24416567|gb|AAH38819.1|
4632417N05Rik protein [Mus musculus]
gi|12852533|dbj|BAB29446.1| unnamed protein product [Mus
musculus]
Length = 394
Score = 269 bits (688), Expect = 1e-70
Identities = 157/347 (45%), Positives = 215/347 (61%), Gaps = 19/347 (5%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+T+++TGG G+ G L L ++GA + FD+ + G++ + GDI D
Sbjct: 9 ETVLITGGGGYFGFRLGCALNQKGARVI-LFDITQPAQNLP----EGIKFVCGDIRCLAD 63
Query: 72 VESALRGAD---CVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCS 128
VE+A + A+ CVFH+A++GMSG+E L T++++VN+ GT ++L ACL G+ RLVY S
Sbjct: 64 VETAFQDAEKVACVFHVASYGMSGREQLNKTQIEEVNVGGTENILRACLERGVPRLVYTS 123
Query: 129 TYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTC 188
T+NV+F GQ I NG+ESLPY P+ H D Y R+KSIAE+ VL+ N FK G L TC
Sbjct: 124 TFNVIFGGQVIRNGDESLPYLPLHLHPDHYSRTKSIAEKKVLEANGLAFKQGDG-ILRTC 182
Query: 189 AVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLD 248
A+RPA IYG GE RHLPRI++ + GL F GD ++V V+NL A ILAS L
Sbjct: 183 AIRPAGIYGAGEQRHLPRIVSYIERGLFRFVYGDPQSLVEFVHVDNLAKAHILASEAL-- 240
Query: 249 DSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQ 308
A KG +A+GQ YFISDG PVN FEF +PL+ L Y P T L + L + +
Sbjct: 241 -KADKGH---VASGQPYFISDGRPVNNFEFFRPLVEGLGYTFPSTRLPLTLIYCLAFLVE 296
Query: 309 GVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVP 355
+ I + R + QPF+ +EV+K GVTHYFS KAK+E+G+ P
Sbjct: 297 MTHFI----VGRLYNFQPFLTRTEVYKTGVTHYFSLEKAKKELGFEP 339
>gb|AAX46671.1| NAD(P) dependent steroid dehydrogenase-like [Bos taurus]
Length = 399
Score = 269 bits (687), Expect = 2e-70
Identities = 157/344 (45%), Positives = 211/344 (60%), Gaps = 18/344 (5%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+T+++TGG G+ G L L G H + FD+ S P I G+R I GDI D
Sbjct: 9 ETVLITGGGGYFGFRLGCALNLLGVHVI-LFDI--SRPAQTI--PEGIRFILGDIRCLSD 63
Query: 72 VESALRGAD--CVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCST 129
+E+A +G D CVFH+A++GMSG+E L + ++++N+ GT ++L AC G+ RLVY ST
Sbjct: 64 IENAFQGVDVACVFHIASYGMSGREQLNRSLIEEINVGGTDNILQACRRRGVPRLVYTST 123
Query: 130 YNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCA 189
+NV+F GQ I NG+ESLPY P+ H D Y R+KSIAE+ VL N + G L TCA
Sbjct: 124 FNVIFGGQVIRNGDESLPYLPLHLHPDHYSRTKSIAEKKVLSANGTALER-GGGVLSTCA 182
Query: 190 VRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDD 249
+RPA IYGPGE RHLPRI++ + GL F GD ++V V+NLV A ILAS L
Sbjct: 183 LRPAGIYGPGEQRHLPRIVSYIEKGLFRFVYGDPKSLVEFVHVDNLVQAHILASEAL--- 239
Query: 250 SAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQG 309
A KG IAAGQ YFISDG PVN FEF +PL+ L Y+ P T L + + +
Sbjct: 240 KANKGH---IAAGQPYFISDGRPVNNFEFFRPLVEGLGYKFPSTRLPLTLIYCFAFLTEM 296
Query: 310 VYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGY 353
+ I L R + QPF+ +E +K G+THYF+ KA++E+GY
Sbjct: 297 THFI----LGRLYNFQPFLTRTEFYKTGITHYFTLEKARKELGY 336
>ref|XP_414167.1| PREDICTED: similar to NAD(P) dependent steroid dehydrogenase-like
[Gallus gallus]
Length = 391
Score = 265 bits (676), Expect = 3e-69
Identities = 150/344 (43%), Positives = 209/344 (60%), Gaps = 16/344 (4%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+T+++TGG G+ G L + ++G +V FD+ + P + G++ +QG++ +
Sbjct: 9 ETVLITGGGGYFGFRLGCTIYKKGV-DVILFDV--TKPLQPV--PEGIKFMQGNVCCLAE 63
Query: 72 VESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCSTYN 131
VE AL+ CVFH+A++GMSG+E L ++ VN+ GT +V+ AC G+ LVY STYN
Sbjct: 64 VEEALKDVICVFHIASYGMSGREQLNRKLIEDVNVKGTENVIQACKSTGVSSLVYTSTYN 123
Query: 132 VVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCAVR 191
V+F GQ I NG+ESLPY P+ H D Y R+KS+AE VLK N N G L TCA+R
Sbjct: 124 VIFGGQIIENGDESLPYLPLHLHPDHYSRTKSLAEMKVLKANGTELGNGKG-VLRTCALR 182
Query: 192 PAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDDSA 251
PA IYGPGE RHLPRI++ + GL F GD ++V V+NLV A ILA L
Sbjct: 183 PAGIYGPGEQRHLPRIVSYIERGLFKFVYGDPLSLVEFVHVDNLVQAHILAFEAL----- 237
Query: 252 GKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQGVY 311
K ++ IAAGQAYFISDG PVN FEF +PL+ L Y+ P L + + + V+
Sbjct: 238 -KANKKHIAAGQAYFISDGRPVNNFEFFRPLVEGLGYKFPTCRLPLSLVYFFAFLTEVVH 296
Query: 312 TILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVP 355
+ + + QP + +EV+K GVTHYFS KA++E+GY P
Sbjct: 297 FL----VGHVYNFQPLLTRTEVYKTGVTHYFSMEKARKELGYEP 336
>emb|CAF93262.1| unnamed protein product [Tetraodon nigroviridis]
Length = 376
Score = 263 bits (671), Expect = 1e-68
Identities = 150/347 (43%), Positives = 207/347 (59%), Gaps = 24/347 (6%)
Query: 13 TLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVR----CIQGDIVR 68
T V+TGG G+ G L L ++GA ++ FD IL + V +QGDI
Sbjct: 6 TFVITGGCGYFGYRLACSLHKKGA-KIILFD--------KILPEENVPEDILSVQGDIRD 56
Query: 69 KEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCS 128
DV A+ GA CVFH+A++GMSGKE L +++VN+ GT ++L AC+ + RLVY S
Sbjct: 57 YADVVKAVAGATCVFHIASYGMSGKEQLNKHMIEEVNVRGTQNILKACIEHKVPRLVYTS 116
Query: 129 TYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTC 188
T+NVVF GQ I NG+ESLPY P+ H D Y R+K++AE VLK N K+ +G +C
Sbjct: 117 TFNVVFGGQVIENGDESLPYLPLHLHPDHYSRTKALAEMAVLKANGTALKDGSG-VFRSC 175
Query: 189 AVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLD 248
A+RPA IYGPGE RHLPRI++ + G+ F G+ + ++V V+NLV A LA+ L
Sbjct: 176 ALRPAGIYGPGEKRHLPRIVSYIEKGIFSFVYGEPSSLVEFVHVDNLVSAHELAAKAL-- 233
Query: 249 DSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQ 308
++R AAGQ YFISDG PVN FEF +PL+ L Y PK L + + +
Sbjct: 234 ----TAERRYSAAGQPYFISDGRPVNNFEFFRPLVEGLGYSFPKLRLPISLVYFFAFLTE 289
Query: 309 GVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVP 355
+++ ++ + QP + +EV+K GVTHYFS KAK E+GY P
Sbjct: 290 ----MIHSFIRHIYNFQPLLTRTEVYKTGVTHYFSMAKAKAELGYEP 332
>ref|XP_546811.1| PREDICTED: similar to NAD(P) dependent steroid dehydrogenase-like
[Canis familiaris]
Length = 383
Score = 256 bits (654), Expect = 1e-66
Identities = 152/325 (46%), Positives = 201/325 (61%), Gaps = 18/325 (5%)
Query: 31 LVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKEDVESALRGAD--CVFHLAAF 88
L ++G H + FD+ SSP +I G+ I+GDI DVE A + AD CVFH+A++
Sbjct: 18 LNQKGFHVI-LFDI--SSPTHSI--PEGIEFIRGDIRHLSDVEKAFQDADVTCVFHIASY 72
Query: 89 GMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCSTYNVVFAGQRIVNGNESLPY 148
GMSG+E L + +++VN+ GT ++L C G+ RLVY ST+NV+F GQ I NG+ESLPY
Sbjct: 73 GMSGREQLNRSLIEEVNVGGTDNILQVCRRRGVPRLVYTSTFNVIFGGQVIRNGDESLPY 132
Query: 149 FPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCAVRPAAIYGPGEDRHLPRII 208
P+ H D Y R+KSIAE+ VL+ + G L TCA+RPA IYGPGE RHLPRI+
Sbjct: 133 LPLHLHPDHYSRTKSIAEKKVLEASGTTLLRSDG-VLRTCALRPAGIYGPGEQRHLPRIV 191
Query: 209 TTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDDSAGKGKQRPIAAGQAYFIS 268
+ + GL F GD ++V V+NLV A ILAS L A KG IA+GQ YFIS
Sbjct: 192 SYIERGLFKFVYGDPRSLVEFVHVDNLVQAHILASEAL---KADKGH---IASGQPYFIS 245
Query: 269 DGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQGVYTILYPWLDRWWLPQPFI 328
DG PVN FEF +PL+ L Y P + + + + V+ I L R + QPF+
Sbjct: 246 DGRPVNNFEFFRPLVEGLGYTFPSVRVPLTLIYCFAFLTEMVHFI----LGRLYNFQPFL 301
Query: 329 LPSEVHKVGVTHYFSYLKAKEEIGY 353
+EV+K GVTHYFS KAK+E+GY
Sbjct: 302 TRTEVYKTGVTHYFSLDKAKKELGY 326
>ref|XP_414919.1| PREDICTED: similar to NAD(P) dependent steroid dehydrogenase-like
[Gallus gallus]
Length = 454
Score = 243 bits (621), Expect = 7e-63
Identities = 145/342 (42%), Positives = 192/342 (55%), Gaps = 18/342 (5%)
Query: 15 VVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKEDVES 74
VVTGG G+ G L L GA V +D+ P + I NGV CIQ D+ + V +
Sbjct: 71 VVTGGGGYFGYKLGCALASSGASVV-LYDIH--KPIWEI--PNGVVCIQADVRDYDAVFA 125
Query: 75 ALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCSTYNVVF 134
A GADCVFH+A++GMSG+E L ++ VNI GT ++DAC I RL+Y ST NVVF
Sbjct: 126 ACEGADCVFHVASYGMSGREQLHREEIETVNINGTRFIIDACKQRNITRLIYTSTVNVVF 185
Query: 135 AGQRIVNGN-ESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCAVRPA 193
G I +G+ E++PYFPI+ HVD Y R+KSIAEQ+VL N P G LYT +RP
Sbjct: 186 GGLPIEDGDEETVPYFPIEKHVDHYSRTKSIAEQMVLAANGTPLA--GGGVLYTSVLRPP 243
Query: 194 AIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDDSAGK 253
IYGP E RHLPR+ + G+L F GD + K +WV VENLV A ILA+ L +
Sbjct: 244 GIYGPEEQRHLPRLAKNIERGILSFKFGDPSAKMNWVHVENLVQAQILAAEALTPE---- 299
Query: 254 GKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQGVYTI 313
+ IA+GQ YFI DG N FE+L PL L P + + + ++ I
Sbjct: 300 --KNYIASGQVYFIHDGEKFNLFEWLAPLFERLGCSKPWIPIPTSLVYASATVMEHLHLI 357
Query: 314 LYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVP 355
L P ++ P + +EV + TH F KA+ ++GY P
Sbjct: 358 LKPLVEL----SPLLTRNEVQNISTTHTFRIDKARRQLGYSP 395
>ref|XP_226509.3| PREDICTED: similar to RIKEN cDNA 4632417N05 [Rattus norvegicus]
Length = 561
Score = 233 bits (595), Expect = 8e-60
Identities = 133/282 (47%), Positives = 178/282 (62%), Gaps = 14/282 (4%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+T+++TGG G+ G L L ++G + FD+ + + G+ ++GDI D
Sbjct: 76 ETVLITGGGGYFGFRLGCALNQKGVRVI-LFDIIEPAQNLP----EGITFVRGDIRCLSD 130
Query: 72 VESALRGAD--CVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCST 129
VE+A + AD CVFH+A++GMSG+E L TR+++VN+ GT ++L ACL G+ LVY ST
Sbjct: 131 VEAAFQDADIACVFHIASYGMSGREQLNKTRIEEVNVGGTENILQACLGRGVPSLVYTST 190
Query: 130 YNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCA 189
+NV+F GQ I NG+ESLPY P+ H D Y R+KSIAE+ VL+ N FK G L TCA
Sbjct: 191 FNVIFGGQVIRNGDESLPYLPLHLHPDHYSRTKSIAEKKVLEANGLAFKQGDG-VLRTCA 249
Query: 190 VRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDD 249
+RPA IYG GE RHLPR+++ + GL F GD ++V V+NL A ILAS L
Sbjct: 250 IRPAGIYGAGEQRHLPRVVSYIERGLFRFVYGDPQSLVEFVHVDNLAKAHILASEAL--- 306
Query: 250 SAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELP 291
A KG IA+GQ YFISDG PVN FEF +PL+ L Y P
Sbjct: 307 KADKGH---IASGQPYFISDGRPVNNFEFFRPLVEGLGYTFP 345
>ref|XP_488079.2| PREDICTED: similar to NAD(P) dependent steroid dehydrogenase-like
[Mus musculus]
Length = 638
Score = 213 bits (542), Expect = 1e-53
Identities = 139/399 (34%), Positives = 198/399 (48%), Gaps = 68/399 (17%)
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKEDVE 73
++VTGG G++G +L L +RG V DLR P + + +G +Q D+ +E +
Sbjct: 130 VLVTGGGGYLGFSLGSSLAKRGT-SVILLDLR--RPQWPL--PSGTEFVQADVRDEEALY 184
Query: 74 SALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCSTYNVV 133
A +G DCVFH+A++GMSG E LQ ++ +N+ GT V++ C+ + RLVY ST NV
Sbjct: 185 QAFQGVDCVFHVASYGMSGAEKLQKQEIESINVGGTKLVINVCVRRRVPRLVYTSTVNVT 244
Query: 134 FAGQRIVNGN-ESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPF--------------- 177
F G+ I GN ES+PYFP+D H+D Y R+K+IA+QL L N P
Sbjct: 245 FGGKPIEQGNEESIPYFPLDKHMDHYSRTKAIADQLTLMANGTPLLEHLHEPLQPGRSLD 304
Query: 178 -----KNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFV 232
G L TC +RP IYGP E RHLPR+ + K L +F GD+ + +WV V
Sbjct: 305 CKLPCLQSGGGTLRTCVLRPPGIYGPEEQRHLPRVASHIKKRLFMFRFGDRKTRMNWVHV 364
Query: 233 ENLVLALILASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPK 292
+NLV A +LA+ GL + KG +A+GQAY+I+DG VN FE++ PL L Y P
Sbjct: 365 QNLVQAHMLAAEGL---TMAKGY---VASGQAYYINDGESVNLFEWMAPLFEKLGYSQPW 418
Query: 293 TSLAVDHALVLGRICQGVYTILYPWL-----------------------DRWWLPQ---- 325
+ + + + ++ L P +R PQ
Sbjct: 419 IQVPTSCVYLTAAVMEYLHLALRPICTIPPLLTRSEVIEEAAPSKKKISERKPPPQEGIR 478
Query: 326 ---------PFILPSEVHKVGVTHYFSYLKAKEEIGYVP 355
P V V VTH F KA+ ++GY P
Sbjct: 479 NQTQQLVFKPQCDGKAVLSVAVTHTFQIAKARTQLGYAP 517
>ref|XP_219290.3| PREDICTED: similar to NAD(P) dependent steroid dehydrogenase-like
[Rattus norvegicus]
Length = 642
Score = 210 bits (534), Expect = 9e-53
Identities = 139/395 (35%), Positives = 198/395 (49%), Gaps = 66/395 (16%)
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKEDVE 73
++VTGG G++G +L L ++GA V DLR P + + +G IQ D+ +E +
Sbjct: 130 VLVTGGGGYLGFSLGSSLAKKGA-SVILLDLR--RPQWPL--PSGTEFIQADVRDEEALY 184
Query: 74 SALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCSTYNVV 133
A +G DCVFH+A++GMSG E LQ ++ +N+ GT V++ C+ + RLVY ST NV
Sbjct: 185 QAFQGVDCVFHVASYGMSGAEKLQKREIESINVGGTKLVINVCVRRRVPRLVYTSTVNVT 244
Query: 134 FAGQRIVNGNE-SLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCAVRP 192
F G+ I GNE S+PYFP+D H+D Y R+K+IA+QL+L N P G L TC +RP
Sbjct: 245 FGGKPIEQGNEDSIPYFPLDKHMDHYSRTKAIADQLILMANGTPLL--GGGTLRTCVLRP 302
Query: 193 AAIYGPGEDRHLPRIITTA----------------------KLGLLLFTIGDQTVKSDWV 230
IYGP E +HLPR+ +A K L +F GD+ +WV
Sbjct: 303 PGIYGPEEQKHLPRVAPSAFIRSGPEMAVQSEWTTIIQSHIKKRLFMFRFGDRRTHMNWV 362
Query: 231 FVENLVLALILASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYEL 290
V+NLV A + A+ GL + KG +A+GQAY+I DG VN FE++ PL L Y
Sbjct: 363 HVQNLVQAHMRAAEGL---TLAKGY---VASGQAYYIHDGESVNLFEWMAPLFEKLGYSQ 416
Query: 291 PKTSLAVDHALV-----------LGRICQ-------------------GVYTILYPWLDR 320
P + + ++C V L+ L
Sbjct: 417 PWIQVPTSCVYLTVPAPTPWSHPTKKVCSAAALCWPPSKKASRRLASAAVMEFLHLALRP 476
Query: 321 WWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVP 355
P + SEV + VTH F KA+ ++GY P
Sbjct: 477 ICTISPLLTRSEVLSMTVTHTFQIAKARAQLGYAP 511
>dbj|BAC42142.1| putative 3-beta-hydroxysteroid dehydrogenase [Arabidopsis thaliana]
gi|28827710|gb|AAO50699.1| putative
3-beta-hydroxysteroid dehydrogenase [Arabidopsis
thaliana] gi|30694041|ref|NP_849779.1| 3-beta
hydroxysteroid dehydrogenase/isomerase family protein
[Arabidopsis thaliana] gi|25405196|pir||F96513
hypothetical protein T3F24.9 [imported] - Arabidopsis
thaliana gi|9993351|gb|AAG11424.1| Similar to steriod
dehydrogenase [Arabidopsis thaliana]
Length = 439
Score = 190 bits (483), Expect = 7e-47
Identities = 131/385 (34%), Positives = 196/385 (50%), Gaps = 36/385 (9%)
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDL---------RDSSPWF 51
M ++E E + VVTGG GF L LVR VR DL ++
Sbjct: 3 MEVTETE----RWCVVTGGRGFAARHLVEMLVRYQMFHVRIADLAPAIVLNPHEETGILG 58
Query: 52 AILKDNGVRCIQGDIVRKEDVESALRGADCVFHLAAFGMS-GKEMLQLTRVDQVNITGTC 110
++ V+ + D+ K V +GA+ VFH+AA S LQ + VN+ GT
Sbjct: 59 EAIRSGRVQYVSADLRNKTQVVKGFQGAEVVFHMAAPDSSINNHQLQYS----VNVQGTT 114
Query: 111 HVLDACLHLGIKRLVYCSTYNVVFAGQR-IVNGNESLPYFPIDHHVDPYGRSKSIAEQLV 169
+V+DAC+ +G+KRL+Y S+ +VVF G +N +ESLPY P H D Y +K+ E L+
Sbjct: 115 NVIDACIEVGVKRLIYTSSPSVVFDGVHGTLNADESLPYPP--KHNDSYSATKAEGEALI 172
Query: 170 LKNNARPFKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDW 229
LK N R + L TC +RP++I+GPG+ +P ++T A+ G F IGD + D+
Sbjct: 173 LKANGR-------SGLLTCCIRPSSIFGPGDKLMVPSLVTAARAGKSKFIIGDGSNFYDF 225
Query: 230 VFVENLVLALILASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYE 289
+VEN+V A + A L A G+ AAGQAYFI++ P+ +EF+ LL L YE
Sbjct: 226 TYVENVVHAHVCAERAL----ASGGEVCAKAAGQAYFITNMEPIKFWEFMSQLLEGLGYE 281
Query: 290 LPKTSLAVDHALVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKE 349
P + + + + + Y +L P + + P + PS V + F KAK+
Sbjct: 282 RPSIKIPASLMMPIAYLVELAYKLLGP----YGMKVPVLTPSRVRLLSCNRTFDSSKAKD 337
Query: 350 EIGYVPMVSSREGMASTISYWQQRK 374
+GY P+V +EG+ TI + K
Sbjct: 338 RLGYSPVVPLQEGIKRTIDSFSHLK 362
>ref|NP_564502.1| 3-beta hydroxysteroid dehydrogenase/isomerase family protein
[Arabidopsis thaliana]
Length = 382
Score = 190 bits (483), Expect = 7e-47
Identities = 131/385 (34%), Positives = 196/385 (50%), Gaps = 36/385 (9%)
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDL---------RDSSPWF 51
M ++E E + VVTGG GF L LVR VR DL ++
Sbjct: 3 MEVTETE----RWCVVTGGRGFAARHLVEMLVRYQMFHVRIADLAPAIVLNPHEETGILG 58
Query: 52 AILKDNGVRCIQGDIVRKEDVESALRGADCVFHLAAFGMS-GKEMLQLTRVDQVNITGTC 110
++ V+ + D+ K V +GA+ VFH+AA S LQ + VN+ GT
Sbjct: 59 EAIRSGRVQYVSADLRNKTQVVKGFQGAEVVFHMAAPDSSINNHQLQYS----VNVQGTT 114
Query: 111 HVLDACLHLGIKRLVYCSTYNVVFAGQR-IVNGNESLPYFPIDHHVDPYGRSKSIAEQLV 169
+V+DAC+ +G+KRL+Y S+ +VVF G +N +ESLPY P H D Y +K+ E L+
Sbjct: 115 NVIDACIEVGVKRLIYTSSPSVVFDGVHGTLNADESLPYPP--KHNDSYSATKAEGEALI 172
Query: 170 LKNNARPFKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDW 229
LK N R + L TC +RP++I+GPG+ +P ++T A+ G F IGD + D+
Sbjct: 173 LKANGR-------SGLLTCCIRPSSIFGPGDKLMVPSLVTAARAGKSKFIIGDGSNFYDF 225
Query: 230 VFVENLVLALILASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYE 289
+VEN+V A + A L A G+ AAGQAYFI++ P+ +EF+ LL L YE
Sbjct: 226 TYVENVVHAHVCAERAL----ASGGEVCAKAAGQAYFITNMEPIKFWEFMSQLLEGLGYE 281
Query: 290 LPKTSLAVDHALVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKE 349
P + + + + + Y +L P + + P + PS V + F KAK+
Sbjct: 282 RPSIKIPASLMMPIAYLVELAYKLLGP----YGMKVPVLTPSRVRLLSCNRTFDSSKAKD 337
Query: 350 EIGYVPMVSSREGMASTISYWQQRK 374
+GY P+V +EG+ TI + K
Sbjct: 338 RLGYSPVVPLQEGIKRTIDSFSHLK 362
>dbj|BAA14290.1| NAD(P)-dependent cholesterol dehydrogenase [Nocardia sp.]
gi|477637|pir||A49781 cholesterol dehydrogenase (EC
1.1.1.-) - Nocardia sp. (strain Ch 2-1)
Length = 364
Score = 189 bits (481), Expect = 1e-46
Identities = 131/363 (36%), Positives = 198/363 (54%), Gaps = 37/363 (10%)
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDN-GVRCIQGDIVRKEDV 72
++VTGG GFVG+ L EL+ RG + VR+FD R SP L D+ G+ I+GDI KE V
Sbjct: 13 VLVTGGSGFVGANLVTELLDRG-YAVRSFD-RAPSP----LGDHAGLEVIEGDICDKETV 66
Query: 73 ESALRGADCVFHLAAFG--MSGKEMLQLTRVDQ--VNITGTCHVLDACLHLGIKRLVYCS 128
+A++ D V H AA M G + + R VN+ GT +++ A G+KR VY +
Sbjct: 67 AAAVKDIDTVIHTAAIIDLMGGASVTEAYRQRSFAVNVEGTKNLVHASQEAGVKRFVYTA 126
Query: 129 TYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTC 188
+ +VV GQ IVNG+E++PY D Y +K +AE+ VL N K+D + TC
Sbjct: 127 SNSVVMGGQDIVNGDETMPY--TTRFNDLYTETKVVAEKFVLAENG---KHD----MLTC 177
Query: 189 AVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLD 248
A+RP+ I+G G+ ++ G + +G++ +K D +V NL+ ILA L+
Sbjct: 178 AIRPSGIWGRGDQTMFRKVFENVLAGHVKVLVGNKNIKLDNSYVHNLIHGFILAGQDLVP 237
Query: 249 DSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQ 308
A GQAYFI+DG P+N FEF +P+L + LP V GR+
Sbjct: 238 GGT--------APGQAYFINDGEPINMFEFARPVLAACGRPLPT-------FYVSGRLVH 282
Query: 309 GVYTILYPWLD-RWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSREGMASTI 367
V + + WL ++ LP+P I P V ++ + +YFS KAK ++GY P+ ++ + MA +
Sbjct: 283 KV-MMAWQWLHFKFALPEPLIEPLAVERLYLNNYFSIAKAKRDLGYEPLFTTEQAMAECM 341
Query: 368 SYW 370
Y+
Sbjct: 342 PYY 344
>gb|AAM62504.1| 3-beta-hydroxysteroid dehydrogenase, putative [Arabidopsis
thaliana]
Length = 380
Score = 188 bits (478), Expect = 3e-46
Identities = 130/385 (33%), Positives = 195/385 (49%), Gaps = 36/385 (9%)
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDL---------RDSSPWF 51
M ++E E + VVTGG GF L LVR VR DL ++
Sbjct: 1 MEVTETE----RWCVVTGGRGFAARHLVEMLVRYQMFHVRIADLAPAIVLDPHEETGILG 56
Query: 52 AILKDNGVRCIQGDIVRKEDVESALRGADCVFHLAAFGMS-GKEMLQLTRVDQVNITGTC 110
++ V+ + D+ K V +GA+ VFH+AA S LQ + VN+ GT
Sbjct: 57 EAIRSGRVQYVSADLRNKTQVVKGFQGAEVVFHMAAPDSSINNHQLQYS----VNVQGTT 112
Query: 111 HVLDACLHLGIKRLVYCSTYNVVFAGQR-IVNGNESLPYFPIDHHVDPYGRSKSIAEQLV 169
+V+DAC+ +G+KRL+Y S+ +VVF G +N +ESLPY P H D Y +K+ E L+
Sbjct: 113 NVIDACIEVGVKRLIYTSSPSVVFDGVHGTLNADESLPYPP--KHNDSYSATKAEGEALI 170
Query: 170 LKNNARPFKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDW 229
LK N R + L TC +RP++I+GPG+ +P ++T A+ G F IGD + D+
Sbjct: 171 LKANGR-------SGLLTCCIRPSSIFGPGDKLMVPSLVTAARAGKSKFIIGDGSNFYDF 223
Query: 230 VFVENLVLALILASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYE 289
+ EN+V A + A L A G+ AAGQAYFI++ P+ +EF+ LL L YE
Sbjct: 224 TYFENVVHAHVCAERAL----ASGGEVCAKAAGQAYFITNMEPIKFWEFMSQLLEGLGYE 279
Query: 290 LPKTSLAVDHALVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKE 349
P + + + + + Y +L P + + P + PS V + F KAK+
Sbjct: 280 RPSIKIPASLMMPIAYLVELAYKLLGP----YGMKVPVLTPSRVRLLSCNRTFDSSKAKD 335
Query: 350 EIGYVPMVSSREGMASTISYWQQRK 374
+GY P+V +EG+ TI + K
Sbjct: 336 RLGYSPVVPLQEGIKRTIDSFSHLK 360
>gb|AAC14524.1| 3-beta-hydroxysteroid dehydrogenase [Arabidopsis thaliana]
gi|25407832|pir||C84658 3-beta-hydroxysteroid
dehydrogenase [imported] - Arabidopsis thaliana
Length = 390
Score = 186 bits (472), Expect = 1e-45
Identities = 132/365 (36%), Positives = 190/365 (51%), Gaps = 34/365 (9%)
Query: 15 VVTGGLGFVGSALCLELVRRGAHEVRAFDLR-----DSSPWFAILKDNGVRC-----IQG 64
VVTGG GF L LVR VR DL D +L D G+R I
Sbjct: 13 VVTGGRGFAARHLVEMLVRYEMFCVRIADLAPAIMLDPQEGNGVL-DEGLRSGRVQYISA 71
Query: 65 DIVRKEDVESALRGADCVFHLAAFGMS-GKEMLQLTRVDQVNITGTCHVLDACLHLGIKR 123
D+ K V A +GA+ VFH+AA S LQ + VN+ GT +V+DAC+ +G+KR
Sbjct: 72 DLRDKSQVVKAFQGAEVVFHMAAPDSSINNHQLQYS----VNVQGTQNVIDACVDVGVKR 127
Query: 124 LVYCSTYNVVFAGQR-IVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAG 182
L+Y S+ +VVF G I+NG ES+ Y PI H+ D Y +K+ E+L++K N R
Sbjct: 128 LIYTSSPSVVFDGVHGILNGTESMAY-PIKHN-DSYSATKAEGEELIMKANGR------- 178
Query: 183 NCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILA 242
N L TC +RP++I+GPG+ +P ++ A+ G F IGD D+ +VEN+ A + A
Sbjct: 179 NGLLTCCIRPSSIFGPGDRLLVPSLVAAARAGKSKFIIGDGNNLYDFTYVENVAHAHVCA 238
Query: 243 SMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALV 302
L A G AAGQAYFI++ P+ +EF+ LL L YE P + +
Sbjct: 239 ERAL----ASGGDVSTKAAGQAYFITNMEPIKFWEFMSQLLDGLGYERPSIKIPAFIMMP 294
Query: 303 LGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSREG 362
+ + + Y +L P + + P + PS V + + F KAK+ +GY P+V +EG
Sbjct: 295 IAHLVELTYKVLGP----YGMTVPQLTPSRVRLLSCSRTFDSTKAKDRLGYAPVVPLQEG 350
Query: 363 MASTI 367
+ TI
Sbjct: 351 IRRTI 355
>ref|XP_640712.1| hypothetical protein DDB0205620 [Dictyostelium discoideum]
gi|60468723|gb|EAL66725.1| hypothetical protein
DDB0205620 [Dictyostelium discoideum]
Length = 349
Score = 186 bits (472), Expect = 1e-45
Identities = 125/364 (34%), Positives = 193/364 (52%), Gaps = 36/364 (9%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
K+ +V GG GF+G + L+ RG V FD+R S +D+ V GDI + ED
Sbjct: 3 KSYLVVGGCGFLGRYIVESLLARGEKNVHVFDIRKS------FEDDRVTFHIGDIRKTED 56
Query: 72 VESALRGADCVFHLAA--FGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCST 129
+ESA +G VFH A+ GM G ++ VN+ GT +++AC+ G+K+LVY S+
Sbjct: 57 LESACKGITTVFHTASPTHGM-GYDIYY-----SVNVIGTERLIEACIKCGVKQLVYTSS 110
Query: 130 YNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCA 189
+VVF G+ IVNG+E+LPY +D H+DPY ++K + E+ VLK G+ L CA
Sbjct: 111 SSVVFNGKDIVNGDETLPY--VDKHIDPYNKTKELGERAVLKAK--------GSNLLVCA 160
Query: 190 VRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDD 249
+RPA I+GP E + P+ + AK G F GD DW +++N+V A ILA+ + +
Sbjct: 161 LRPAGIFGPREVQGWPQFLKAAKEGKNKFMFGDGNNLCDWTYIDNVVHAHILAADNMTTN 220
Query: 250 SAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQG 309
S PI +G YFI++ P+ ++ + YE PK + + +
Sbjct: 221 S-------PI-SGSVYFITNDEPIPFWDMPIFAYEAFGYERPKMKIPFTIMYCIAWMIDL 272
Query: 310 VYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSREGMASTISY 369
+ +L P++ P I + T YF+ KAK+E+ Y P+VS R+GM T +
Sbjct: 273 ITLLLSPFVKL----HPTISLFRIIYTNSTRYFNIEKAKKELKYKPIVSLRDGMEKTKEW 328
Query: 370 WQQR 373
+ Q+
Sbjct: 329 FLQQ 332
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 822,506,303
Number of Sequences: 2540612
Number of extensions: 35051683
Number of successful extensions: 107936
Number of sequences better than 10.0: 2625
Number of HSP's better than 10.0 without gapping: 315
Number of HSP's successfully gapped in prelim test: 2310
Number of HSP's that attempted gapping in prelim test: 104593
Number of HSP's gapped (non-prelim): 3084
length of query: 479
length of database: 863,360,394
effective HSP length: 132
effective length of query: 347
effective length of database: 527,999,610
effective search space: 183215864670
effective search space used: 183215864670
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0335a.3


