

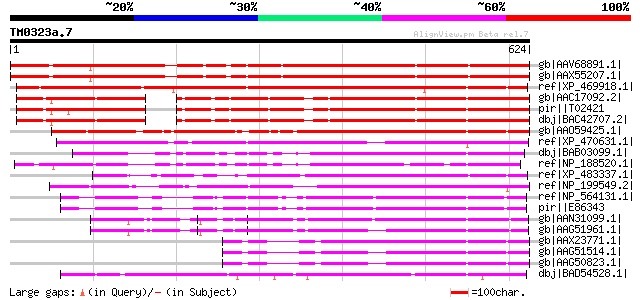
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0323a.7
(624 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAV68891.1| hypothetical protein AT5G66600 [Arabidopsis thali... 651 0.0
gb|AAX55207.1| hypothetical protein At5g66600 [Arabidopsis thali... 651 0.0
ref|XP_469918.1| hypothetical protein [Oryza sativa (japonica cu... 502 e-140
gb|AAC17092.2| unknown protein [Arabidopsis thaliana] gi|1522405... 451 e-125
pir||T02421 hypothetical protein At2g23700 [imported] - Arabidop... 451 e-125
dbj|BAC42707.2| unknown protein [Arabidopsis thaliana] 449 e-125
gb|AAO59425.1| putative ternary complex factor MIP1 [Antirrhinum... 440 e-122
ref|XP_470631.1| Hypothetical protein [Oryza sativa (japonica cu... 383 e-105
dbj|BAB03099.1| unnamed protein product [Arabidopsis thaliana] 325 2e-87
ref|NP_188520.1| expressed protein [Arabidopsis thaliana] 295 2e-78
ref|XP_483337.1| ternary complex factor-like [Oryza sativa (japo... 277 8e-73
ref|NP_199549.2| expressed protein [Arabidopsis thaliana] 245 4e-63
ref|NP_564131.1| expressed protein [Arabidopsis thaliana] 243 1e-62
pir||E86343 T22I11.12 protein - Arabidopsis thaliana gi|8886996|... 243 1e-62
gb|AAN31099.1| At1g76620/F14G6_22 [Arabidopsis thaliana] gi|1841... 235 4e-60
gb|AAG51961.1| unknown protein; 93089-95433 [Arabidopsis thalian... 213 1e-53
gb|AAX23771.1| hypothetical protein At1g43020 [Arabidopsis thali... 202 2e-50
gb|AAG51514.1| hypothetical protein [Arabidopsis thaliana] 184 1e-44
gb|AAG50823.1| hypothetical protein [Arabidopsis thaliana] gi|25... 184 1e-44
dbj|BAD54528.1| ternary complex factor MIP1-like [Oryza sativa (... 180 1e-43
>gb|AAV68891.1| hypothetical protein AT5G66600 [Arabidopsis thaliana]
Length = 614
Score = 651 bits (1679), Expect = 0.0
Identities = 360/632 (56%), Positives = 444/632 (69%), Gaps = 37/632 (5%)
Query: 1 MLGLSFTPRHKRSNSV--PDKNRVENDN-AESLKVSAQRMKLDEGYITECAQARKKGAPT 57
ML L HKRS S P+K RVE D + S ++QRMKLD G E K
Sbjct: 12 MLDLRIVQNHKRSKSASFPEKKRVEGDKTSNSSHEASQRMKLDMGRSNES----KHNQYH 67
Query: 58 SEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPS---LVNSSEMIMPKPATELIKE 114
S ++LKQE LE RLQDQF+VRC LEKALGY++ S L ++++ MPKPAT+LIK+
Sbjct: 68 SNTETSLKQEITHLETRLQDQFKVRCALEKALGYRTASSYVLTETNDIAMPKPATDLIKD 127
Query: 115 IAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYP-LTTPRAQVVKASLPEVL 173
+AVLEMEV++LEQ+LLSLYRKAF+QQ+SSVSP + + K P +TTPR ++ + +
Sbjct: 128 VAVLEMEVIHLEQYLLSLYRKAFEQQISSVSPNLENKKPKSPPVTTPRRRLDFSEDDDTP 187
Query: 174 TKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSA 233
+K + TV D N K+ + +D R HS S + +
Sbjct: 188 SKTDQHTVPLLDDN-------------QNQSKKTEIAAVDRDQMDPSFRRSHSQRSAFGS 234
Query: 234 FTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLS 293
R + P + K+ R+CHSQPL + N+ISLAEHLGTRISDHV P TPNKLS
Sbjct: 235 --RKASPEDSWGKASRSCHSQPLYVQN-----GDNLISLAEHLGTRISDHV-PETPNKLS 286
Query: 294 EDMVKCISAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDAR 353
E MVKC+S IYCKLA+PP L GL SP+SSLSS SAFS DQ D SPGF N+SSFD R
Sbjct: 287 EGMVKCMSEIYCKLAEPPSVLHRGLSSPNSSLSS-SAFSPSDQYDTSSPGFGNSSSFDVR 345
Query: 354 LDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKH 413
LDNSFH EG ++FSGPYS++VEV IYR+ K ++E L Q F+SLI RLEEVDPRKLKH
Sbjct: 346 LDNSFHVEGEKDFSGPYSSIVEVLCIYRDAKKASEVEDLLQNFKSLISRLEEVDPRKLKH 405
Query: 414 EEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCR 473
EEKLAFWIN+HNALVMHAFLAYGIPQNNVKRV LLLKAAYN+GG T+SA+ IQS+IL C+
Sbjct: 406 EEKLAFWINVHNALVMHAFLAYGIPQNNVKRVLLLLKAAYNIGGHTISAEAIQSSILGCK 465
Query: 474 MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQ 533
MS PGQWLR+ F+SR KF++GD R YAI++PEPL HFAL SG+HSDPAVRVYTPKR+ Q
Sbjct: 466 MSHPGQWLRLLFASR-KFKAGDERLAYAIDHPEPLLHFALTSGSHSDPAVRVYTPKRIQQ 524
Query: 534 ELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVK 593
ELE +K+EYIR L +R+ Q++L+PKLVETF KDS +CP G+ EM+ S+PES RK VK
Sbjct: 525 ELETSKEEYIRMNLSIRK--QRILLPKLVETFAKDSGLCPAGLTEMVNRSIPESSRKCVK 582
Query: 594 KCH-QLAKSRKCIEWIPHNFTFRYLISKDVLK 624
+C +K RK I+WIPH+FTFRYLI ++ K
Sbjct: 583 RCQSSTSKPRKTIDWIPHSFTFRYLIFREAAK 614
>gb|AAX55207.1| hypothetical protein At5g66600 [Arabidopsis thaliana]
gi|10177541|dbj|BAB10936.1| unnamed protein product
[Arabidopsis thaliana] gi|15240016|ref|NP_201461.1|
expressed protein [Arabidopsis thaliana]
Length = 614
Score = 651 bits (1679), Expect = 0.0
Identities = 360/632 (56%), Positives = 444/632 (69%), Gaps = 37/632 (5%)
Query: 1 MLGLSFTPRHKRSNSV--PDKNRVENDN-AESLKVSAQRMKLDEGYITECAQARKKGAPT 57
ML L HKRS S P+K RVE D + S ++QRMKLD G E K
Sbjct: 12 MLDLRIVQNHKRSKSASFPEKKRVEGDKTSNSSHEASQRMKLDMGRSNES----KHNQYH 67
Query: 58 SEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPS---LVNSSEMIMPKPATELIKE 114
S ++LKQE LE RLQDQF+VRC LEKALGY++ S L ++++ MPKPAT+LIK+
Sbjct: 68 SNTETSLKQEITHLETRLQDQFKVRCALEKALGYRTASSYVLTETNDIAMPKPATDLIKD 127
Query: 115 IAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYP-LTTPRAQVVKASLPEVL 173
+AVLEMEV++LEQ+LLSLYRKAF+QQ+SSVSP + + K P +TTPR ++ + +
Sbjct: 128 VAVLEMEVIHLEQYLLSLYRKAFEQQISSVSPNLENKKPKSPPVTTPRRRLDFSEDDDTP 187
Query: 174 TKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSA 233
+K + TV D N K+ + +D R HS S + +
Sbjct: 188 SKTDQHTVPLLDDN-------------QNQSKKTEIAAVDRDQMDPSFRRSHSQRSAFGS 234
Query: 234 FTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLS 293
R + P + K+ R+CHSQPL + N+ISLAEHLGTRISDHV P TPNKLS
Sbjct: 235 --RKASPEDSWGKASRSCHSQPLYVQN-----GDNLISLAEHLGTRISDHV-PETPNKLS 286
Query: 294 EDMVKCISAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDAR 353
E MVKC+S IYCKLA+PP L GL SP+SSLSS SAFS DQ D SPGF N+SSFD R
Sbjct: 287 EGMVKCMSEIYCKLAEPPSVLHRGLSSPNSSLSS-SAFSPSDQYDTSSPGFGNSSSFDVR 345
Query: 354 LDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKH 413
LDNSFH EG ++FSGPYS++VEV IYR+ K ++E L Q F+SLI RLEEVDPRKLKH
Sbjct: 346 LDNSFHVEGEKDFSGPYSSIVEVLCIYRDAKKASEVEDLLQNFKSLISRLEEVDPRKLKH 405
Query: 414 EEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCR 473
EEKLAFWIN+HNALVMHAFLAYGIPQNNVKRV LLLKAAYN+GG T+SA+ IQS+IL C+
Sbjct: 406 EEKLAFWINVHNALVMHAFLAYGIPQNNVKRVLLLLKAAYNIGGHTISAEAIQSSILGCK 465
Query: 474 MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQ 533
MS PGQWLR+ F+SR KF++GD R YAI++PEPL HFAL SG+HSDPAVRVYTPKR+ Q
Sbjct: 466 MSHPGQWLRLLFASR-KFKAGDERLAYAIDHPEPLLHFALTSGSHSDPAVRVYTPKRIQQ 524
Query: 534 ELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVK 593
ELE +K+EYIR L +R+ Q++L+PKLVETF KDS +CP G+ EM+ S+PES RK VK
Sbjct: 525 ELETSKEEYIRMNLSIRK--QRILLPKLVETFAKDSGLCPAGLTEMVNRSIPESSRKCVK 582
Query: 594 KCH-QLAKSRKCIEWIPHNFTFRYLISKDVLK 624
+C +K RK I+WIPH+FTFRYLI ++ K
Sbjct: 583 RCQSSTSKPRKTIDWIPHSFTFRYLILREAAK 614
>ref|XP_469918.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|40539066|gb|AAR87323.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 644
Score = 502 bits (1293), Expect = e-140
Identities = 282/625 (45%), Positives = 398/625 (63%), Gaps = 18/625 (2%)
Query: 9 RHKRSNSVPDKNRVENDNAESLKVSAQRMKLDEGYITECAQARKKGAPTSEIHSTLKQET 68
RHKRS S + ++ S K Q K D + + + P S L++E
Sbjct: 21 RHKRSKSDLEDMCAKDALYASDKTCVQP-KPDAVKVKVKSDINAEVQPGRGAQSFLRKEI 79
Query: 69 LQLERRLQDQFEVRCTLEKALGYKSPSL-VN-SSEMIMPKPATELIKEIAVLEMEVVYLE 126
LQLE+ L+DQ +R LEKALG + + VN S+E MPK A ELI+EIA LE+EV +E
Sbjct: 80 LQLEKHLKDQQVMRGALEKALGPNATAAPVNVSNENPMPKDAKELIREIATLELEVKNME 139
Query: 127 QHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQSNDH 186
Q+LL+LYRKAF+QQ+ + SPP + K P+ A +S P L K ++
Sbjct: 140 QYLLTLYRKAFEQQVPTFSPPDHRGAPKPPVP---AMAAVSSQPVQLQKSPSVKASRKNN 196
Query: 187 ELEAMRKEH-----DRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYS-AFTRSSPP 240
+AM + R D + +C+ + +S V RC S++S +R SP
Sbjct: 197 RADAMLRSSYPPPSRRTLNDPVMTDCSTSGCSSRLGESDVLRCQSAVSYRGICSSRISPS 256
Query: 241 AEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCI 300
+ LA++LR+CHSQP S +E ++ +ISLAE+LGT ++DH+ P TPN LSE+MV+C+
Sbjct: 257 EDSLARALRSCHSQPFSFLEEGESTAAGVISLAEYLGTNVADHI-PETPNNLSEEMVRCM 315
Query: 301 SAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHG 360
+ +YCKLADPP+ SP+SS SS SA S GD+WSP ++ S+ D+RL N FH
Sbjct: 316 AGVYCKLADPPLVHHGSSSSPTSSFSSTSAISPQYLGDIWSPNYKRESTLDSRLINPFHV 375
Query: 361 EGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFW 420
EG++EFSGPY+TMVEV I R+ + ++E L Q ++ ++ RLE VD R++ +EEK+AFW
Sbjct: 376 EGLKEFSGPYNTMVEVPLICRDSRRLKEVEDLLQTYKLILYRLETVDLRRMTNEEKIAFW 435
Query: 421 INIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQW 480
+NIHNA +MHA+L YG+PQNN+K+ LL+KAA + GR+++ IQS +L C PGQW
Sbjct: 436 VNIHNAQLMHAYLKYGVPQNNLKKTSLLVKAACKIAGRSINVAVIQSMVLGCNTYCPGQW 495
Query: 481 LRMFFSSRTKFRSGDGR---QTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEV 537
LR R K + G Q +A+ EPL FALCSG+HSDPAVRVYTPKR+F +LE
Sbjct: 496 LRTLLHPRIKSKVGKVGHVWQAFAVAQSEPLLRFALCSGSHSDPAVRVYTPKRLFHQLEA 555
Query: 538 AKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQ 597
AK+E+IRAT+G+ K+QK+L+PKLVE + KD + G+++M+Q LPESLR +++KC Q
Sbjct: 556 AKEEFIRATVGI-WKEQKILLPKLVEAYAKDVKLSSQGLVDMVQRYLPESLRMAMQKCQQ 614
Query: 598 LAKSRKCIEWIPHNFTFRYLISKDV 622
++S K IEW+PHN FRYL+S+D+
Sbjct: 615 -SRSSKIIEWVPHNLNFRYLLSRDL 638
>gb|AAC17092.2| unknown protein [Arabidopsis thaliana] gi|15224054|ref|NP_179950.1|
expressed protein [Arabidopsis thaliana]
Length = 707
Score = 451 bits (1160), Expect = e-125
Identities = 239/425 (56%), Positives = 311/425 (72%), Gaps = 24/425 (5%)
Query: 201 DNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFT-RSSPPAEPLAKSLRACHSQPLSMM 259
+ L+ +C + E++ +DS V RC SSL+Q S F R SPP + S+ ACHSQPLS+
Sbjct: 306 NRLKDQC---FIEKEDIDSCVRRCQSSLNQRSTFNNRISPPED----SVFACHSQPLSIH 358
Query: 260 EYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLL 319
EY + SN SLAEH+GTRISDH+ +TPNKLSE+M+KC SAIY KLADPP ++ G
Sbjct: 359 EYIQN-GSNDASLAEHMGTRISDHIF-MTPNKLSEEMIKCASAIYSKLADPP-SINHGFS 415
Query: 320 SPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWI 379
SPSSS SS S FS DQ DMWSP FR NSSFD + EFSGPYS+M+EVS I
Sbjct: 416 SPSSSPSSTSEFSPQDQYDMWSPSFRKNSSFDDQF----------EFSGPYSSMIEVSHI 465
Query: 380 YREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQ 439
+R K D++ +++ F L+ +LE VDPRKL H+EKLAFWIN+HNALVMH FLA GIPQ
Sbjct: 466 HRNR-KRRDLDLMNRNFSLLLKQLESVDPRKLTHQEKLAFWINVHNALVMHTFLANGIPQ 524
Query: 440 NNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQT 499
NN KR LL K AY +GGR VS + IQS ILR +M RPGQWL++ + KFR+GD Q
Sbjct: 525 NNGKRFLLLSKPAYKIGGRMVSLEAIQSYILRIKMPRPGQWLKLLLIPK-KFRTGDEHQE 583
Query: 500 YAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVP 559
Y++E+ EPL +FALCSGNHSDPA+RV+TPK ++QELE AK+EYIRAT GV +KDQK+++P
Sbjct: 584 YSLEHSEPLLYFALCSGNHSDPAIRVFTPKGIYQELETAKEEYIRATFGV-KKDQKLVLP 642
Query: 560 KLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLIS 619
K++E+F+KDS + +MEMIQE LPE+++K++KK + + +EW PHNF FRYLI+
Sbjct: 643 KIIESFSKDSGLGQAALMEMIQECLPETMKKTIKKLNSGRSRKSIVEWTPHNFVFRYLIA 702
Query: 620 KDVLK 624
+++++
Sbjct: 703 RELVR 707
Score = 140 bits (354), Expect = 9e-32
Identities = 87/163 (53%), Positives = 116/163 (70%), Gaps = 12/163 (7%)
Query: 9 RHKRSNS--VPDKNRVENDNA-ESLKVSAQRMKLDEGYITECA----QARKKGAPTSEIH 61
RHKRS S VP+K ++E++N+ +S ++QR+KLD + C + +K +P +
Sbjct: 12 RHKRSKSCTVPEKKKLEDENSIDSSLDASQRLKLD---LPRCGDKSFEMKKDLSPDVKFK 68
Query: 62 STLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEME 121
S+LKQE +LE+RLQ+QF+VR LEKALGYK+PS + PKP TELIKEIAVLE+E
Sbjct: 69 SSLKQEIQELEKRLQNQFDVRGALEKALGYKTPSRDIKGDST-PKPPTELIKEIAVLELE 127
Query: 122 VVYLEQHLLSLYRKAFDQQLSSVSPPT-KEESLKYPLTTPRAQ 163
V +LEQ+LLSLYRKAFDQQ SSVSPPT K++S P +T R +
Sbjct: 128 VSHLEQYLLSLYRKAFDQQTSSVSPPTSKQQSSCSPKSTLRGK 170
>pir||T02421 hypothetical protein At2g23700 [imported] - Arabidopsis thaliana
Length = 722
Score = 451 bits (1160), Expect = e-125
Identities = 239/425 (56%), Positives = 311/425 (72%), Gaps = 24/425 (5%)
Query: 201 DNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFT-RSSPPAEPLAKSLRACHSQPLSMM 259
+ L+ +C + E++ +DS V RC SSL+Q S F R SPP + S+ ACHSQPLS+
Sbjct: 321 NRLKDQC---FIEKEDIDSCVRRCQSSLNQRSTFNNRISPPED----SVFACHSQPLSIH 373
Query: 260 EYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLL 319
EY + SN SLAEH+GTRISDH+ +TPNKLSE+M+KC SAIY KLADPP ++ G
Sbjct: 374 EYIQN-GSNDASLAEHMGTRISDHIF-MTPNKLSEEMIKCASAIYSKLADPP-SINHGFS 430
Query: 320 SPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWI 379
SPSSS SS S FS DQ DMWSP FR NSSFD + EFSGPYS+M+EVS I
Sbjct: 431 SPSSSPSSTSEFSPQDQYDMWSPSFRKNSSFDDQF----------EFSGPYSSMIEVSHI 480
Query: 380 YREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQ 439
+R K D++ +++ F L+ +LE VDPRKL H+EKLAFWIN+HNALVMH FLA GIPQ
Sbjct: 481 HRNR-KRRDLDLMNRNFSLLLKQLESVDPRKLTHQEKLAFWINVHNALVMHTFLANGIPQ 539
Query: 440 NNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQT 499
NN KR LL K AY +GGR VS + IQS ILR +M RPGQWL++ + KFR+GD Q
Sbjct: 540 NNGKRFLLLSKPAYKIGGRMVSLEAIQSYILRIKMPRPGQWLKLLLIPK-KFRTGDEHQE 598
Query: 500 YAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVP 559
Y++E+ EPL +FALCSGNHSDPA+RV+TPK ++QELE AK+EYIRAT GV +KDQK+++P
Sbjct: 599 YSLEHSEPLLYFALCSGNHSDPAIRVFTPKGIYQELETAKEEYIRATFGV-KKDQKLVLP 657
Query: 560 KLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLIS 619
K++E+F+KDS + +MEMIQE LPE+++K++KK + + +EW PHNF FRYLI+
Sbjct: 658 KIIESFSKDSGLGQAALMEMIQECLPETMKKTIKKLNSGRSRKSIVEWTPHNFVFRYLIA 717
Query: 620 KDVLK 624
+++++
Sbjct: 718 RELVR 722
Score = 131 bits (329), Expect = 7e-29
Identities = 85/177 (48%), Positives = 116/177 (65%), Gaps = 25/177 (14%)
Query: 9 RHKRSNS--VPDKNRVENDNA-ESLKVSAQRMKLDEGYITECA----QARKKGAPTSEIH 61
RHKRS S VP+K ++E++N+ +S ++QR+KLD + C + +K +P +
Sbjct: 12 RHKRSKSCTVPEKKKLEDENSIDSSLDASQRLKLD---LPRCGDKSFEMKKDLSPDVKFK 68
Query: 62 STLKQETL--------------QLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKP 107
S+LKQE+ +LE+RLQ+QF+VR LEKALGYK+PS + +P
Sbjct: 69 SSLKQESKIICLIHIVGYIQIQELEKRLQNQFDVRGALEKALGYKTPSRDIKGDSTPKQP 128
Query: 108 ATELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPT-KEESLKYPLTTPRAQ 163
TELIKEIAVLE+EV +LEQ+LLSLYRKAFDQQ SSVSPPT K++S P +T R +
Sbjct: 129 PTELIKEIAVLELEVSHLEQYLLSLYRKAFDQQTSSVSPPTSKQQSSCSPKSTLRGK 185
>dbj|BAC42707.2| unknown protein [Arabidopsis thaliana]
Length = 707
Score = 449 bits (1156), Expect = e-125
Identities = 238/425 (56%), Positives = 311/425 (73%), Gaps = 24/425 (5%)
Query: 201 DNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFT-RSSPPAEPLAKSLRACHSQPLSMM 259
+ L+ +C + E++ +DS V RC SSL+Q S F R SPP + S+ ACHSQPLS+
Sbjct: 306 NRLKDQC---FIEKEDIDSCVRRCQSSLNQRSTFNNRISPPED----SVFACHSQPLSIH 358
Query: 260 EYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLL 319
EY + SN SLAEH+GTRISDH+ +TPNKLSE+M+KC SAIY KLADPP ++ G
Sbjct: 359 EYIQN-GSNDASLAEHMGTRISDHIF-MTPNKLSEEMIKCASAIYSKLADPP-SINHGFS 415
Query: 320 SPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWI 379
SPSSS SS S FS DQ DMWSP FR NSSFD + EFSGPYS+M+EVS I
Sbjct: 416 SPSSSPSSTSEFSPQDQYDMWSPSFRKNSSFDDQF----------EFSGPYSSMIEVSHI 465
Query: 380 YREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQ 439
+R K D++ +++ F L+ +LE VDPRKL H+EKLAFWIN+HNALVMH FLA GIPQ
Sbjct: 466 HRNR-KRRDLDLMNRNFSLLLKQLESVDPRKLTHQEKLAFWINVHNALVMHTFLANGIPQ 524
Query: 440 NNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQT 499
NN KR LL K AY +GGR VS + IQS ILR +M RPGQWL++ + KFR+GD Q
Sbjct: 525 NNGKRFLLLSKPAYKIGGRMVSLEAIQSYILRIKMPRPGQWLKLLLIPK-KFRTGDEHQE 583
Query: 500 YAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVP 559
Y++E+ EPL +FALCSGNHSDPA+RV+TPK ++QELE AK+EYIRAT GV +KDQ++++P
Sbjct: 584 YSLEHSEPLLYFALCSGNHSDPAIRVFTPKGIYQELETAKEEYIRATFGV-KKDQELVLP 642
Query: 560 KLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLIS 619
K++E+F+KDS + +MEMIQE LPE+++K++KK + + +EW PHNF FRYLI+
Sbjct: 643 KIIESFSKDSGLGQAALMEMIQECLPETMKKTIKKLNSGRSRKSIVEWTPHNFVFRYLIA 702
Query: 620 KDVLK 624
+++++
Sbjct: 703 RELVR 707
Score = 140 bits (354), Expect = 9e-32
Identities = 87/163 (53%), Positives = 116/163 (70%), Gaps = 12/163 (7%)
Query: 9 RHKRSNS--VPDKNRVENDNA-ESLKVSAQRMKLDEGYITECA----QARKKGAPTSEIH 61
RHKRS S VP+K ++E++N+ +S ++QR+KLD + C + +K +P +
Sbjct: 12 RHKRSKSCTVPEKKKLEDENSIDSSLDASQRLKLD---LPRCGDKSFEMKKDLSPDVKFK 68
Query: 62 STLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEME 121
S+LKQE +LE+RLQ+QF+VR LEKALGYK+PS + PKP TELIKEIAVLE+E
Sbjct: 69 SSLKQEIQELEKRLQNQFDVRGALEKALGYKTPSRDIKGDST-PKPPTELIKEIAVLELE 127
Query: 122 VVYLEQHLLSLYRKAFDQQLSSVSPPT-KEESLKYPLTTPRAQ 163
V +LEQ+LLSLYRKAFDQQ SSVSPPT K++S P +T R +
Sbjct: 128 VSHLEQYLLSLYRKAFDQQTSSVSPPTSKQQSSCSPKSTLRGK 170
>gb|AAO59425.1| putative ternary complex factor MIP1 [Antirrhinum majus]
Length = 555
Score = 440 bits (1131), Expect = e-122
Identities = 263/575 (45%), Positives = 365/575 (62%), Gaps = 30/575 (5%)
Query: 51 RKKGAPTSEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATE 110
+K+ +E S LK+E LQL++ L+ Q VR LEKAL + P N + + +PA
Sbjct: 10 KKRQLSPNEAQSLLKEEILQLQKELEGQTVVRSALEKALNCQ-PLCYNPTYESLSQPAEN 68
Query: 111 LIKEIAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLP 170
LIKEIA+LE+EV YLE++LLSLYR++F ++LS++ K P + K +
Sbjct: 69 LIKEIALLELEVEYLEKYLLSLYRRSFTKRLSTLQAVDKRPK-------PNVETHKRTFS 121
Query: 171 EVLTKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQ 230
EV K +V+ D + E N D KE N + EE+ DSG+ R SSLSQ
Sbjct: 122 EV-PKTNLASVRE-DSVISCSTLE---NTTDMFTKERNDIFEEEQLYDSGICRSQSSLSQ 176
Query: 231 YSAFT-RSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTP 289
+SA + R SP E LA+ + + HS PL M+E A +++ S AE+LG+ P
Sbjct: 177 HSACSFRVSPSFESLARGVDSYHSLPLWMLERAEDATAHANS-AEYLGSE--------AP 227
Query: 290 NKLSEDMVKCISAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSS 349
N LSE+M+KCIS IYC L+DPP+ G S S LS P+ FS Q S N+S
Sbjct: 228 NYLSEEMIKCISTIYCHLSDPPL-FNHGFNSVSL-LSPPTTFSPQAQHGKCS---EENTS 282
Query: 350 FDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPR 409
F + ++N F+ E +EF+G +MVEV + R+ +E+L Q +R LI +L EVDP
Sbjct: 283 FGSWMNNPFNVEESKEFNGSLYSMVEVQGLLRDSQSLDSVEELLQNYRFLISKLGEVDPG 342
Query: 410 KLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTI 469
KLKH+EKLAFWIN+HN+LVMHAFL YGIPQ N+KR+ L LKAAYNVGG T+S DTIQS+I
Sbjct: 343 KLKHDEKLAFWINVHNSLVMHAFLVYGIPQGNMKRISLALKAAYNVGGHTISVDTIQSSI 402
Query: 470 LRCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPK 529
LRCR+ RP QWL+ F + KF++ D R+ YAI + EP FALCSG +SD VR+YT K
Sbjct: 403 LRCRLPRPSQWLQSLFFPKQKFKACDPRKVYAIRHSEPRLRFALCSGCNSDAPVRIYTSK 462
Query: 530 RVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLR 589
+VFQELE+AK+EYI+ + V K+Q++LVPK VE + K+ + P G+ EM+Q S+P+SLR
Sbjct: 463 KVFQELEIAKEEYIQMNVSV-HKEQRLLVPKNVEYYAKEMGLSPQGIAEMLQHSMPDSLR 521
Query: 590 KSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDVLK 624
K+ +Q K K I+++P NFTFR+L++ ++++
Sbjct: 522 KNFSHNYQ-GKLWKKIDYVPQNFTFRFLLTNELVR 555
>ref|XP_470631.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|20330763|gb|AAM19126.1| Hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 590
Score = 383 bits (984), Expect = e-105
Identities = 223/574 (38%), Positives = 336/574 (57%), Gaps = 37/574 (6%)
Query: 57 TSEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIA 116
T+E+ +L QE LE+RL DQF +R LEKALGYK ++ +S+E +PKP ELIKEIA
Sbjct: 35 TTELPCSLIQEVQHLEKRLNDQFAMRRALEKALGYKPCAIHSSNESCIPKPTEELIKEIA 94
Query: 117 VLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKE 176
VLE+EV+ LEQHLL+LYRKAFDQQ+ SVS E K + + +S + T
Sbjct: 95 VLELEVICLEQHLLALYRKAFDQQICSVSSSCDMEINKQSARSFSGILTGSSELDFSTPR 154
Query: 177 EYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFT- 235
++ +QS+ + ++ L E +K +G+ R HSSL Q S +
Sbjct: 155 KHQLLQSSGMVMAR------KSTPTTLTSETRTSHYNDK---TGIGRSHSSLLQRSICSA 205
Query: 236 RSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSED 295
R SP A LA++L+ CH+ PLS +E + I+SLA+ LGTRI+DHV P TPNK+SED
Sbjct: 206 RVSPSANNLARALKPCHTLPLSFVEEGKCMDPGIVSLADILGTRIADHV-PQTPNKISED 264
Query: 296 MVKCISAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLD 355
M+KCI++IY ++ D P SP SS SS S S GD+WSP R +A D
Sbjct: 265 MIKCIASIYIRIRDFNAVQHPFFPSPCSSFSSASGLSSKYTGDIWSPRCRKEGYIEAWQD 324
Query: 356 NSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEE 415
++ FS Y +++EVS + + + D++ + K++SL+ LE D +K+EE
Sbjct: 325 DALGTGESRYFSQQYDSVIEVSALCKGAQRSADVKDMLHKYKSLVQLLESADLNGMKNEE 384
Query: 416 KLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMS 475
K+AFWIN+HNA++MH +Y + G+ V+ + I+ IL CR+
Sbjct: 385 KIAFWINVHNAMMMH--------------------LSYLISGQRVNPELIEYHILCCRVH 424
Query: 476 RPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQEL 535
P QWLR+ + K + + Q +A++ PEPL HFAL SG+HSDP VR+Y P+R+ Q+L
Sbjct: 425 SPTQWLRLLLYPKWKSKEKEDLQGFAVDRPEPLVHFALSSGSHSDPVVRLYRPERLLQQL 484
Query: 536 EVAKDEYIRATLGV------RRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLR 589
E A+DE++RA +GV R + +L+PKL+E +++D+ + ++ ++ LPE LR
Sbjct: 485 EAARDEFVRANVGVRGGRRGRGRRVLLLLPKLLEPYSRDAGLGAHDLLRAVESCLPEPLR 544
Query: 590 KSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDVL 623
+ ++ + +EW PHN FRYL++++++
Sbjct: 545 PAAQQAARSRGGGGGVEWRPHNPAFRYLLARELV 578
>dbj|BAB03099.1| unnamed protein product [Arabidopsis thaliana]
Length = 524
Score = 325 bits (834), Expect = 2e-87
Identities = 215/546 (39%), Positives = 308/546 (56%), Gaps = 91/546 (16%)
Query: 76 QDQFEVRCTLEKALGYKSPSLVNSSE-MIMPKPATELIKEIAVLEMEVVYLEQHLLSLYR 134
+D+ VR ++ LG K + +E +PK A +L++EIA LE++V+YLE +LL LYR
Sbjct: 67 EDEVLVRREMKTVLGRKLFLEDSKNEDSSIPKEAKKLVEEIAGLELQVMYLETYLLLLYR 126
Query: 135 KAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQSNDHELEAMRKE 194
+ F+ +++S K E + ++ LE +
Sbjct: 127 RFFNNKITS-------------------------------KLESEEKERSEDLLECTKL- 154
Query: 195 HDRNELDNLRKE-CNGGWPEEKHLDSGVYRCHSSLSQYSAFT-RSSPPAEPLAKSLRACH 252
+D+ +K C+ P++ DSG++R HSSLS S ++ R SP A +
Sbjct: 155 -----IDSPKKGVCS---PQKLVEDSGIFRSHSSLSHCSGYSFRMSPHAMDSSYH----R 202
Query: 253 SQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPM 312
S P SM+E + + E +GT +S++V +PN LSE+MVKCIS + +L DP
Sbjct: 203 SLPFSMLEQS--------DIDELIGTYVSENVHK-SPNSLSEEMVKCISELCRQLVDP-- 251
Query: 313 TLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYST 372
G L SSP FR E ++ S PY
Sbjct: 252 ----GSLDNDLESSSP---------------FRGK-------------EPLKIISRPYDK 279
Query: 373 MVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAF 432
++ V I R+ K +E + FRSL+ +LE V+PRKL HEEKLAFWINIHN+LVMH+
Sbjct: 280 LLMVKSISRDSEKLNAVEPALKHFRSLVNKLEGVNPRKLNHEEKLAFWINIHNSLVMHSI 339
Query: 433 LAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFR 492
L YG P+N++KRV LLKAAYNVGGR+++ DTIQ++IL CR+SRPG R F+SR+K R
Sbjct: 340 LVYGNPKNSMKRVSGLLKAAYNVGGRSLNLDTIQTSILGCRVSRPGLVFRFLFASRSKGR 399
Query: 493 SGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRK 552
+GD + YAI + E L HFALCSG+ SDP+VR+YTPK V ELE ++EY+R+ LG+ K
Sbjct: 400 AGDLGRDYAITHRESLLHFALCSGSLSDPSVRIYTPKNVMMELECGREEYVRSNLGI-SK 458
Query: 553 DQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNF 612
D K+L+PKLVE + KD+++C GV++MI + LP R ++KC R I+WI H+F
Sbjct: 459 DNKILLPKLVEIYAKDTELCNVGVLDMIGKCLPCEARDRIQKCRNKKHGRFSIDWIAHDF 518
Query: 613 TFRYLI 618
F L+
Sbjct: 519 RFGLLL 524
>ref|NP_188520.1| expressed protein [Arabidopsis thaliana]
Length = 815
Score = 295 bits (756), Expect = 2e-78
Identities = 222/617 (35%), Positives = 325/617 (51%), Gaps = 117/617 (18%)
Query: 7 TPRHKRSNS--VPDKNRVENDNAESLKVSAQRMKLDEGYITECAQAR-----KKGAPTSE 59
T HKRS S VP K+ ++E+L + +DE E Q + +K P
Sbjct: 5 TSNHKRSRSDPVPRKSLKNKKSSEAL----YNIHMDELLQREIEQLKSLLKSRKLHPGGR 60
Query: 60 IHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLE 119
S+ + + L E + + E++ L + L + +SS +PK A +L++EIA LE
Sbjct: 61 RVSSDEIDLLLQEDEVLVRREMKTVLGRKLFLEDSKNEDSS---IPKEAKKLVEEIAGLE 117
Query: 120 MEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYP 179
++V+YLE +LL LYR+ F+ +++S K E
Sbjct: 118 LQVMYLETYLLLLYRRFFNNKITS-------------------------------KLESE 146
Query: 180 TVQSNDHELEAMRKEHDRNELDNLRKE-CNGGWPEEKHLDSGVYRCHSSLSQYSAFT-RS 237
+ ++ LE + +D+ +K C+ P++ DSG++R HSSLS S ++ R
Sbjct: 147 EKERSEDLLECTKL------IDSPKKGVCS---PQKLVEDSGIFRSHSSLSHCSGYSFRM 197
Query: 238 SPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMV 297
SP A + S P SM+E + + E +GT +S++V +PN LSE+MV
Sbjct: 198 SPHAMDSSYH----RSLPFSMLEQS--------DIDELIGTYVSENVHK-SPNSLSEEMV 244
Query: 298 KCISAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNS 357
KCIS + +L DP G L SSP FR
Sbjct: 245 KCISELCRQLVDP------GSLDNDLESSSP---------------FRGK---------- 273
Query: 358 FHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKL 417
E ++ S PY ++ V I R+ K +E + FRSL+ +LE V+PRKL HEEKL
Sbjct: 274 ---EPLKIISRPYDKLLMVKSISRDSEKLNAVEPALKHFRSLVNKLEGVNPRKLNHEEKL 330
Query: 418 AFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRP 477
AFWINIHN+LVMH+ L YG P+N++KRV LLKAAYNVGGR+++ DTIQ++IL CR+
Sbjct: 331 AFWINIHNSLVMHSILVYGNPKNSMKRVSGLLKAAYNVGGRSLNLDTIQTSILGCRV--- 387
Query: 478 GQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEV 537
R F+SR+K R+GD + YAI + E L HFALCSG+ SDP+ V ELE
Sbjct: 388 ---FRFLFASRSKGRAGDLGRDYAITHRESLLHFALCSGSLSDPS-------NVMMELEC 437
Query: 538 AKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQ 597
++EY+R+ LG+ KD K+L+PKLVE + KD+++C GV++MI + LP R ++KC
Sbjct: 438 GREEYVRSNLGI-SKDNKILLPKLVEIYAKDTELCNVGVLDMIGKCLPCEARDRIQKCRN 496
Query: 598 LAKSRKCIEWIPHNFTF 614
R I+WI H+F F
Sbjct: 497 KKHGRFSIDWIAHDFRF 513
>ref|XP_483337.1| ternary complex factor-like [Oryza sativa (japonica
cultivar-group)] gi|42408754|dbj|BAD09990.1| ternary
complex factor-like [Oryza sativa (japonica
cultivar-group)]
Length = 538
Score = 277 bits (708), Expect = 8e-73
Identities = 187/525 (35%), Positives = 272/525 (51%), Gaps = 78/525 (14%)
Query: 100 SEMIMPKPATELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTT 159
SE + K +L+ EIA LE EVV E HLLSLYR AFDQ L VS
Sbjct: 88 SEKTISKSIADLVWEIAALEEEVVRKELHLLSLYRAAFDQHL-GVS-------------- 132
Query: 160 PRAQVVKASLPEVLTKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDS 219
P V T+ + Q H+ + + L N+++ + P +
Sbjct: 133 ----------PRVSTQVD----QEIHHQKSRKKADEGALRLRNIKESASYNLPTVSTVSD 178
Query: 220 GVYRCHSSLSQYSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTR 279
+ +RSS LA L A S+AE+
Sbjct: 179 SKH----------GLSRSSSGHSSLANFLSA--------------------SIAEY---- 204
Query: 280 ISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGL--LSPSSSLSSPSAFSIGDQG 337
+P KLSED+V+CISA+YCKLA P LS S SS S FS+ +
Sbjct: 205 -----VPKISCKLSEDIVRCISAVYCKLASQPSQNLADFETLSTPSFSSSSSTFSLKHRV 259
Query: 338 DMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFR 397
D WSP N + +S + + E S Y+ M+ IY + K+ K+ + R
Sbjct: 260 DSWSPRCHYNVN-----TSSDKYDSLNEKSEQYNGMIICPRIYMDAEKFEYASKMLETVR 314
Query: 398 SLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGG 457
SLI RLE++DP K+ HEE+L FWINIHNALVMHAF+AYG+ + +K ++LKAAYNVGG
Sbjct: 315 SLIKRLEKIDPTKMAHEEQLCFWINIHNALVMHAFMAYGLQEKRMKNTDMILKAAYNVGG 374
Query: 458 RTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGN 517
+V+A IQ++I+ C+ R W+R F+ K SG YA+ PEPL+HFAL +G
Sbjct: 375 LSVNAQIIQNSIIGCQSHRTSVWVRTLFTPLKKSASGSSIHPYALHPPEPLAHFALSTGA 434
Query: 518 HSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVM 577
SDP VR+YT K+V +L+ A+ E+I+A++ VR+ Q + +PK++ + KD+ + ++
Sbjct: 435 ISDPPVRLYTAKKVNHQLDQARTEFIQASVIVRK--QTIFLPKVLHHYAKDAALELPDLV 492
Query: 578 EMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDV 622
EM E +PE+ +K +++C + + KC+EWIP +FRY I + +
Sbjct: 493 EMACEIMPEAQQKEIRQCLR-RRIDKCVEWIPFKSSFRYTIHRSL 536
>ref|NP_199549.2| expressed protein [Arabidopsis thaliana]
Length = 618
Score = 245 bits (625), Expect = 4e-63
Identities = 174/585 (29%), Positives = 294/585 (49%), Gaps = 78/585 (13%)
Query: 49 QARKKGAPTSEIHSTLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPA 108
Q K +S ++L+++ QL RLQ + +R LE+A+G S SL + A
Sbjct: 77 QMLTKNNVSSNDRASLERDVEQLHLRLQQEKSMRMVLERAMGRASSSL-SPGHRHFAGQA 135
Query: 109 TELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKAS 168
ELI EI +LE EV E H+LSLYR F+Q +S P+++ S
Sbjct: 136 NELITEIELLEAEVTNREHHVLSLYRSIFEQTVSRA--PSEQSS---------------- 177
Query: 169 LPEVLTKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSL 228
++ S H ++ ++ D N + N N +P + H+ +
Sbjct: 178 -----------SISSPAHHIKQPPRKQDPNVISNAFCSSNN-FPLKPW--------HAMV 217
Query: 229 SQYSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARG-CSSNIISLAEHLGTRISDHVLPV 287
+ + ++S + R C S A+ + +++ + DH+
Sbjct: 218 TLKDSSRKTSKKDQSSQFQFRNCIPSTTSCSSQAKSHFLKDSVTVKSPSQRTLKDHLYQC 277
Query: 288 TPNKLSEDMVKCISAIYCKLADPPMTLPPG--LLSPSSSLSSPSAFSIGDQGDMWSPGFR 345
PNKLSEDMVKC+S++Y L M+ P +LS SS+ + +I ++ WS
Sbjct: 278 -PNKLSEDMVKCMSSVYFWLCCSAMSADPEKRILSRSSTSNVIIPKNIMNEDRAWS---- 332
Query: 346 NNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEE 405
+MVEVSWI + ++ + +R L+ +LE
Sbjct: 333 ------------------------CRSMVEVSWISSDKKRFSQVTYAINNYRLLVEQLER 368
Query: 406 VDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTI 465
V +++ KLAFWINI+NAL+MHA+LAYG+P ++++R+ L K+AYN+GG ++A+TI
Sbjct: 369 VTINQMEGNAKLAFWINIYNALLMHAYLAYGVPAHSLRRLALFHKSAYNIGGHIINANTI 428
Query: 466 QSTILRCRMSRPGQWLRMFFSS--RTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAV 523
+ +I + R G+WL S+ R K + ++++ PEPL FALC G SDP +
Sbjct: 429 EYSIFCFQTPRNGRWLETIISTALRKKPAEDKVKSMFSLDKPEPLVCFALCIGALSDPVL 488
Query: 524 RVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQES 583
+ YT V +EL+ +K E++ A + V+ + +KVL+PK++E FTK++ + +M + ++
Sbjct: 489 KAYTASNVKEELDASKREFLGANVVVKMQ-KKVLLPKIIERFTKEASLSFDDLMRWLIDN 547
Query: 584 LPESLRKSVKKCHQ----LAKSRKCIEWIPHNFTFRYLISKDVLK 624
E L +S++KC Q K+ + +EW+P++ FRY+ SKD+++
Sbjct: 548 ADEKLGESIQKCVQGKPNNKKASQVVEWLPYSSKFRYVFSKDLME 592
>ref|NP_564131.1| expressed protein [Arabidopsis thaliana]
Length = 505
Score = 243 bits (621), Expect = 1e-62
Identities = 190/561 (33%), Positives = 283/561 (49%), Gaps = 104/561 (18%)
Query: 62 STLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEME 121
ST + +L +E+RL + C + PK + +L KEIA LE E
Sbjct: 37 STYTKPSLSIEKRLLTYQDSNCN------------------VTPKSSEDLRKEIASLEFE 78
Query: 122 VVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTV 181
++ EQ+LLSLYR AFD+Q+SS SP T+ + + L K E V
Sbjct: 79 ILRTEQYLLSLYRTAFDEQVSSFSPHTETSLVSN---------------QFLPKSEQSDV 123
Query: 182 QSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPA 241
S V+ H S S + PP
Sbjct: 124 TS-------------------------------------VFSYHYQASPASECSSLCPPR 146
Query: 242 EPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLG-TRISDHVLPVTPNKLSEDMVKCI 300
A SL+A LS E +R SS+ +L + LG T I D++ P++LSED+++CI
Sbjct: 147 SFQA-SLKA-----LSAREKSRYVSSSHTTLGDLLGSTLIVDNI--ANPSRLSEDILRCI 198
Query: 301 SAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHG 360
++YC L+ SPSS PS+ S + ++FD+ NS H
Sbjct: 199 CSVYCTLSSKARINSCLQASPSS----PSSVS-------------SKATFDSL--NSRHE 239
Query: 361 EGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFW 420
E +E + P ++E ++ + + + Q FRSL+ +LE+VDP ++K EEKLAFW
Sbjct: 240 ER-KEANVPGVVVIESLELHLDDGSFNHAAVMLQNFRSLVQKLEKVDPSRMKREEKLAFW 298
Query: 421 INIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQW 480
INIHNAL MHA+LAYG +N R +LKAAY+VGG +V+ IQS+IL R
Sbjct: 299 INIHNALTMHAYLAYGT--HNRARNTSVLKAAYDVGGYSVNPYIIQSSILGIRPHFSQPL 356
Query: 481 LRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKD 540
L+ FS K ++ + + YA+E PE L+HFAL SG +DP VRVYT VF++L +K+
Sbjct: 357 LQTLFSPSRKSKTCNVKHIYALEYPEALAHFALSSGFSTDPPVRVYTADCVFRDLRKSKE 416
Query: 541 EYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAK 600
E+IR + + + K+L+PK+V + KD + P +ME + LP+S +++ +K L K
Sbjct: 417 EFIRNNVRI-HNETKILLPKIVHYYAKDMSLEPSALMETTVKCLPDSTKRTAQKL--LKK 473
Query: 601 SRKCIEWIPHNFTFRYLISKD 621
+ IE+ P N +FRY+I ++
Sbjct: 474 KSRNIEYSPENSSFRYVIIQE 494
>pir||E86343 T22I11.12 protein - Arabidopsis thaliana gi|8886996|gb|AAF80656.1|
Contains similarity to an unknown protein F14G6.22
gi|6642679 from Arabidopsis thaliana gb|AC015450. ESTs
gb|AI994240 and gb|T42814 come from this gene
Length = 504
Score = 243 bits (621), Expect = 1e-62
Identities = 190/561 (33%), Positives = 283/561 (49%), Gaps = 104/561 (18%)
Query: 62 STLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEME 121
ST + +L +E+RL + C + PK + +L KEIA LE E
Sbjct: 36 STYTKPSLSIEKRLLTYQDSNCN------------------VTPKSSEDLRKEIASLEFE 77
Query: 122 VVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTV 181
++ EQ+LLSLYR AFD+Q+SS SP T+ + + L K E V
Sbjct: 78 ILRTEQYLLSLYRTAFDEQVSSFSPHTETSLVSN---------------QFLPKSEQSDV 122
Query: 182 QSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSPPA 241
S V+ H S S + PP
Sbjct: 123 TS-------------------------------------VFSYHYQASPASECSSLCPPR 145
Query: 242 EPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLG-TRISDHVLPVTPNKLSEDMVKCI 300
A SL+A LS E +R SS+ +L + LG T I D++ P++LSED+++CI
Sbjct: 146 SFQA-SLKA-----LSAREKSRYVSSSHTTLGDLLGSTLIVDNI--ANPSRLSEDILRCI 197
Query: 301 SAIYCKLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHG 360
++YC L+ SPSS PS+ S + ++FD+ NS H
Sbjct: 198 CSVYCTLSSKARINSCLQASPSS----PSSVS-------------SKATFDSL--NSRHE 238
Query: 361 EGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFW 420
E +E + P ++E ++ + + + Q FRSL+ +LE+VDP ++K EEKLAFW
Sbjct: 239 ER-KEANVPGVVVIESLELHLDDGSFNHAAVMLQNFRSLVQKLEKVDPSRMKREEKLAFW 297
Query: 421 INIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQW 480
INIHNAL MHA+LAYG +N R +LKAAY+VGG +V+ IQS+IL R
Sbjct: 298 INIHNALTMHAYLAYGT--HNRARNTSVLKAAYDVGGYSVNPYIIQSSILGIRPHFSQPL 355
Query: 481 LRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKD 540
L+ FS K ++ + + YA+E PE L+HFAL SG +DP VRVYT VF++L +K+
Sbjct: 356 LQTLFSPSRKSKTCNVKHIYALEYPEALAHFALSSGFSTDPPVRVYTADCVFRDLRKSKE 415
Query: 541 EYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAK 600
E+IR + + + K+L+PK+V + KD + P +ME + LP+S +++ +K L K
Sbjct: 416 EFIRNNVRI-HNETKILLPKIVHYYAKDMSLEPSALMETTVKCLPDSTKRTAQKL--LKK 472
Query: 601 SRKCIEWIPHNFTFRYLISKD 621
+ IE+ P N +FRY+I ++
Sbjct: 473 KSRNIEYSPENSSFRYVIIQE 493
>gb|AAN31099.1| At1g76620/F14G6_22 [Arabidopsis thaliana]
gi|18411161|ref|NP_565137.1| expressed protein
[Arabidopsis thaliana] gi|16974598|gb|AAL31203.1|
At1g76620/F14G6_22 [Arabidopsis thaliana]
Length = 527
Score = 235 bits (599), Expect = 4e-60
Identities = 159/403 (39%), Positives = 227/403 (55%), Gaps = 21/403 (5%)
Query: 226 SSLSQYSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGT-RISDHV 284
+SL+ Y A+ + SL+A LS E R S SL E LG+ I DH
Sbjct: 135 TSLTHYQAYQKPISYPRSFNTSLKA-----LSSREGTRVVSGTH-SLGELLGSSHIVDHN 188
Query: 285 LPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLL--SPSSSLSSPSAFSIGDQGDMWSP 342
+ PNKLSED+++CIS++YC L+ + SP SS +S S + D WS
Sbjct: 189 NFINPNKLSEDIMRCISSVYCTLSRGSTSTTSTCFPASPVSSNASTIFSSKFNYEDKWS- 247
Query: 343 GFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICR 402
N +S D L+ H + + ++E ++ + +G + Q FRSL+
Sbjct: 248 --LNGASEDHFLN---HCQDQDNVLPCGVVVIEALRVHLDDGSFGYAALMLQNFRSLVQN 302
Query: 403 LEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSA 462
LE+VDP ++K EEKLAFWINIHNALVMHA+LAYG +N R +LKAAY++GG ++
Sbjct: 303 LEKVDPSRMKREEKLAFWINIHNALVMHAYLAYG--THNRARNTSVLKAAYDIGGYRINP 360
Query: 463 DTIQSTIL--RCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSD 520
IQS+IL R + P L+ FS K ++ R YA+E PE L+HFA+ SG +D
Sbjct: 361 YIIQSSILGIRPHYTSPSPLLQTLFSPSRKSKTCSVRHIYALEYPEALAHFAISSGAFTD 420
Query: 521 PAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMI 580
P VRVYT R+F++L AK EYIR+ + V K K+L+PK+ + + KD M +ME
Sbjct: 421 PTVRVYTADRIFRDLRQAKQEYIRSNVRV-YKGTKILLPKIFQHYVKDMSMDVSKLMEAT 479
Query: 581 QESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDVL 623
+ LPE RK +KC + KS K EW+P N +FRY+I+ +++
Sbjct: 480 SQCLPEDARKIAEKCLKEKKS-KNFEWLPENLSFRYVIAGELV 521
Score = 60.8 bits (146), Expect = 1e-07
Identities = 68/216 (31%), Positives = 100/216 (45%), Gaps = 44/216 (20%)
Query: 98 NSSEMIMPKPATELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQL---------------- 141
NSS ++PK + EL KEIA +E+E++++E++LLSLYRK+F+QQL
Sbjct: 70 NSSLCLIPKSSEELKKEIASIEIEILHMERYLLSLYRKSFEQQLPNSFSNLSVTTTLPRS 129
Query: 142 --SSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQSNDHEL-EAMRKEH--D 196
+S + T ++ + P++ PR+ SL + L+ E V S H L E + H D
Sbjct: 130 VTTSPTSLTHYQAYQKPISYPRS--FNTSL-KALSSREGTRVVSGTHSLGELLGSSHIVD 186
Query: 197 RNELDNLRKECNGGWPEEKHLDSGVYRCHSS----LSQYSAFTRSSP-PAEPLAKSLRAC 251
N N K L + RC SS LS+ S T S+ PA P++ +
Sbjct: 187 HNNFINPNK-----------LSEDIMRCISSVYCTLSRGSTSTTSTCFPASPVSSNASTI 235
Query: 252 HSQPLSMMEYARGCSSNIISLAEHLG-TRISDHVLP 286
S S Y S N S L + D+VLP
Sbjct: 236 FS---SKFNYEDKWSLNGASEDHFLNHCQDQDNVLP 268
>gb|AAG51961.1| unknown protein; 93089-95433 [Arabidopsis thaliana]
gi|25372914|pir||D96794 unknown protein F14G6.22
[imported] - Arabidopsis thaliana
Length = 509
Score = 213 bits (543), Expect = 1e-53
Identities = 150/403 (37%), Positives = 215/403 (53%), Gaps = 39/403 (9%)
Query: 226 SSLSQYSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGT-RISDHV 284
+SL+ Y A+ + SL+A LS E R S SL E LG+ I DH
Sbjct: 135 TSLTHYQAYQKPISYPRSFNTSLKA-----LSSREGTRVVSGTH-SLGELLGSSHIVDHN 188
Query: 285 LPVTPNKLSEDMVKCISAIYCKLADPPMTLPPGLL--SPSSSLSSPSAFSIGDQGDMWSP 342
+ PNKLSED+++CIS++YC L+ + SP SS +S S + D WS
Sbjct: 189 NFINPNKLSEDIMRCISSVYCTLSRGSTSTTSTCFPASPVSSNASTIFSSKFNYEDKWS- 247
Query: 343 GFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICR 402
N +S D L+ H + + ++E ++ + +G + Q FRSL+
Sbjct: 248 --LNGASEDHFLN---HCQDQDNVLPCGVVVIEALRVHLDDGSFGYAALMLQNFRSLVQN 302
Query: 403 LEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSA 462
LE+VDP ++K EEKLAFWINIHNALVMHA AY++GG ++
Sbjct: 303 LEKVDPSRMKREEKLAFWINIHNALVMHA--------------------AYDIGGYRINP 342
Query: 463 DTIQSTILRCR--MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSD 520
IQS+IL R + P L+ FS K ++ R YA+E PE L+HFA+ SG +D
Sbjct: 343 YIIQSSILGIRPHYTSPSPLLQTLFSPSRKSKTCSVRHIYALEYPEALAHFAISSGAFTD 402
Query: 521 PAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMI 580
P VRVYT R+F++L AK EYIR+ + V K K+L+PK+ + + KD M +ME
Sbjct: 403 PTVRVYTADRIFRDLRQAKQEYIRSNVRV-YKGTKILLPKIFQHYVKDMSMDVSKLMEAT 461
Query: 581 QESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDVL 623
+ LPE RK +KC + KS K EW+P N +FRY+I+ +++
Sbjct: 462 SQCLPEDARKIAEKCLKEKKS-KNFEWLPENLSFRYVIAGELV 503
Score = 60.8 bits (146), Expect = 1e-07
Identities = 68/216 (31%), Positives = 100/216 (45%), Gaps = 44/216 (20%)
Query: 98 NSSEMIMPKPATELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQL---------------- 141
NSS ++PK + EL KEIA +E+E++++E++LLSLYRK+F+QQL
Sbjct: 70 NSSLCLIPKSSEELKKEIASIEIEILHMERYLLSLYRKSFEQQLPNSFSNLSVTTTLPRS 129
Query: 142 --SSVSPPTKEESLKYPLTTPRAQVVKASLPEVLTKEEYPTVQSNDHEL-EAMRKEH--D 196
+S + T ++ + P++ PR+ SL + L+ E V S H L E + H D
Sbjct: 130 VTTSPTSLTHYQAYQKPISYPRS--FNTSL-KALSSREGTRVVSGTHSLGELLGSSHIVD 186
Query: 197 RNELDNLRKECNGGWPEEKHLDSGVYRCHSS----LSQYSAFTRSSP-PAEPLAKSLRAC 251
N N K L + RC SS LS+ S T S+ PA P++ +
Sbjct: 187 HNNFINPNK-----------LSEDIMRCISSVYCTLSRGSTSTTSTCFPASPVSSNASTI 235
Query: 252 HSQPLSMMEYARGCSSNIISLAEHLG-TRISDHVLP 286
S S Y S N S L + D+VLP
Sbjct: 236 FS---SKFNYEDKWSLNGASEDHFLNHCQDQDNVLP 268
>gb|AAX23771.1| hypothetical protein At1g43020 [Arabidopsis thaliana]
gi|49660055|gb|AAT68318.1| hypothetical protein
At1g43020 [Arabidopsis thaliana]
Length = 445
Score = 202 bits (515), Expect = 2e-50
Identities = 126/368 (34%), Positives = 194/368 (52%), Gaps = 53/368 (14%)
Query: 257 SMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPP 316
S + +G S SLA+ LG ++PNKLSE++++ I I+ KL+D
Sbjct: 126 SKTDKIKGSDSGHPSLADLLGLNT------LSPNKLSEEILRSICVIHYKLSD------- 172
Query: 317 GLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEV 376
+ + + NS + E +E V +
Sbjct: 173 -------------------------------NGHNRLVKNSKNEEYGQELG------VGI 195
Query: 377 SWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYG 436
+Y + +E + Q FRSL+ +LE+VDP +L EEKLAFWINIHNALVMH ++ YG
Sbjct: 196 HKLYLDDYNLKSVESMLQNFRSLVQKLEKVDPARLGREEKLAFWINIHNALVMHEYIVYG 255
Query: 437 IPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDG 496
I ++ + + LKAA+N+GG V+A IQS+IL R LR FS ++ G
Sbjct: 256 IGEDTTSTL-MNLKAAFNIGGEWVNAYDIQSSILGIRPCHSPSRLRTLFSPAKSSKTSSG 314
Query: 497 RQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKV 556
R TYA++ EPL HFAL +G +DP VRVYT + +FQEL A+D YI+ ++G K+ K+
Sbjct: 315 RHTYALDYAEPLLHFALSTGASTDPMVRVYTSEGIFQELRQARDSYIQTSVGF-EKETKI 373
Query: 557 LVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRY 616
L+PK++ + KD+ + G + + E L ES R ++++ K +CI W+ FRY
Sbjct: 374 LLPKIIYNYAKDTSLDMGELFSTVSECLMESQRTAMRRIVN-KKQERCIRWVHDESKFRY 432
Query: 617 LISKDVLK 624
+I ++++
Sbjct: 433 VIHSELVR 440
Score = 38.5 bits (88), Expect = 0.66
Identities = 22/42 (52%), Positives = 29/42 (68%), Gaps = 3/42 (7%)
Query: 110 ELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQL---SSVSPPT 148
ELI EIA L+ E++ LE++LLSLYR +F L SS+ P T
Sbjct: 61 ELIGEIAELDTEILQLERYLLSLYRSSFGDHLPDNSSLPPCT 102
>gb|AAG51514.1| hypothetical protein [Arabidopsis thaliana]
Length = 426
Score = 184 bits (466), Expect = 1e-44
Identities = 121/368 (32%), Positives = 183/368 (48%), Gaps = 72/368 (19%)
Query: 257 SMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPP 316
S + +G S SLA+ LG ++PNKLSE++++ I I+ KL+D
Sbjct: 126 SKTDKIKGSDSGHPSLADLLGLNT------LSPNKLSEEILRSICVIHYKLSD------- 172
Query: 317 GLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEV 376
+ + + NS + E +E V +
Sbjct: 173 -------------------------------NGHNRLVKNSKNEEYGQELG------VGI 195
Query: 377 SWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYG 436
+Y + +E + Q FRSL+ +LE+VDP +L EEKLAFWINIHNALVMHA
Sbjct: 196 HKLYLDDYNLKSVESMLQNFRSLVQKLEKVDPARLGREEKLAFWINIHNALVMHA----- 250
Query: 437 IPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDG 496
A+N+GG V+A IQS+IL R LR FS ++ G
Sbjct: 251 ---------------AFNIGGEWVNAYDIQSSILGIRPCHSPSRLRTLFSPAKSSKTSSG 295
Query: 497 RQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKV 556
R TYA++ EPL HFAL +G +DP VRVYT + +FQEL A+D YI+ ++G K+ K+
Sbjct: 296 RHTYALDYAEPLLHFALSTGASTDPMVRVYTSEGIFQELRQARDSYIQTSVGF-EKETKI 354
Query: 557 LVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRY 616
L+PK++ + KD+ + G + + E L ES R ++++ K +CI W+ FRY
Sbjct: 355 LLPKIIYNYAKDTSLDMGELFSTVSECLMESQRTAMRRIVN-KKQERCIRWVHDESKFRY 413
Query: 617 LISKDVLK 624
+I ++++
Sbjct: 414 VIHSELVR 421
Score = 38.5 bits (88), Expect = 0.66
Identities = 22/42 (52%), Positives = 29/42 (68%), Gaps = 3/42 (7%)
Query: 110 ELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQL---SSVSPPT 148
ELI EIA L+ E++ LE++LLSLYR +F L SS+ P T
Sbjct: 61 ELIGEIAELDTEILQLERYLLSLYRSSFGDHLPDNSSLPPCT 102
>gb|AAG50823.1| hypothetical protein [Arabidopsis thaliana] gi|25405107|pir||G96497
hypothetical protein F2H10.4 [imported] - Arabidopsis
thaliana
Length = 406
Score = 184 bits (466), Expect = 1e-44
Identities = 121/368 (32%), Positives = 183/368 (48%), Gaps = 72/368 (19%)
Query: 257 SMMEYARGCSSNIISLAEHLGTRISDHVLPVTPNKLSEDMVKCISAIYCKLADPPMTLPP 316
S + +G S SLA+ LG ++PNKLSE++++ I I+ KL+D
Sbjct: 106 SKTDKIKGSDSGHPSLADLLGLNT------LSPNKLSEEILRSICVIHYKLSD------- 152
Query: 317 GLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEV 376
+ + + NS + E +E V +
Sbjct: 153 -------------------------------NGHNRLVKNSKNEEYGQELG------VGI 175
Query: 377 SWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYG 436
+Y + +E + Q FRSL+ +LE+VDP +L EEKLAFWINIHNALVMHA
Sbjct: 176 HKLYLDDYNLKSVESMLQNFRSLVQKLEKVDPARLGREEKLAFWINIHNALVMHA----- 230
Query: 437 IPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDG 496
A+N+GG V+A IQS+IL R LR FS ++ G
Sbjct: 231 ---------------AFNIGGEWVNAYDIQSSILGIRPCHSPSRLRTLFSPAKSSKTSSG 275
Query: 497 RQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKV 556
R TYA++ EPL HFAL +G +DP VRVYT + +FQEL A+D YI+ ++G K+ K+
Sbjct: 276 RHTYALDYAEPLLHFALSTGASTDPMVRVYTSEGIFQELRQARDSYIQTSVGF-EKETKI 334
Query: 557 LVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRY 616
L+PK++ + KD+ + G + + E L ES R ++++ K +CI W+ FRY
Sbjct: 335 LLPKIIYNYAKDTSLDMGELFSTVSECLMESQRTAMRRIVN-KKQERCIRWVHDESKFRY 393
Query: 617 LISKDVLK 624
+I ++++
Sbjct: 394 VIHSELVR 401
Score = 38.5 bits (88), Expect = 0.66
Identities = 22/42 (52%), Positives = 29/42 (68%), Gaps = 3/42 (7%)
Query: 110 ELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQL---SSVSPPT 148
ELI EIA L+ E++ LE++LLSLYR +F L SS+ P T
Sbjct: 41 ELIGEIAELDTEILQLERYLLSLYRSSFGDHLPDNSSLPPCT 82
>dbj|BAD54528.1| ternary complex factor MIP1-like [Oryza sativa (japonica
cultivar-group)] gi|53792826|dbj|BAD53859.1| ternary
complex factor MIP1-like [Oryza sativa (japonica
cultivar-group)]
Length = 654
Score = 180 bits (456), Expect = 1e-43
Identities = 157/587 (26%), Positives = 277/587 (46%), Gaps = 40/587 (6%)
Query: 62 STLKQETLQLERRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELIKEIAVLEME 121
S L+QE +L+R+L+++ +++ L A+ + ++ S + P A ELI IA LE
Sbjct: 75 SELEQEVKKLQRQLEEEIDLQLALTDAITNNATLILEPSAKL-PNKAQELIISIASLENT 133
Query: 122 VVYLEQHLLSLYRKAFDQQLSSVSPPTK--EESLKYPLTTPRAQVVKASLPEVLTKEEYP 179
V LE+ L L QL + T+ E + +Y T + L L +
Sbjct: 134 VSKLEKDLNDLC-----YQLCHLRNNTRLAENNSRYLETLAEENNSRGLLSTSLQYQPPS 188
Query: 180 TVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRCHSSLSQYSAFTRSSP 239
T + E + ++ E +++++ G +++++
Sbjct: 189 TCKCTGEEDISTLRDIKLGESESMQENLFPGLEDQQNIQKESEGREIVSQDGLLEEHQDV 248
Query: 240 PAEPLAKSLRACHSQPLSMMEYARGCSSNIISL-----AEHLGTRISDHVLPVTPNKLSE 294
P+ L + Q ME G II + + I+ +V PNKLSE
Sbjct: 249 PSNRLLEKHWNEEMQESYPMENG-GREYQIIDALSFDQSHQRKSSINGNVWNGNPNKLSE 307
Query: 295 DMVKCISAIYCKLADPPMTLPP-----GLLSPSSSLSSPSAFSIGDQGDMWSPGFRNNSS 349
+MV+C+ I+ +L+D + P +S + LS + S+ D M S + S
Sbjct: 308 EMVRCMRDIFLRLSDSSSEISPKGSSVNSISSTERLSGCTLTSVSDSSLMAS--VMQSPS 365
Query: 350 FDARLDN----------SFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSL 399
D+ D+ + +G+ V G Y ++ EVSW+Y + + + FR+L
Sbjct: 366 VDSNHDSIDEVRYFDPYNVNGKEVRRDIGNYCSVAEVSWMYVGKEQLAYASEALKNFRNL 425
Query: 400 ICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRT 459
+ +L +VDP + E+LAFWIN++N L+MHA+LAYG+P+N++K L+ KA Y VGG++
Sbjct: 426 VEQLSKVDPTCMTCAERLAFWINLYNTLIMHAYLAYGVPENDIKLFSLMQKACYIVGGQS 485
Query: 460 VSADTIQSTILRCR--MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGN 517
SA I+ IL+ + + RP L + + KFR + + Y+I++ EPL F L G
Sbjct: 486 FSAAEIEFVILKMKTPVHRPQLSLML---ALHKFRVTEEHKKYSIDDAEPLVLFGLSCGM 542
Query: 518 HSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTK---DSDMCPG 574
S PAVR+++ V QEL+ + +YIRA++G+ K++VPKL++++ K + +
Sbjct: 543 FSSPAVRIFSAGNVRQELQESMRDYIRASVGI-NDSGKLIVPKLLQSYAKGTVEDSLLAD 601
Query: 575 GVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLISKD 621
+ + + +++ + Q + +P + FRYL D
Sbjct: 602 WICRHLTPNQVAAVQDTSSSRKQRLLGVRSFSVVPFDSKFRYLFLPD 648
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,079,086,476
Number of Sequences: 2540612
Number of extensions: 45781701
Number of successful extensions: 125435
Number of sequences better than 10.0: 156
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 125043
Number of HSP's gapped (non-prelim): 282
length of query: 624
length of database: 863,360,394
effective HSP length: 134
effective length of query: 490
effective length of database: 522,918,386
effective search space: 256230009140
effective search space used: 256230009140
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0323a.7


