

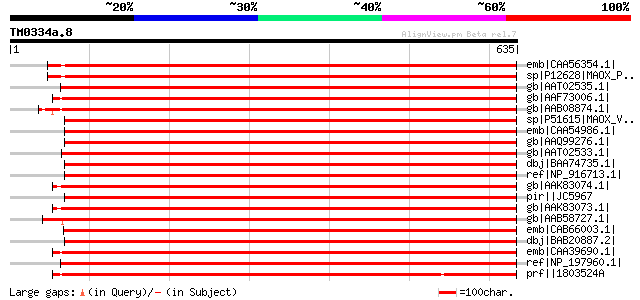
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0334a.8
(635 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA56354.1| NADP dependent malic enzyme [Phaseolus vulgaris] 1064 0.0
sp|P12628|MAOX_PHAVU NADP-dependent malic enzyme (NADP-ME) gi|16... 1053 0.0
gb|AAT02535.1| NADP-dependent malic enzyme 3 [Hydrilla verticill... 991 0.0
gb|AAF73006.1| NADP-dependent malic protein [Ricinus communis] 986 0.0
gb|AAB08874.1| malate dehydrogenase [Vitis vinifera] 978 0.0
sp|P51615|MAOX_VITVI NADP-dependent malic enzyme (NADP-ME) gi|51... 974 0.0
emb|CAA54986.1| malate dehydrogenase (oxaloacetate decarboxylati... 972 0.0
gb|AAQ99276.1| NADP malic enzyme [Oryza sativa (japonica cultiva... 971 0.0
gb|AAT02533.1| NADP-dependent malic enzyme 1 [Hydrilla verticill... 970 0.0
dbj|BAA74735.1| NADP-malic enzyme [Aloe arborescens] 969 0.0
ref|NP_916713.1| P0022F10.12 [Oryza sativa (japonica cultivar-gr... 967 0.0
gb|AAK83074.1| putative cytosolic NADP-malic enzyme [Flaveria pr... 966 0.0
pir||JC5967 malate dehydrogenase (oxaloacetate-decarboxylating) ... 966 0.0
gb|AAK83073.1| putative cytosolic NADP-malic enzyme [Flaveria pr... 966 0.0
gb|AAB58727.1| NADP-malic enzyme [Lycopersicon esculentum] gi|74... 964 0.0
emb|CAB66003.1| NADP-dependent malate dehydrogenase (decarboxyla... 962 0.0
dbj|BAB20887.2| NADP dependent malic enzyme [Oryza sativa (japon... 962 0.0
emb|CAA39690.1| malic enzyme [Populus trichocarpa] gi|1346485|sp... 957 0.0
ref|NP_197960.1| malate oxidoreductase, putative [Arabidopsis th... 954 0.0
prf||1803524A malic enzyme 952 0.0
>emb|CAA56354.1| NADP dependent malic enzyme [Phaseolus vulgaris]
Length = 589
Score = 1064 bits (2751), Expect = 0.0
Identities = 528/587 (89%), Positives = 559/587 (94%), Gaps = 3/587 (0%)
Query: 48 AMSRNGGEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNK 107
++ NGGE V +D+ N GGVRDLYGEDSATED LITPWTFSVASG +LLRDPRYNK
Sbjct: 5 SLKENGGEVSVNKDYSNG---GGVRDLYGEDSATEDHLITPWTFSVASGCSLLRDPRYNK 61
Query: 108 GLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLF 167
GLAFTE ERDAHYLRGLLPP+VF QELQEKR MHN+RQYEVPLH+YMA+MDLQERNERLF
Sbjct: 62 GLAFTEGERDAHYLRGLLPPSVFNQELQEKRLMHNLRQYEVPLHRYMALMDLQERNERLF 121
Query: 168 YKILIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQ 227
YK+LIDNVEELLP+VYTP VGEACQKYGSI++RPQGLYISLKEKGKILEVLKNWPEK+IQ
Sbjct: 122 YKLLIDNVEELLPVVYTPTVGEACQKYGSIFRRPQGLYISLKEKGKILEVLKNWPEKSIQ 181
Query: 228 VIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDE 287
VIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNE LLNDE
Sbjct: 182 VIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNEKLLNDE 241
Query: 288 FYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVF 347
FY GLRQ+RATG+EY ELLDEFMRAVKQNYGEKVL+QFEDFANHNAF+LL++YSSSHLVF
Sbjct: 242 FYIGLRQRRATGQEYAELLDEFMRAVKQNYGEKVLVQFEDFANHNAFDLLEKYSSSHLVF 301
Query: 348 NDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVE 407
NDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALE+SKQTKAPVE
Sbjct: 302 NDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEVSKQTKAPVE 361
Query: 408 ETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGK 467
ETRKK+WLVDSKGLIV SRLESLQ FKKPWAHEHEPVK LL+AVKAIKPTVLIGSSG GK
Sbjct: 362 ETRKKIWLVDSKGLIVSSRLESLQQFKKPWAHEHEPVKGLLEAVKAIKPTVLIGSSGAGK 421
Query: 468 TFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGK 527
TFTKEVVE MASLNEKPLILALSNPTSQSECTAEEAYTWSKG+AIFASGSPFDPV+Y GK
Sbjct: 422 TFTKEVVETMASLNEKPLILALSNPTSQSECTAEEAYTWSKGRAIFASGSPFDPVEYEGK 481
Query: 528 VYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTN 587
++ PGQANNAYIFPGFGLGLI+SGAIRVRDEMLLAASEALAAQVS+ENYDKGLIYPPFTN
Sbjct: 482 LFVPGQANNAYIFPGFGLGLIMSGAIRVRDEMLLAASEALAAQVSEENYDKGLIYPPFTN 541
Query: 588 IRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
IRKISA+IAA VAAK Y+LGLASHLPRPKDLVKYAESCMYSPGYRSY
Sbjct: 542 IRKISANIAAKVAAKAYDLGLASHLPRPKDLVKYAESCMYSPGYRSY 588
>sp|P12628|MAOX_PHAVU NADP-dependent malic enzyme (NADP-ME) gi|169327|gb|AAA19575.1|
NADP-dependent malic enzyme
Length = 589
Score = 1053 bits (2722), Expect = 0.0
Identities = 523/587 (89%), Positives = 555/587 (94%), Gaps = 3/587 (0%)
Query: 48 AMSRNGGEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNK 107
++ NGGE V +D+ N GGVRDLYGEDSATED LITPWTFSVASG +LLRDPRYNK
Sbjct: 5 SLKENGGEVSVKKDYSNG---GGVRDLYGEDSATEDHLITPWTFSVASGCSLLRDPRYNK 61
Query: 108 GLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLF 167
GLAFTE ERDAHYLRGLLPP+VF QELQEKR MHN+RQYEVPLH+YMA+MDLQERNERLF
Sbjct: 62 GLAFTEGERDAHYLRGLLPPSVFNQELQEKRLMHNLRQYEVPLHRYMALMDLQERNERLF 121
Query: 168 YKILIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQ 227
YK+LIDNV ELLP+VYTP VGEACQKYGSI++RPQGLYISLKEKGKILEVLKNWPEK+IQ
Sbjct: 122 YKLLIDNVAELLPVVYTPTVGEACQKYGSIFRRPQGLYISLKEKGKILEVLKNWPEKSIQ 181
Query: 228 VIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDE 287
VIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNE LLNDE
Sbjct: 182 VIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNEKLLNDE 241
Query: 288 FYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVF 347
FY GLRQ+RATG+EY LDEFMRAVKQNYGEKVL+QFEDFANHNAF+LL++YSSSHLVF
Sbjct: 242 FYIGLRQRRATGQEYATFLDEFMRAVKQNYGEKVLVQFEDFANHNAFDLLEKYSSSHLVF 301
Query: 348 NDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVE 407
NDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIA+E+SKQTKAPVE
Sbjct: 302 NDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIAVEVSKQTKAPVE 361
Query: 408 ETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGK 467
ETRKK+WLVDSKGLIV SRLESLQ FKKPWAHEHEPVK LL+AVKAIKPTVLIGSSG GK
Sbjct: 362 ETRKKIWLVDSKGLIVSSRLESLQQFKKPWAHEHEPVKGLLEAVKAIKPTVLIGSSGAGK 421
Query: 468 TFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGK 527
TFTKEVVE MASLNEKPLILALSNPTSQSECTAEEAYTWSKG+AIFASGSPFDPV+Y GK
Sbjct: 422 TFTKEVVETMASLNEKPLILALSNPTSQSECTAEEAYTWSKGRAIFASGSPFDPVEYEGK 481
Query: 528 VYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTN 587
++ PGQANNAYIFPGFGLGLI+SGAIRVRDEMLLAASEALAAQVS+ENYDKGLIYPPFTN
Sbjct: 482 LFVPGQANNAYIFPGFGLGLIMSGAIRVRDEMLLAASEALAAQVSEENYDKGLIYPPFTN 541
Query: 588 IRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
IRKISA+IAA VAAK Y+LGLASHL RPKDLVKYAESCMYSPGYRSY
Sbjct: 542 IRKISANIAAKVAAKAYDLGLASHLKRPKDLVKYAESCMYSPGYRSY 588
>gb|AAT02535.1| NADP-dependent malic enzyme 3 [Hydrilla verticillata]
Length = 575
Score = 991 bits (2563), Expect = 0.0
Identities = 483/571 (84%), Positives = 531/571 (92%)
Query: 64 NSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRG 123
NS GGVRD YGED ATE+QL+TPWTFSVASG TLLRDPR+NKGLAFTEKERDAHYLRG
Sbjct: 4 NSNACGGVRDPYGEDCATEEQLVTPWTFSVASGYTLLRDPRHNKGLAFTEKERDAHYLRG 63
Query: 124 LLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVY 183
LLPP TQE QEK+ MHN+R Y+VPL +YMAMMDLQERN+RLFYK+LIDNVEELLP+VY
Sbjct: 64 LLPPVCITQERQEKKIMHNLRNYQVPLQRYMAMMDLQERNQRLFYKLLIDNVEELLPVVY 123
Query: 184 TPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDL 243
TP VGEACQKYG IY+RPQGLYISLKEKGKILEVLKNWPEK IQVIVVTDGERILGLGDL
Sbjct: 124 TPTVGEACQKYGCIYRRPQGLYISLKEKGKILEVLKNWPEKNIQVIVVTDGERILGLGDL 183
Query: 244 GCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYE 303
G QGMGIPVGKL+LYTALGG+RPSSCLP+TIDVGTNNETLLN+EFY GLRQKRATGKEY
Sbjct: 184 GTQGMGIPVGKLALYTALGGIRPSSCLPITIDVGTNNETLLNEEFYIGLRQKRATGKEYA 243
Query: 304 ELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLL 363
+LL EFM +VKQNYGE+VL+QFEDFANHNAF LL +YS +HLVFNDDIQGTASVVLAG++
Sbjct: 244 DLLHEFMISVKQNYGERVLVQFEDFANHNAFELLSKYSKTHLVFNDDIQGTASVVLAGVV 303
Query: 364 ASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIV 423
A+LKLVGG LA+HTFLFLGAGEAGTGIAELIALEISKQ KAPVEETRKK+WLVDSKGLIV
Sbjct: 304 AALKLVGGALAEHTFLFLGAGEAGTGIAELIALEISKQIKAPVEETRKKIWLVDSKGLIV 363
Query: 424 RSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEK 483
SR ESLQHFKKPWAHEHEP+K L+DAVK IKPTVL+GSSGVG+ FTKEV+EAMAS NEK
Sbjct: 364 SSRKESLQHFKKPWAHEHEPIKHLVDAVKTIKPTVLVGSSGVGRAFTKEVIEAMASFNEK 423
Query: 484 PLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGF 543
P+ILALSNPTSQSECTAEEAYTW++G+AIFASGSPFDPV+Y GK + PGQANNAYIFPGF
Sbjct: 424 PIILALSNPTSQSECTAEEAYTWTQGRAIFASGSPFDPVEYEGKTFVPGQANNAYIFPGF 483
Query: 544 GLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKV 603
GLGLII+GAIRV D+ML+AASEALA QVS+EN++KGLIYPPF+NIRKISA+IAA+VAAK
Sbjct: 484 GLGLIIAGAIRVHDDMLIAASEALANQVSEENFEKGLIYPPFSNIRKISANIAANVAAKA 543
Query: 604 YELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
YELGLASHLPRP +LVKYAESCMYSP YR+Y
Sbjct: 544 YELGLASHLPRPANLVKYAESCMYSPVYRTY 574
>gb|AAF73006.1| NADP-dependent malic protein [Ricinus communis]
Length = 641
Score = 986 bits (2549), Expect = 0.0
Identities = 487/581 (83%), Positives = 536/581 (91%), Gaps = 1/581 (0%)
Query: 54 GEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTE 113
G V+ D K S +GGVRD+YGED+ATEDQ +TPW+ SVASG +LLRDP +NKGLAF +
Sbjct: 61 GASVLDLDPK-STVAGGVRDVYGEDTATEDQFVTPWSLSVASGYSLLRDPHHNKGLAFND 119
Query: 114 KERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILID 173
KERDAHYLRGLLPPA+ +QELQ K+ MH IRQY++PL KYMAMMDLQERNERLFYK+LI
Sbjct: 120 KERDAHYLRGLLPPAIVSQELQVKKMMHIIRQYQLPLQKYMAMMDLQERNERLFYKLLIQ 179
Query: 174 NVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTD 233
NVEE+LPIVYTP VGEACQKYGSI+ RPQGLYISLKEKG+ILEVL+NWPEK IQVIVVTD
Sbjct: 180 NVEEMLPIVYTPTVGEACQKYGSIFGRPQGLYISLKEKGRILEVLRNWPEKNIQVIVVTD 239
Query: 234 GERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLR 293
GERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNE LLNDEFY GLR
Sbjct: 240 GERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNEKLLNDEFYIGLR 299
Query: 294 QKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQG 353
Q+RATG+EY ELL EFM AVKQNYGE+VL+QFEDFANHNAF+LL +Y ++HLVFNDDIQG
Sbjct: 300 QRRATGQEYAELLHEFMTAVKQNYGERVLVQFEDFANHNAFDLLAKYGTTHLVFNDDIQG 359
Query: 354 TASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKV 413
TASVVLAGL+A+LKLVGG+LADH FLFLGAGEAGTGIAELIALE+SKQT PVEETRKK+
Sbjct: 360 TASVVLAGLVAALKLVGGSLADHRFLFLGAGEAGTGIAELIALEMSKQTNMPVEETRKKI 419
Query: 414 WLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEV 473
WLVDSKGLIV SR++SLQHFK+PWAHEHEP+K LLDAV IKPTVLIG+SGVG+TFTKEV
Sbjct: 420 WLVDSKGLIVSSRMDSLQHFKRPWAHEHEPIKTLLDAVNDIKPTVLIGTSGVGRTFTKEV 479
Query: 474 VEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQ 533
VEAMAS NEKP+ILALSNPTSQSECTAEEAYTWS+G+AIFASGSPF PV+Y GKVY PGQ
Sbjct: 480 VEAMASFNEKPIILALSNPTSQSECTAEEAYTWSQGRAIFASGSPFAPVEYEGKVYVPGQ 539
Query: 534 ANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISA 593
ANNAYIFPGFGLGLI+SG IRV D+MLLAASEALAAQV+QEN+DKGLIYPPFTNIRKISA
Sbjct: 540 ANNAYIFPGFGLGLIMSGTIRVHDDMLLAASEALAAQVTQENFDKGLIYPPFTNIRKISA 599
Query: 594 HIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
+IAA+VAAK YELGLAS LP+PKDLVKYAESCMYSP YRSY
Sbjct: 600 NIAANVAAKAYELGLASRLPQPKDLVKYAESCMYSPAYRSY 640
>gb|AAB08874.1| malate dehydrogenase [Vitis vinifera]
Length = 640
Score = 978 bits (2527), Expect = 0.0
Identities = 485/604 (80%), Positives = 548/604 (90%), Gaps = 9/604 (1%)
Query: 37 GSCGEPKLNIAAMSRN------GGEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWT 90
G GE LN++A+ + G V+ D K S SGGVRD+ GED+ATEDQ++TPWT
Sbjct: 39 GRAGE--LNVSAVMESTLKELRDGNTVLEVDSK-SAVSGGVRDVQGEDAATEDQIVTPWT 95
Query: 91 FSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPL 150
+VASG +LLR+P +NKGLAFTEKERD HYLRGLLPPAV +Q+LQ K+ M NIRQY VPL
Sbjct: 96 ITVASGYSLLRNPHHNKGLAFTEKERDHHYLRGLLPPAVVSQDLQVKKLMANIRQYTVPL 155
Query: 151 HKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKE 210
KYMAMMDLQERNE+LFYK+L+DNVEELLP+VYTP VGEACQKYG I +RPQGL+ISL E
Sbjct: 156 QKYMAMMDLQERNEKLFYKLLMDNVEELLPVVYTPTVGEACQKYGGILRRPQGLFISLNE 215
Query: 211 KGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCL 270
KGKILEVLKNWPEK IQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPS+CL
Sbjct: 216 KGKILEVLKNWPEKNIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSACL 275
Query: 271 PVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFAN 330
PVTIDVGTNN+ LL+DEFY GLRQKRATG+EY EL+ EFM AVKQNYGEKVL+QFEDFAN
Sbjct: 276 PVTIDVGTNNQKLLDDEFYIGLRQKRATGQEYAELIHEFMCAVKQNYGEKVLVQFEDFAN 335
Query: 331 HNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGI 390
HNAF+LL RY ++HLVFNDDIQGTASVVLAGL+++L LVGGTLA+HTFLFLGAGEAGTGI
Sbjct: 336 HNAFDLLARYGTTHLVFNDDIQGTASVVLAGLISALNLVGGTLAEHTFLFLGAGEAGTGI 395
Query: 391 AELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDA 450
AELIALE+SKQT+AP+EETRKK+WLVDSKGLIV SR++SLQHFKKPWAHEHEP+K L+DA
Sbjct: 396 AELIALEMSKQTQAPLEETRKKIWLVDSKGLIVSSRMDSLQHFKKPWAHEHEPIKNLVDA 455
Query: 451 VKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGK 510
VKAIKPTVLIGSSGVG+TFTKEVVEAMAS NEKP+ILALSNPTSQSECTAEEAYTWS+G+
Sbjct: 456 VKAIKPTVLIGSSGVGRTFTKEVVEAMASFNEKPIILALSNPTSQSECTAEEAYTWSEGR 515
Query: 511 AIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQ 570
AIFASGSPFDPV+Y GKV+ PGQ+NNAYIFPGFGLGLIISGAIRV D+MLLAASEALA Q
Sbjct: 516 AIFASGSPFDPVEYNGKVFVPGQSNNAYIFPGFGLGLIISGAIRVHDDMLLAASEALAKQ 575
Query: 571 VSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPG 630
+QEN+DKG+IYPPF+NIRKISAHIAA+VAAK YELGLA+ LP+P++LV+YAE+CMYSP
Sbjct: 576 ATQENFDKGMIYPPFSNIRKISAHIAANVAAKAYELGLATRLPQPENLVEYAENCMYSPA 635
Query: 631 YRSY 634
YRS+
Sbjct: 636 YRSF 639
>sp|P51615|MAOX_VITVI NADP-dependent malic enzyme (NADP-ME) gi|515759|gb|AAA67087.1|
malate dehydrogenase (NADP+)
Length = 591
Score = 974 bits (2517), Expect = 0.0
Identities = 476/566 (84%), Positives = 525/566 (92%)
Query: 69 GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPA 128
GGV DLYGED ATEDQL+TPWT SVASG +LLRDPR+NKGLAF +KERDAHYL GLLPP
Sbjct: 25 GGVEDLYGEDFATEDQLVTPWTVSVASGYSLLRDPRHNKGLAFNDKERDAHYLCGLLPPV 84
Query: 129 VFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVG 188
V TQELQE++ M++IRQY+VPL KYMAMMDLQERNERLFYK+LIDNVEELLP+VYTP VG
Sbjct: 85 VSTQELQERKLMNSIRQYQVPLQKYMAMMDLQERNERLFYKLLIDNVEELLPVVYTPTVG 144
Query: 189 EACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGM 248
EACQKYGSI++RPQGLYISLKEKGKILEVLKNWPE+ IQVIVVTDGERILGLGDLGCQGM
Sbjct: 145 EACQKYGSIFRRPQGLYISLKEKGKILEVLKNWPERRIQVIVVTDGERILGLGDLGCQGM 204
Query: 249 GIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDE 308
GIPVGKLSLYTALGGVRPS+CLP+TIDVGTNNE LL +EFY GL+Q+RATGKEY E L E
Sbjct: 205 GIPVGKLSLYTALGGVRPSACLPITIDVGTNNEKLLANEFYIGLKQRRATGKEYSEFLQE 264
Query: 309 FMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKL 368
FM VKQNYGEKVLIQFEDFANHNAF+LL +Y ++HL FNDDIQGTASVVLAG++++L+L
Sbjct: 265 FMSPVKQNYGEKVLIQFEDFANHNAFDLLAKYGTTHLAFNDDIQGTASVVLAGIVSALRL 324
Query: 369 VGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE 428
+GGTLADH FLFLGAGEAGTGIAELIALE+SKQTK P+EETRKK+WLVDSKGLIV SR +
Sbjct: 325 LGGTLADHKFLFLGAGEAGTGIAELIALEMSKQTKCPIEETRKKIWLVDSKGLIVGSRKD 384
Query: 429 SLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILA 488
SLQ FKKPWAHEHEPVK LLDAVK IKPTVLIGSSGVGK FTKEV+EAMAS NEKPLILA
Sbjct: 385 SLQQFKKPWAHEHEPVKDLLDAVKVIKPTVLIGSSGVGKAFTKEVIEAMASCNEKPLILA 444
Query: 489 LSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLI 548
LSNPTSQSECTAEEAYTW++G+AIFASGSPFDPV+Y GK + PGQANNAYIFPG G+GL+
Sbjct: 445 LSNPTSQSECTAEEAYTWTQGRAIFASGSPFDPVEYNGKTFVPGQANNAYIFPGLGMGLV 504
Query: 549 ISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGL 608
ISGAIRV DEMLLAASEALA QV+QEN+DKGLIYPPF+NIRKISAHIAA+VAAK YELGL
Sbjct: 505 ISGAIRVHDEMLLAASEALARQVTQENFDKGLIYPPFSNIRKISAHIAANVAAKAYELGL 564
Query: 609 ASHLPRPKDLVKYAESCMYSPGYRSY 634
A+ LP+P++LVKYAESCMYSP YRSY
Sbjct: 565 ATRLPQPENLVKYAESCMYSPVYRSY 590
>emb|CAA54986.1| malate dehydrogenase (oxaloacetate decarboxylating) (NADP+)
[Flaveria pringlei] gi|547886|sp|P36444|MAOC_FLAPR
NADP-dependent malic enzyme, chloroplast precursor
(NADP-ME)
Length = 647
Score = 973 bits (2514), Expect = 0.0
Identities = 472/566 (83%), Positives = 524/566 (92%)
Query: 69 GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPA 128
GGV D+YGEDSATED ITPW+ SVASG +LLRDP +NKGLAFTEKERDAHYLRGLLPP
Sbjct: 81 GGVEDVYGEDSATEDHFITPWSVSVASGYSLLRDPHHNKGLAFTEKERDAHYLRGLLPPV 140
Query: 129 VFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVG 188
V +LQ K+ MHNIRQYEVPL +Y AMMDLQERNERLFYK+LI+N+EELLPIVYTP VG
Sbjct: 141 VVNHDLQVKKMMHNIRQYEVPLQRYQAMMDLQERNERLFYKLLIENIEELLPIVYTPTVG 200
Query: 189 EACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGM 248
EACQKYG+I+K PQGLYISLK+KGK+LE+LKNWP+K IQVIVVTDGERILGLGDLGCQGM
Sbjct: 201 EACQKYGTIFKNPQGLYISLKDKGKVLEILKNWPQKKIQVIVVTDGERILGLGDLGCQGM 260
Query: 249 GIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDE 308
GIPVGKLSLYTALGG+RPS+CLP+TIDVGTNNE +LNDEFY GLRQ+RA+GKEY EL++E
Sbjct: 261 GIPVGKLSLYTALGGIRPSACLPITIDVGTNNEKMLNDEFYIGLRQRRASGKEYAELMNE 320
Query: 309 FMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKL 368
FM AVKQNYGEKVLIQFEDFANHNAF+LL++Y ++HLVFNDDIQGTASVVLAGL+++LKL
Sbjct: 321 FMSAVKQNYGEKVLIQFEDFANHNAFDLLEKYRTTHLVFNDDIQGTASVVLAGLISALKL 380
Query: 369 VGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE 428
VGG+LADH FLFLGAGEAGTGIAELIALEISKQT AP+EETRKK+WLVDSKGLIVRSRL+
Sbjct: 381 VGGSLADHKFLFLGAGEAGTGIAELIALEISKQTNAPLEETRKKIWLVDSKGLIVRSRLD 440
Query: 429 SLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILA 488
SLQHFKKPWAH+HEPV LDAVKAIKPTVLIGSSG G+TFTKEVVEAM+S NEKP+ILA
Sbjct: 441 SLQHFKKPWAHDHEPVNKFLDAVKAIKPTVLIGSSGAGQTFTKEVVEAMSSFNEKPIILA 500
Query: 489 LSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLI 548
LSNPTSQSECTAE+AYTWS+G+ IFASGSPF PV+Y GKVY GQ+NNAYIFPGFGLGLI
Sbjct: 501 LSNPTSQSECTAEQAYTWSEGRTIFASGSPFAPVEYNGKVYVSGQSNNAYIFPGFGLGLI 560
Query: 549 ISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGL 608
ISGAIRV DEMLLAASEALA QV+QE++D GLIYPPFTNIRKISAHIAA VAAK YELGL
Sbjct: 561 ISGAIRVHDEMLLAASEALAEQVTQEHFDNGLIYPPFTNIRKISAHIAAKVAAKAYELGL 620
Query: 609 ASHLPRPKDLVKYAESCMYSPGYRSY 634
AS LP+P++LV YAESCMYSP YR+Y
Sbjct: 621 ASRLPQPENLVAYAESCMYSPKYRNY 646
>gb|AAQ99276.1| NADP malic enzyme [Oryza sativa (japonica cultivar-group)]
gi|54287505|gb|AAV31249.1| NADP malic enzyme [Oryza
sativa (japonica cultivar-group)]
Length = 570
Score = 971 bits (2510), Expect = 0.0
Identities = 470/566 (83%), Positives = 527/566 (93%)
Query: 69 GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPA 128
GGV D YGED ATEDQLITPW+FSVASG TLLRDPR+NKGLAF+E ERDAHYLRGLLPP+
Sbjct: 4 GGVEDTYGEDRATEDQLITPWSFSVASGYTLLRDPRHNKGLAFSEAERDAHYLRGLLPPS 63
Query: 129 VFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVG 188
+ +QELQEK+ MHN+R Y VPL +Y+AMMDLQERNERLFYK+LIDNVEELLP+VYTP VG
Sbjct: 64 IVSQELQEKKLMHNLRNYTVPLQRYIAMMDLQERNERLFYKLLIDNVEELLPVVYTPTVG 123
Query: 189 EACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGM 248
EACQKYGSIY+RPQGLYISLK+KGKILEVLKNWPE++IQVIVVTDGERILGLGDLGCQGM
Sbjct: 124 EACQKYGSIYRRPQGLYISLKDKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGCQGM 183
Query: 249 GIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDE 308
GIPVGKLSLYTALGGVRPS+CLP+TIDVGTNNE+LLNDEFY GL+Q+RATG+EY ELL+E
Sbjct: 184 GIPVGKLSLYTALGGVRPSACLPITIDVGTNNESLLNDEFYIGLKQRRATGEEYHELLEE 243
Query: 309 FMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKL 368
FM AVKQNYGEKVL+QFEDFANHNAF+LL +YS SHLVFNDDIQGTASVVLAGL+A+LK+
Sbjct: 244 FMTAVKQNYGEKVLVQFEDFANHNAFDLLAKYSKSHLVFNDDIQGTASVVLAGLIAALKV 303
Query: 369 VGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE 428
VGGTLADHT+LFLGAGEAGTGIAELIALE+SKQT+ P+ + RKKVWLVDS+GLIV SR E
Sbjct: 304 VGGTLADHTYLFLGAGEAGTGIAELIALEMSKQTEIPINDCRKKVWLVDSRGLIVESRKE 363
Query: 429 SLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILA 488
SLQHFK+P+AHEHEPVK LL+AV++IKPTVLIG+SGVGKTFT+EVVEAMA+ NEKP+I A
Sbjct: 364 SLQHFKQPFAHEHEPVKTLLEAVQSIKPTVLIGTSGVGKTFTQEVVEAMAAFNEKPVIFA 423
Query: 489 LSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLI 548
LSNPTS SECTAEEAYTW+KG A+FASGSPFD V+Y GK Y PGQ+NNAYIFPGFGLG++
Sbjct: 424 LSNPTSHSECTAEEAYTWTKGSAVFASGSPFDAVEYEGKTYVPGQSNNAYIFPGFGLGVV 483
Query: 549 ISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGL 608
ISGAIRV D+MLLAASEALA QVS++N+ +GLI+PPFTNIRKISAHIAA VAAK YELGL
Sbjct: 484 ISGAIRVHDDMLLAASEALAEQVSEDNFARGLIFPPFTNIRKISAHIAAKVAAKAYELGL 543
Query: 609 ASHLPRPKDLVKYAESCMYSPGYRSY 634
AS LPRP DLVKYAESCMY+P YR Y
Sbjct: 544 ASRLPRPDDLVKYAESCMYTPAYRCY 569
>gb|AAT02533.1| NADP-dependent malic enzyme 1 [Hydrilla verticillata]
Length = 654
Score = 970 bits (2507), Expect = 0.0
Identities = 473/570 (82%), Positives = 524/570 (90%)
Query: 65 SEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGL 124
S SGGVRD YGED ATEDQL+TPWT SVASGN+LLRDPR NKGLAFTEKERDAHYLRGL
Sbjct: 84 SAVSGGVRDTYGEDRATEDQLLTPWTASVASGNSLLRDPRLNKGLAFTEKERDAHYLRGL 143
Query: 125 LPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYT 184
LPP TQE+QEK+ M N+RQY+VPL +YMAMMDLQERNERLFYK+LIDNVEELLP+VYT
Sbjct: 144 LPPVFLTQEIQEKKLMTNLRQYQVPLQRYMAMMDLQERNERLFYKLLIDNVEELLPVVYT 203
Query: 185 PVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLG 244
P VGEACQKYGSI++RPQGLYISL+EKGKILEVLKNWPE+ I+VIVVTDGERILGLGDLG
Sbjct: 204 PTVGEACQKYGSIFRRPQGLYISLREKGKILEVLKNWPERNIEVIVVTDGERILGLGDLG 263
Query: 245 CQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEE 304
CQGMGIPVGKL+LYTALGGVRPS+CLPVTIDVGTNNE LL DEFY GLRQKRA GKEY E
Sbjct: 264 CQGMGIPVGKLALYTALGGVRPSACLPVTIDVGTNNENLLQDEFYIGLRQKRARGKEYAE 323
Query: 305 LLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLA 364
L++EFM AVKQNYGE+VL+QFEDFANHNAF LL +YS SHLVFNDDIQGTASVVLAGL+A
Sbjct: 324 LIEEFMSAVKQNYGERVLVQFEDFANHNAFELLTKYSKSHLVFNDDIQGTASVVLAGLIA 383
Query: 365 SLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVR 424
SLKLVGGTLA+HTFLF GAGEAGTGIAELIAL++SK+T P+EETRKK+WLVD+KGLIV
Sbjct: 384 SLKLVGGTLAEHTFLFFGAGEAGTGIAELIALQMSKETGVPIEETRKKIWLVDTKGLIVS 443
Query: 425 SRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKP 484
SR ESLQHFK+PWAH+HEPV+ LLDAVK IKPTVLIGSSGV FT++VVEAMAS NEKP
Sbjct: 444 SRKESLQHFKQPWAHDHEPVQDLLDAVKMIKPTVLIGSSGVPNKFTQDVVEAMASFNEKP 503
Query: 485 LILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFG 544
+I ALSNPTS SECTAE+AYTWSKG+AIFASGSPFDPV+Y G+ Y GQANNAYIFPGFG
Sbjct: 504 VIFALSNPTSNSECTAEQAYTWSKGRAIFASGSPFDPVEYEGQTYTSGQANNAYIFPGFG 563
Query: 545 LGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVY 604
LGL++SGAIRV D+MLLAASEALA QV++EN++KGLIYPPFTNIRKISA+IAA VAAK Y
Sbjct: 564 LGLVMSGAIRVHDDMLLAASEALAEQVTEENFEKGLIYPPFTNIRKISANIAAKVAAKAY 623
Query: 605 ELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
ELGL+ HLPRP++LV+YAESCMYSP YRSY
Sbjct: 624 ELGLSGHLPRPENLVEYAESCMYSPVYRSY 653
>dbj|BAA74735.1| NADP-malic enzyme [Aloe arborescens]
Length = 585
Score = 969 bits (2505), Expect = 0.0
Identities = 476/566 (84%), Positives = 522/566 (92%)
Query: 69 GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPA 128
GGV D YGED ATE+QL+TPWT SVASG +LLRDP +NKGLAFTEKERDAHYLRGLLPP
Sbjct: 19 GGVEDAYGEDQATEEQLVTPWTISVASGYSLLRDPHHNKGLAFTEKERDAHYLRGLLPPV 78
Query: 129 VFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVG 188
+QELQEK+ M ++RQY+VPL +YMAMMDLQERNERLFYK+LIDNVEELLP+VYTP VG
Sbjct: 79 CISQELQEKKLMRSLRQYQVPLQRYMAMMDLQERNERLFYKLLIDNVEELLPVVYTPTVG 138
Query: 189 EACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGM 248
EACQKYG I++RPQGLYISLKEKGKILEVLKNWPE++IQVIVVTDGERILGLGDLG QGM
Sbjct: 139 EACQKYGCIFRRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGSQGM 198
Query: 249 GIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDE 308
GIPVGKLSLYTALGG+ PSSCLPVTIDVGTNNE LLNDEFY GLRQKRATG+EY ELL E
Sbjct: 199 GIPVGKLSLYTALGGIHPSSCLPVTIDVGTNNEQLLNDEFYIGLRQKRATGQEYAELLHE 258
Query: 309 FMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKL 368
FM AVKQNYGEKV++Q+EDFANHNAF LL +YS SHLVFNDDIQGTASVVLAGL+A+LK+
Sbjct: 259 FMTAVKQNYGEKVIVQYEDFANHNAFELLAKYSKSHLVFNDDIQGTASVVLAGLVAALKV 318
Query: 369 VGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE 428
VG TLA+HTFLFLGAGEAGTGIAELIALE+S+QTKAP+EETRKK+WLVDSKGLIV SR E
Sbjct: 319 VGRTLAEHTFLFLGAGEAGTGIAELIALEMSRQTKAPIEETRKKIWLVDSKGLIVSSRKE 378
Query: 429 SLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILA 488
SLQHFKKPWAHEHEPVK LLDAVK IKPTVLIGSSGVG+TFTKEV EAM S NEKP+ILA
Sbjct: 379 SLQHFKKPWAHEHEPVKTLLDAVKTIKPTVLIGSSGVGQTFTKEVAEAMPSFNEKPVILA 438
Query: 489 LSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLI 548
LSNPTSQSECTAE+AY+WS+G+AIFASGSPFDPV+Y GK++ PGQANNAYIFPG GLGL+
Sbjct: 439 LSNPTSQSECTAEQAYSWSEGRAIFASGSPFDPVEYNGKLFVPGQANNAYIFPGLGLGLV 498
Query: 549 ISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGL 608
ISGAIRV DEMLLAASEALA QV++EN+ GLIYPPFT IRKISAHIAA+VAAK YELGL
Sbjct: 499 ISGAIRVHDEMLLAASEALAQQVTEENFANGLIYPPFTIIRKISAHIAANVAAKAYELGL 558
Query: 609 ASHLPRPKDLVKYAESCMYSPGYRSY 634
A+HLPRP +LVKYAES MYSP YRSY
Sbjct: 559 ATHLPRPANLVKYAESRMYSPLYRSY 584
>ref|NP_916713.1| P0022F10.12 [Oryza sativa (japonica cultivar-group)]
Length = 593
Score = 967 bits (2499), Expect = 0.0
Identities = 467/566 (82%), Positives = 524/566 (92%)
Query: 69 GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPA 128
GGV D YGED ATE+QL+TPWT SVASG LLRDPRYNKGLAF E+ER+ HYLRGLLPPA
Sbjct: 27 GGVGDTYGEDCATEEQLVTPWTVSVASGYNLLRDPRYNKGLAFNERERETHYLRGLLPPA 86
Query: 129 VFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVG 188
+ +QELQE++ MHNIRQY++PL KYMAMMDLQE NERLFYK+LIDNVEELLP+VYTP VG
Sbjct: 87 IVSQELQERKIMHNIRQYQLPLQKYMAMMDLQEGNERLFYKLLIDNVEELLPVVYTPTVG 146
Query: 189 EACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGM 248
EACQKYGSI+ RPQGLYISLKEKGKILEVLKNWPE++IQVIVVTDGERILGLGDLGCQGM
Sbjct: 147 EACQKYGSIFSRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGCQGM 206
Query: 249 GIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDE 308
GIPVGKL+LYTALGGVRPS+CLP+T+DVGTNNE LLNDEFY GLRQKRAT +EY + L E
Sbjct: 207 GIPVGKLALYTALGGVRPSACLPITLDVGTNNEALLNDEFYIGLRQKRATAQEYADFLHE 266
Query: 309 FMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKL 368
FM AVKQNYGEKVLIQFEDFANHNAF LL +Y ++HLVFNDDIQGTASVVL+GL+A+LKL
Sbjct: 267 FMTAVKQNYGEKVLIQFEDFANHNAFELLAKYGTTHLVFNDDIQGTASVVLSGLVAALKL 326
Query: 369 VGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE 428
VGG+L++H++LFLGAGEAGTGIAELIALEIS+QTKAP+EE RKK+WLVDSKGLIV SR E
Sbjct: 327 VGGSLSEHSYLFLGAGEAGTGIAELIALEISRQTKAPIEECRKKIWLVDSKGLIVSSRKE 386
Query: 429 SLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILA 488
+LQHFKKPWAHEHEPV LLDAVK IKPTVLIG+SG G+TFT+EVVEA++S NE+P+I A
Sbjct: 387 TLQHFKKPWAHEHEPVGNLLDAVKTIKPTVLIGTSGKGQTFTQEVVEAISSFNERPVIFA 446
Query: 489 LSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLI 548
LSNPTSQSECTAE+AYTWSKG+A+FASGSPFDPV+Y GK+Y PGQANNAYIFPGFGLG++
Sbjct: 447 LSNPTSQSECTAEQAYTWSKGRAVFASGSPFDPVEYDGKIYVPGQANNAYIFPGFGLGVV 506
Query: 549 ISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGL 608
+SGAIRV D+MLLAASEALA QV+QEN+DKGL YPPF+NIRKISAHIAA+VAAK YELGL
Sbjct: 507 MSGAIRVHDDMLLAASEALAQQVTQENFDKGLTYPPFSNIRKISAHIAANVAAKAYELGL 566
Query: 609 ASHLPRPKDLVKYAESCMYSPGYRSY 634
AS PRPKDLVKYAESCMYSP YR+Y
Sbjct: 567 ASRRPRPKDLVKYAESCMYSPLYRNY 592
>gb|AAK83074.1| putative cytosolic NADP-malic enzyme [Flaveria pringlei]
Length = 589
Score = 966 bits (2497), Expect = 0.0
Identities = 471/581 (81%), Positives = 532/581 (91%), Gaps = 3/581 (0%)
Query: 54 GEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTE 113
GE VV +S GGV D+YGED A+EDQLITPWT SVASG TLLRDP +NKGLAFT+
Sbjct: 11 GESVVDH---SSAVGGGVDDVYGEDRASEDQLITPWTVSVASGYTLLRDPHHNKGLAFTQ 67
Query: 114 KERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILID 173
+ERD+HYLRGLLPPAV TQELQEK+ M NIR YEVPLH+Y+AMM+L+ERNERLFYK+LID
Sbjct: 68 RERDSHYLRGLLPPAVATQELQEKKLMQNIRSYEVPLHRYVAMMELEERNERLFYKLLID 127
Query: 174 NVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTD 233
NVEELLP+VYTP VGEACQKYGSI+KRPQGLYISLKEKGKILEVL+NWPE+ IQVIVVTD
Sbjct: 128 NVEELLPVVYTPTVGEACQKYGSIFKRPQGLYISLKEKGKILEVLRNWPERNIQVIVVTD 187
Query: 234 GERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLR 293
GERILGLGDLGCQGMGIPVGKL+LYTALGGVRPS+CLP+TIDVGTNN+ LL+DEFY GLR
Sbjct: 188 GERILGLGDLGCQGMGIPVGKLALYTALGGVRPSACLPITIDVGTNNQKLLDDEFYIGLR 247
Query: 294 QKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQG 353
QKRATGKEY +LL+EFM AVKQNYGEKVL+QFEDFANHNAF LL +Y +SHLVFNDDIQG
Sbjct: 248 QKRATGKEYYDLLEEFMSAVKQNYGEKVLVQFEDFANHNAFELLAKYRTSHLVFNDDIQG 307
Query: 354 TASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKV 413
TASVVLAGL+ASLKL+GG+LADHTFLFLGAGEAGTGIAELIALEIS +T P++E RKK+
Sbjct: 308 TASVVLAGLVASLKLLGGSLADHTFLFLGAGEAGTGIAELIALEISTKTYIPIDEARKKI 367
Query: 414 WLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEV 473
WLVDSKGLIV SR E+LQHFKKPWAHEHEP+ LLDAVKAIKP+VLIG+SGVG+TFT++V
Sbjct: 368 WLVDSKGLIVSSRKETLQHFKKPWAHEHEPLSTLLDAVKAIKPSVLIGTSGVGQTFTQDV 427
Query: 474 VEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQ 533
VEAMA+ NE+PLI+ALSNPTSQ+ECTAE+AYTW++G+AIF+SGSPFDP +Y G ++ PGQ
Sbjct: 428 VEAMAAFNERPLIMALSNPTSQAECTAEQAYTWTQGRAIFSSGSPFDPYEYKGNLFIPGQ 487
Query: 534 ANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISA 593
ANNAYIFPG G GL+ISGAIRV DEMLLAASEALA QV+QE+YDKG+IYPP TNIRKISA
Sbjct: 488 ANNAYIFPGLGFGLVISGAIRVHDEMLLAASEALANQVTQEHYDKGMIYPPLTNIRKISA 547
Query: 594 HIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
HIAA+VAAK Y+LGLA+ LPRP DLVKYAESCMY+P YRSY
Sbjct: 548 HIAANVAAKAYDLGLATRLPRPADLVKYAESCMYTPNYRSY 588
>pir||JC5967 malate dehydrogenase (oxaloacetate-decarboxylating) (NADP) (EC
1.1.1.40) - aloe gi|2911148|dbj|BAA24950.1| NADP-malic
enzyme [Aloe arborescens]
Length = 592
Score = 966 bits (2497), Expect = 0.0
Identities = 474/566 (83%), Positives = 520/566 (91%)
Query: 69 GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPA 128
GGV D+YGED ATE+Q +TPWT SVASG +LLRDP +NKGLAF+EKERDAHYLRGLLPP
Sbjct: 26 GGVGDVYGEDRATEEQQVTPWTVSVASGYSLLRDPHHNKGLAFSEKERDAHYLRGLLPPT 85
Query: 129 VFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVG 188
+QE+Q K+ +HN+RQY+VPL +YMAMMDLQE NERLFYK+LID+VEELLP+VYTP VG
Sbjct: 86 CISQEIQVKKMLHNLRQYQVPLQRYMAMMDLQEMNERLFYKLLIDHVEELLPVVYTPTVG 145
Query: 189 EACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGM 248
EACQKYG I++RPQGLYISLKEKGKILEVLKNWPE+ IQVIVVTDGERILGLGDLGCQGM
Sbjct: 146 EACQKYGCIFRRPQGLYISLKEKGKILEVLKNWPERNIQVIVVTDGERILGLGDLGCQGM 205
Query: 249 GIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDE 308
GIPVGKLSLYTALGG+RPS+CLPVTIDVGTNNE LL DEFY GLRQKRATG+EY ELL E
Sbjct: 206 GIPVGKLSLYTALGGIRPSACLPVTIDVGTNNEQLLKDEFYIGLRQKRATGQEYAELLHE 265
Query: 309 FMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKL 368
FM AVKQNYGEKVLIQFEDFANHNAF LL +YS+SHLVFNDDIQGTASVVLAGL+A+LKL
Sbjct: 266 FMAAVKQNYGEKVLIQFEDFANHNAFELLAKYSTSHLVFNDDIQGTASVVLAGLVAALKL 325
Query: 369 VGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE 428
VGGTLA+HTFLFLGAGEAGTGIAELIALE+SKQTKAPVEETRKK+WLVDSKGLIV SR +
Sbjct: 326 VGGTLAEHTFLFLGAGEAGTGIAELIALEMSKQTKAPVEETRKKIWLVDSKGLIVSSRKD 385
Query: 429 SLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILA 488
+LQHFKKPWAHEHEPV LL AVK IKPTVLIGSSGVG+TFTKEV+EAM+S EKP+ILA
Sbjct: 386 TLQHFKKPWAHEHEPVDTLLGAVKTIKPTVLIGSSGVGRTFTKEVIEAMSSFTEKPVILA 445
Query: 489 LSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLI 548
LSNPTSQSECTAEEAYTWSKG+AIFASGSPFDPV Y GK++ PGQANNAYIFPG GLGL+
Sbjct: 446 LSNPTSQSECTAEEAYTWSKGRAIFASGSPFDPVLYNGKLFVPGQANNAYIFPGLGLGLV 505
Query: 549 ISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGL 608
ISGAIRV D+MLLAASEALA QV+QEN+ GLIYPPF+ IRKISA IAA+VAAK YELGL
Sbjct: 506 ISGAIRVHDDMLLAASEALAQQVTQENFANGLIYPPFSIIRKISAQIAANVAAKAYELGL 565
Query: 609 ASHLPRPKDLVKYAESCMYSPGYRSY 634
A+ LPRP DLVKYAESCMY+P YRSY
Sbjct: 566 ATRLPRPADLVKYAESCMYTPAYRSY 591
>gb|AAK83073.1| putative cytosolic NADP-malic enzyme [Flaveria pringlei]
Length = 589
Score = 966 bits (2496), Expect = 0.0
Identities = 471/581 (81%), Positives = 530/581 (91%), Gaps = 3/581 (0%)
Query: 54 GEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTE 113
GE VV +S GGV D+YGED A+EDQLITPWT SVASG TLLRDP +NKGLAFTE
Sbjct: 11 GESVVDP---SSAVGGGVDDVYGEDRASEDQLITPWTVSVASGYTLLRDPHHNKGLAFTE 67
Query: 114 KERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILID 173
+ERD+HYLRGLLPPAV TQELQEK+ M NIR YEVPLH+Y+AMM+L+ERNERLFYK+LID
Sbjct: 68 RERDSHYLRGLLPPAVATQELQEKKLMQNIRSYEVPLHRYVAMMELEERNERLFYKLLID 127
Query: 174 NVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTD 233
NVEELLP+VYTP VGEACQKYGSI+KRPQGLYISLKEKGKILEVL+NWPE+ IQVIVVTD
Sbjct: 128 NVEELLPVVYTPTVGEACQKYGSIFKRPQGLYISLKEKGKILEVLRNWPERNIQVIVVTD 187
Query: 234 GERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLR 293
GERILGLGDLGCQGMGIPVGKL+LYTALGGVRPS+CLP+TIDVGTNN+ LL+DEFY GLR
Sbjct: 188 GERILGLGDLGCQGMGIPVGKLALYTALGGVRPSACLPITIDVGTNNQKLLDDEFYIGLR 247
Query: 294 QKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQG 353
QKRATGKEY +LL+EFM AVKQNYGEKVL+QFEDFANHNAF LL +Y +SHLVFNDDIQG
Sbjct: 248 QKRATGKEYYDLLEEFMSAVKQNYGEKVLVQFEDFANHNAFELLAKYRTSHLVFNDDIQG 307
Query: 354 TASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKV 413
TASVVLAGL+ASLKL+GG+LADHTFLFLGAGEAGTGIAELIALEIS + P++E RKK+
Sbjct: 308 TASVVLAGLVASLKLLGGSLADHTFLFLGAGEAGTGIAELIALEISTKANIPIDEARKKI 367
Query: 414 WLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEV 473
WLVDSKGLIV SR E+LQHFKKPWAHEHEP+ LLDAVKAIKP+VLIG+SGVG+TFT+ V
Sbjct: 368 WLVDSKGLIVSSRKETLQHFKKPWAHEHEPLSTLLDAVKAIKPSVLIGTSGVGQTFTQNV 427
Query: 474 VEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQ 533
VEAMA+ NE+PLI+ALSNPTSQ+ECTAE+AYTW++G+AIF+SGSPFDP +Y G ++ PGQ
Sbjct: 428 VEAMAAFNERPLIMALSNPTSQAECTAEQAYTWTQGRAIFSSGSPFDPYEYNGNLFIPGQ 487
Query: 534 ANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISA 593
ANNAYIFPG G GL+ISGAIRV DEMLLAASEALA QV+QE+YDKG+IYPP TNIRKISA
Sbjct: 488 ANNAYIFPGLGFGLVISGAIRVHDEMLLAASEALANQVTQEHYDKGMIYPPLTNIRKISA 547
Query: 594 HIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
HIAA+VAAK Y+LGLA+ LPRP DLVKYAESCMY+P YRSY
Sbjct: 548 HIAANVAAKAYDLGLATRLPRPTDLVKYAESCMYTPNYRSY 588
>gb|AAB58727.1| NADP-malic enzyme [Lycopersicon esculentum] gi|7431231|pir||T06401
malate dehydrogenase (oxaloacetate-decarboxylating)
(NADP) (EC 1.1.1.40) precursor - tomato
Length = 640
Score = 964 bits (2492), Expect = 0.0
Identities = 478/609 (78%), Positives = 542/609 (88%), Gaps = 16/609 (2%)
Query: 42 PKLNIAAMSRNG--GEGVVGEDFKN--------------SEESGGVRDLYGEDSATEDQL 85
P + +AA++ NG G+G V +N S +GGV+D+YGEDSATEDQ
Sbjct: 31 PMVVVAAVNSNGKPGDGSVSVLVENALTESPAPVEMEVKSTVTGGVQDVYGEDSATEDQS 90
Query: 86 ITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQ 145
ITPWT SVASG +LLR+P YNKGLAF+E+ERD HYLRGLLPP V + +LQ K+ M++IR+
Sbjct: 91 ITPWTLSVASGFSLLRNPHYNKGLAFSERERDTHYLRGLLPPVVISHDLQVKKMMNSIRK 150
Query: 146 YEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLY 205
Y+VPL +YMAMMDLQE NERLFYK+LIDNVEELLPIVYTP VGEACQKYGSI++RPQGL+
Sbjct: 151 YDVPLQRYMAMMDLQEMNERLFYKLLIDNVEELLPIVYTPTVGEACQKYGSIFRRPQGLF 210
Query: 206 ISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVR 265
ISLKEKGKI EVLKNWPEK IQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGG+R
Sbjct: 211 ISLKEKGKIHEVLKNWPEKKIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGIR 270
Query: 266 PSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQF 325
PS+CLPVTIDVGTNNE LLNDEFY GLRQ+RATG+EY ELLDEFM AVKQNYGEKVLIQF
Sbjct: 271 PSACLPVTIDVGTNNENLLNDEFYIGLRQRRATGQEYSELLDEFMYAVKQNYGEKVLIQF 330
Query: 326 EDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGE 385
EDFANHNAF+LL +Y +SHLVFNDDIQGTASVVLAGL+A+L LVGG+L++HTFLFLGAGE
Sbjct: 331 EDFANHNAFDLLAKYGTSHLVFNDDIQGTASVVLAGLMAALNLVGGSLSEHTFLFLGAGE 390
Query: 386 AGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVK 445
AGTGIAELIALE+SKQT P+EETRKK+W+VDSKGLIV+SR+E LQHFK+PWAH+HEPV+
Sbjct: 391 AGTGIAELIALEMSKQTGIPLEETRKKIWMVDSKGLIVKSRMEMLQHFKRPWAHDHEPVQ 450
Query: 446 ALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEAYT 505
L++AVK+IKPTVLIGSSG G+TFTKEVV+AMA+ NEKP+I ALSNPTSQSECTAEEAY+
Sbjct: 451 ELVNAVKSIKPTVLIGSSGAGRTFTKEVVQAMATFNEKPIIFALSNPTSQSECTAEEAYS 510
Query: 506 WSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASE 565
WS+G+AIFASGSPF PV+Y GKVYA GQANNAYIFPGFGLGLIISGAIRV D+MLL ASE
Sbjct: 511 WSEGRAIFASGSPFAPVEYNGKVYASGQANNAYIFPGFGLGLIISGAIRVHDDMLLVASE 570
Query: 566 ALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAESC 625
ALA +VSQEN++KGLIYPPF+NIRKISAHI A VAAK YELGLA+ LP+PKDLV YAESC
Sbjct: 571 ALADEVSQENFEKGLIYPPFSNIRKISAHIRAKVAAKAYELGLATRLPQPKDLVAYAESC 630
Query: 626 MYSPGYRSY 634
MYSP YRSY
Sbjct: 631 MYSPAYRSY 639
>emb|CAB66003.1| NADP-dependent malate dehydrogenase (decarboxylating) [Apium
graveolens]
Length = 570
Score = 962 bits (2488), Expect = 0.0
Identities = 464/567 (81%), Positives = 523/567 (91%)
Query: 68 SGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPP 127
+GG +D+YGED A EDQL+TPWT SVASG TLLRDP +NKGLAFTEKERDAHYLRGLLPP
Sbjct: 3 TGGDQDVYGEDCAAEDQLVTPWTISVASGYTLLRDPHHNKGLAFTEKERDAHYLRGLLPP 62
Query: 128 AVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVV 187
A+ +Q+ QEK+ M N+R YEVPLH+YMAMM+LQERNERLFYK+LIDNVEELLP+VYTP V
Sbjct: 63 AIISQQQQEKKLMQNLRSYEVPLHRYMAMMELQERNERLFYKLLIDNVEELLPVVYTPTV 122
Query: 188 GEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQG 247
GEACQKYGSI++RPQGLYISLKEKGKILEVLKNWPEK IQVIVVTDGERILGLGDLGCQG
Sbjct: 123 GEACQKYGSIFRRPQGLYISLKEKGKILEVLKNWPEKNIQVIVVTDGERILGLGDLGCQG 182
Query: 248 MGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLD 307
MGIPVGKL+LYTALGGVRPS+CLP+TIDVGTNNE+LLN+EFY GL+Q+RATGKEY +LL
Sbjct: 183 MGIPVGKLALYTALGGVRPSACLPITIDVGTNNESLLNNEFYIGLKQRRATGKEYADLLQ 242
Query: 308 EFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLK 367
EFM AVKQNYGEKVL+QFEDFANHNAF LL +YS++HLVFNDDIQGTASVVL+G+++SLK
Sbjct: 243 EFMTAVKQNYGEKVLVQFEDFANHNAFELLSKYSTTHLVFNDDIQGTASVVLSGIVSSLK 302
Query: 368 LVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRL 427
L+GG LADHTFLFLGAGEAGTGIAELIALE+SKQTK +EETRKK+WLVDSKGLIV R
Sbjct: 303 LIGGMLADHTFLFLGAGEAGTGIAELIALEMSKQTKIHLEETRKKIWLVDSKGLIVSHRK 362
Query: 428 ESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLIL 487
ESLQHFKKPWAHEHEP L+DAVKAIKPTVLIG+SG GKTFTKEVVEAMAS N+KPLI+
Sbjct: 363 ESLQHFKKPWAHEHEPCTTLIDAVKAIKPTVLIGTSGQGKTFTKEVVEAMASFNKKPLIM 422
Query: 488 ALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGL 547
ALSNPTSQ+ECTAEEAY WS+G+A+FASGSPFDPVKY ++Y PGQANNAYIFPG GLGL
Sbjct: 423 ALSNPTSQAECTAEEAYNWSEGRAVFASGSPFDPVKYNNQLYIPGQANNAYIFPGLGLGL 482
Query: 548 IISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELG 607
++SGAIR+ DEMLLAASEALA QV+QE+YDKG+ +PPF+NIR ISA+IAA VAAK Y+LG
Sbjct: 483 VMSGAIRMHDEMLLAASEALACQVTQEHYDKGMTFPPFSNIRTISANIAAKVAAKAYDLG 542
Query: 608 LASHLPRPKDLVKYAESCMYSPGYRSY 634
LA+ LPRP+DLVK+AESCMYSP YR Y
Sbjct: 543 LATRLPRPEDLVKFAESCMYSPNYRIY 569
>dbj|BAB20887.2| NADP dependent malic enzyme [Oryza sativa (japonica
cultivar-group)]
Length = 593
Score = 962 bits (2488), Expect = 0.0
Identities = 465/566 (82%), Positives = 522/566 (92%)
Query: 69 GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPA 128
GGV D YGED ATE+QL+TPWT SVASG LLRDPRYNKGLAF E+ER+ HYLRGLLPPA
Sbjct: 27 GGVGDTYGEDCATEEQLVTPWTVSVASGYNLLRDPRYNKGLAFNERERETHYLRGLLPPA 86
Query: 129 VFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVG 188
+ +QELQE++ MHNIRQY++PL KYMAMMDLQE NERLFYK+LIDNVEELLP+VYTP VG
Sbjct: 87 IVSQELQERKIMHNIRQYQLPLQKYMAMMDLQEGNERLFYKLLIDNVEELLPVVYTPTVG 146
Query: 189 EACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGM 248
EACQKYGSI+ RPQGLYISLKEKGKILEVLKNWPE++IQVIVVTDGERILGLGDLGCQGM
Sbjct: 147 EACQKYGSIFSRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGCQGM 206
Query: 249 GIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDE 308
GIPVGKL+LYTALGGVRPS+CLP+T+DVGTNNE L NDEFY GLRQKRAT +EY + L E
Sbjct: 207 GIPVGKLALYTALGGVRPSACLPITLDVGTNNEALPNDEFYIGLRQKRATAQEYADFLHE 266
Query: 309 FMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKL 368
FM AVKQNYGEKVLIQFEDFANHNAF LL +Y ++HLVFNDDIQGTASVVL+GL+A+LKL
Sbjct: 267 FMTAVKQNYGEKVLIQFEDFANHNAFELLAKYGTTHLVFNDDIQGTASVVLSGLVAALKL 326
Query: 369 VGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE 428
VGG+L++H++LFLG GEAGTGIAELIALEIS+QTKAP+EE RKK+WLVDSKGLIV SR E
Sbjct: 327 VGGSLSEHSYLFLGVGEAGTGIAELIALEISRQTKAPIEECRKKIWLVDSKGLIVSSRKE 386
Query: 429 SLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILA 488
+LQHFKKPWAHEHEPV LLDAVK IKPTVLIG+SG G+TFT+EVVEA++S NE+P+I A
Sbjct: 387 TLQHFKKPWAHEHEPVGNLLDAVKTIKPTVLIGTSGKGQTFTQEVVEAISSFNERPVIFA 446
Query: 489 LSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLI 548
LSNPTSQSECTAE+AYTWSKG+A+FASGSPFDPV+Y GK+Y PGQANNAYIFPGFGLG++
Sbjct: 447 LSNPTSQSECTAEQAYTWSKGRAVFASGSPFDPVEYDGKIYVPGQANNAYIFPGFGLGVV 506
Query: 549 ISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGL 608
+SGAIRV D+MLLAASEALA QV+QEN+DKGL YPPF+NIRKISAHIAA+VAAK YELGL
Sbjct: 507 MSGAIRVHDDMLLAASEALAQQVTQENFDKGLTYPPFSNIRKISAHIAANVAAKAYELGL 566
Query: 609 ASHLPRPKDLVKYAESCMYSPGYRSY 634
AS PRPKDLVKYAESCMYSP YR+Y
Sbjct: 567 ASRRPRPKDLVKYAESCMYSPLYRNY 592
>emb|CAA39690.1| malic enzyme [Populus trichocarpa] gi|1346485|sp|P34105|MAOX_POPTR
NADP-dependent malic enzyme (NADP-ME)
Length = 591
Score = 957 bits (2475), Expect = 0.0
Identities = 469/581 (80%), Positives = 527/581 (89%), Gaps = 1/581 (0%)
Query: 54 GEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTE 113
G V+ D K S GGV D+YGED ATEDQL+TPWT SVASG TLLRDP +NKGLAFTE
Sbjct: 11 GASVLDMDPK-STVGGGVEDVYGEDRATEDQLVTPWTISVASGYTLLRDPHHNKGLAFTE 69
Query: 114 KERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILID 173
KERDAHYLRGLLPP +Q+LQEK+ M+ IRQY++PL KY AMM+L+ERNERLFYK+LID
Sbjct: 70 KERDAHYLRGLLPPTTISQQLQEKKLMNTIRQYQLPLQKYTAMMELEERNERLFYKLLID 129
Query: 174 NVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTD 233
NVEELLP+VYTP VGEACQKYGSI+KRPQGLYISLKEKGK+L+VLKNWP+K+IQVIVVTD
Sbjct: 130 NVEELLPVVYTPTVGEACQKYGSIFKRPQGLYISLKEKGKVLDVLKNWPQKSIQVIVVTD 189
Query: 234 GERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLR 293
GERILGLGDLGCQG+GIPVGKLSLYTALGGVRPS+CLPVTIDVGTNNE LL DEFY GLR
Sbjct: 190 GERILGLGDLGCQGIGIPVGKLSLYTALGGVRPSACLPVTIDVGTNNEQLLKDEFYIGLR 249
Query: 294 QKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQG 353
Q+RATG+EY ELL EFM AVKQNYGEKVLIQFEDFANHNAF+LL +Y ++HLVFNDDIQG
Sbjct: 250 QRRATGQEYSELLHEFMTAVKQNYGEKVLIQFEDFANHNAFDLLAKYGTTHLVFNDDIQG 309
Query: 354 TASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKV 413
TA+VVLAGL+++LKL+GG+LADHTFLFLGAGEAGTGIAELIALE+S+++K P+EETRKK+
Sbjct: 310 TAAVVLAGLISALKLLGGSLADHTFLFLGAGEAGTGIAELIALEMSRRSKTPLEETRKKI 369
Query: 414 WLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEV 473
WL DSKGLIV SR ESLQHFKKPWAHEHEPVK LL+ VKAIKP VLIG+SGVGKTFTKEV
Sbjct: 370 WLTDSKGLIVSSRKESLQHFKKPWAHEHEPVKGLLEVVKAIKPIVLIGTSGVGKTFTKEV 429
Query: 474 VEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQ 533
+EAMAS NEKPLILALSNPTSQSECTA+EAYTW+KGKAIFASGSPFDPV+Y GKV+ PGQ
Sbjct: 430 IEAMASFNEKPLILALSNPTSQSECTAQEAYTWTKGKAIFASGSPFDPVEYEGKVFVPGQ 489
Query: 534 ANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISA 593
+NNAYIFPG GLGL+ISGAIRV D+MLLAA+EALA Q+ +E KGLIYPP +NIRKIS
Sbjct: 490 SNNAYIFPGLGLGLVISGAIRVHDDMLLAAAEALAGQIKEEYLAKGLIYPPLSNIRKISV 549
Query: 594 HIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
IAA+VAAK YELGLA+ LPRP++LVK+AESCMYSP YR Y
Sbjct: 550 QIAANVAAKAYELGLATRLPRPENLVKHAESCMYSPAYRYY 590
>ref|NP_197960.1| malate oxidoreductase, putative [Arabidopsis thaliana]
gi|5107826|gb|AAD40139.1| similar to malate
dehydrogenases; Pfam PF00390, Score=1290.5. E=0, N=1
[Arabidopsis thaliana]
Length = 588
Score = 954 bits (2466), Expect = 0.0
Identities = 464/571 (81%), Positives = 523/571 (91%)
Query: 64 NSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRG 123
+S GG+ D+YGEDSAT DQL+TPW SVASG TL+RDPRYNKGLAFT+KERDAHY+ G
Sbjct: 17 SSGVGGGISDVYGEDSATLDQLVTPWVTSVASGYTLMRDPRYNKGLAFTDKERDAHYITG 76
Query: 124 LLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVY 183
LLPP V +Q++QE++ MHN+RQY VPL +YMA+MDLQERNERLFYK+LIDNVEELLP+VY
Sbjct: 77 LLPPVVLSQDVQERKVMHNLRQYTVPLQRYMALMDLQERNERLFYKLLIDNVEELLPVVY 136
Query: 184 TPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDL 243
TP VGEACQKYGSIY+RPQGLYISLKEKGKILEVLKNWP++ IQVIVVTDGERILGLGDL
Sbjct: 137 TPTVGEACQKYGSIYRRPQGLYISLKEKGKILEVLKNWPQRGIQVIVVTDGERILGLGDL 196
Query: 244 GCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYE 303
GCQGMGIPVGKLSLYTALGG+RPS+CLP+TIDVGTNNE LLN+EFY GL+QKRA G+EY
Sbjct: 197 GCQGMGIPVGKLSLYTALGGIRPSACLPITIDVGTNNEKLLNNEFYIGLKQKRANGEEYA 256
Query: 304 ELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLL 363
E L EFM AVKQNYGEKVL+QFEDFANH+AF LL +Y SSHLVFNDDIQGTASVVLAGL+
Sbjct: 257 EFLQEFMCAVKQNYGEKVLVQFEDFANHHAFELLSKYCSSHLVFNDDIQGTASVVLAGLI 316
Query: 364 ASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIV 423
A+ K++G +LADHTFLFLGAGEAGTGIAELIAL+ISK+T P++ETRKK+WLVDSKGLIV
Sbjct: 317 AAQKVLGKSLADHTFLFLGAGEAGTGIAELIALKISKETGKPIDETRKKIWLVDSKGLIV 376
Query: 424 RSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEK 483
R ESLQHFK+PWAH+H+PVK LL AV AIKPTVLIG+SGVGKTFTKEVVEAMA+LNEK
Sbjct: 377 SERKESLQHFKQPWAHDHKPVKELLAAVNAIKPTVLIGTSGVGKTFTKEVVEAMATLNEK 436
Query: 484 PLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGF 543
PLILALSNPTSQ+ECTAEEAYTW+KG+AIFASGSPFDPV+Y GK + PGQANN YIFPG
Sbjct: 437 PLILALSNPTSQAECTAEEAYTWTKGRAIFASGSPFDPVQYDGKKFTPGQANNCYIFPGL 496
Query: 544 GLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKV 603
GLGLI+SGAIRVRD+MLLAASEALA+QV++EN+ GLIYPPF NIRKISA+IAASV AK
Sbjct: 497 GLGLIMSGAIRVRDDMLLAASEALASQVTEENFANGLIYPPFANIRKISANIAASVGAKT 556
Query: 604 YELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
YELGLAS+LPRPKDLVK AESCMYSP YR++
Sbjct: 557 YELGLASNLPRPKDLVKMAESCMYSPVYRNF 587
>prf||1803524A malic enzyme
Length = 589
Score = 952 bits (2460), Expect = 0.0
Identities = 469/581 (80%), Positives = 525/581 (89%), Gaps = 3/581 (0%)
Query: 54 GEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTE 113
G V+ D K S GGV D+YGED ATEDQL+TPWT SVASG TLLRDP +NKGLAFTE
Sbjct: 11 GASVLDMDPK-STVGGGVEDVYGEDRATEDQLVTPWTISVASGYTLLRDPHHNKGLAFTE 69
Query: 114 KERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILID 173
KERDAHYLRGLLPP +Q+LQEK+ M+ IRQY++PL KY AMM+L+ERNERLFYK+LID
Sbjct: 70 KERDAHYLRGLLPPTTISQQLQEKKLMNTIRQYQLPLQKYTAMMELEERNERLFYKLLID 129
Query: 174 NVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTD 233
NVEELLP+VYTP VGEACQKYGSI+KRPQGLYISLKEKGK+L+VLKNWP+K+IQVIVVTD
Sbjct: 130 NVEELLPVVYTPTVGEACQKYGSIFKRPQGLYISLKEKGKVLDVLKNWPQKSIQVIVVTD 189
Query: 234 GERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLR 293
GERILGLGDLGCQG+GIPVGKLSLYTALGGVRPS+CLPVTIDVGTNNE LL DEFY GLR
Sbjct: 190 GERILGLGDLGCQGIGIPVGKLSLYTALGGVRPSACLPVTIDVGTNNEQLLKDEFYIGLR 249
Query: 294 QKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQG 353
Q+RATG+EY ELL EFM AVKQNYGEKVLIQFEDFANHNAF+LL +Y ++HLVFNDDIQG
Sbjct: 250 QRRATGQEYSELLHEFMTAVKQNYGEKVLIQFEDFANHNAFDLLAKYGTTHLVFNDDIQG 309
Query: 354 TASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKV 413
TA+VVLAGL+++LKL+GG+LADHTFLFLGAGEAGTGIAELIALE+S+++K P+EETRKK+
Sbjct: 310 TAAVVLAGLISALKLIGGSLADHTFLFLGAGEAGTGIAELIALEMSRRSKTPLEETRKKI 369
Query: 414 WLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEV 473
WL DSKGLIV SR ESLQHFKKPWAHEHEPVK LL+ VKAIKP VLIG+SGVGKTFTKEV
Sbjct: 370 WLTDSKGLIVSSRKESLQHFKKPWAHEHEPVKGLLEVVKAIKPLVLIGTSGVGKTFTKEV 429
Query: 474 VEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQ 533
+EAMAS NEKPLILALSNPTSQSECTAEEAYTW KGKAIFASGSPFDPV+Y GKV+ PGQ
Sbjct: 430 IEAMASFNEKPLILALSNPTSQSECTAEEAYTWGKGKAIFASGSPFDPVEYEGKVFVPGQ 489
Query: 534 ANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISA 593
+NNAYIFP GLGL+ISGAIRV D+MLLAA+EALA Q+ +E KGLIYPP +NIRKIS
Sbjct: 490 SNNAYIFP--GLGLVISGAIRVHDDMLLAAAEALAGQIKEEYLAKGLIYPPLSNIRKISV 547
Query: 594 HIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
IAA+VAAK YELGLA+ LPRP++LVK+AESCMYSP YR Y
Sbjct: 548 QIAANVAAKAYELGLATRLPRPENLVKHAESCMYSPAYRYY 588
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,090,765,604
Number of Sequences: 2540612
Number of extensions: 48456807
Number of successful extensions: 129570
Number of sequences better than 10.0: 639
Number of HSP's better than 10.0 without gapping: 540
Number of HSP's successfully gapped in prelim test: 99
Number of HSP's that attempted gapping in prelim test: 127326
Number of HSP's gapped (non-prelim): 853
length of query: 635
length of database: 863,360,394
effective HSP length: 134
effective length of query: 501
effective length of database: 522,918,386
effective search space: 261982111386
effective search space used: 261982111386
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0334a.8


