

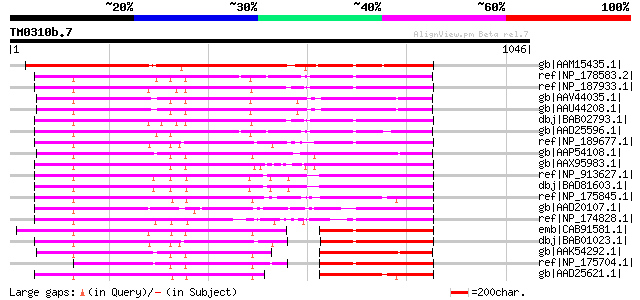
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.7
(1046 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM15435.1| unknown protein [Arabidopsis thaliana] gi|2019761... 667 0.0
ref|NP_178583.2| hypothetical protein [Arabidopsis thaliana] 583 e-165
ref|NP_187933.1| hypothetical protein [Arabidopsis thaliana] 583 e-164
gb|AAV44035.1| putative helicase [Oryza sativa (japonica cultiva... 579 e-163
gb|AAU44208.1| unknown protein [Oryza sativa (japonica cultivar-... 579 e-163
dbj|BAB02793.1| helicase-like protein [Arabidopsis thaliana] 575 e-162
gb|AAD25596.1| putative helicase [Arabidopsis thaliana] gi|25353... 570 e-161
ref|NP_189677.1| hypothetical protein [Arabidopsis thaliana] 560 e-158
gb|AAP54108.1| putative helicase [Oryza sativa (japonica cultiva... 555 e-156
gb|AAX95983.1| hypothetical protein LOC_Os11g13920 [Oryza sativa... 509 e-142
ref|NP_913627.1| putative helicase [Oryza sativa (japonica culti... 504 e-141
dbj|BAD81603.1| helicase-like protein [Oryza sativa (japonica cu... 504 e-141
ref|NP_175845.1| hypothetical protein [Arabidopsis thaliana] 499 e-139
gb|AAD20107.1| putative helicase [Arabidopsis thaliana] gi|25353... 494 e-138
ref|NP_174828.1| AT hook motif-containing protein-related [Arabi... 475 e-132
emb|CAB91581.1| putative protein [Arabidopsis thaliana] gi|15228... 392 e-107
dbj|BAB01023.1| helicase-like protein [Arabidopsis thaliana] 385 e-105
gb|AAK54292.1| putative helicase [Oryza sativa (japonica cultiva... 363 2e-98
ref|NP_175704.1| hypothetical protein [Arabidopsis thaliana] gi|... 355 4e-96
gb|AAD25621.1| Hypothetical protein [Arabidopsis thaliana] gi|... 346 3e-93
>gb|AAM15435.1| unknown protein [Arabidopsis thaliana] gi|20197614|gb|AAM15154.1|
unknown protein [Arabidopsis thaliana]
Length = 1308
Score = 667 bits (1720), Expect = 0.0
Identities = 358/843 (42%), Positives = 519/843 (61%), Gaps = 49/843 (5%)
Query: 33 MFGFSFIPYSQLINLSIDMFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINL 92
M G ++ S + + M DAMA+C+ G+P+LFIT TCN W EI ++ + L
Sbjct: 299 MCGKRYVLPSSFVGGARYMQQKYMDAMALCQHYGYPHLFITFTCNPKWPEITRYLNARGL 358
Query: 93 KPDERPYIVCRVFKMKVDNLIADLKKENIFGEVDAGLPHAHILLWLHGESKLTSPHDIDK 152
++R IV RV+KMK+D+LI D+K N+FG AGLPHAHILL+L E+K+ + D+DK
Sbjct: 359 SAEDRADIVTRVYKMKLDHLIWDIKNNNVFGPSRAGLPHAHILLFLEKEAKIPTTADVDK 418
Query: 153 KISAELPDPQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDN 212
I AE+PD +P L++VV ++HGPCG AN CM+ GKC+K FPK F +ST I+ +
Sbjct: 419 FIWAEIPDKYTHPILHEVVKEMMIHGPCGLANKNSPCMEKGKCSKKFPKDFTSSTHINKD 478
Query: 213 GYPHYRRRQSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVN 272
G+P Y+RR G V KNGI LD+R V+PYNP LL++Y AHINVE+ ++S +KYLFKY+N
Sbjct: 479 GFPIYKRRDDGRFVEKNGILLDNRFVIPYNPTLLLKYQAHINVEWVSQSKIVKYLFKYIN 538
Query: 273 KGSDRVTAEITTTAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIHESENQQSVLF 332
KG+D+VTA I E +G +EIK++YDCRY+SPCEA W F F++H S Q++++
Sbjct: 539 KGNDKVTARI---KEGTG----PNEIKRWYDCRYISPCEAAWRIFGFDLHHSSTSQNIIY 591
Query: 333 GDHEDITDVLDK---------------KGLYHNVFGVDLTYSEFPSKFVYNEKFIIWELR 377
D ED+ DVL+K K + + TY EFPS FV+ +K + W LR
Sbjct: 592 DDDEDVEDVLNKEENQTSQFLAFLKTNKKIAEDPEARKFTYIEFPSHFVWKKKQMEWTLR 651
Query: 378 KQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLS 437
++ S+ R+ ++ P G+ +Y+R+LL +G T +E I+TV G ++ T++ ACYAL LL
Sbjct: 652 QRSVSVGRVYHVTPSAGQRFYLRILLNKVKGPTCYEDIKTVGGILYPTFKDACYALSLLE 711
Query: 438 DDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAEGKMKK*CLLK 497
DD++YI I EAS+ GSG +R LF L+ + S+S P HVW +TW L++ +++ LL+
Sbjct: 712 DDQEYIDTIIEASQWGSGRYMRRLFAQLLTSESLSTPYHVWLNTWNQLSDDQIQNQALLE 771
Query: 498 IDNILQSNGKSLADFTCFSNYDLSRVEHYQNKFGVDEMTYNSEEMQDMHANLVSFLTDEQ 557
I+ +L+++G +L ++ + + N DE+ Y+ H L S LT EQ
Sbjct: 772 IEKLLRNSGSTLRNYESMPYPENNNFPAAMNTLIQDELCYDRHACAQDHLRLFSKLTVEQ 831
Query: 558 RKCA**NHGCSNQIFRYFLLCLRLWRNEKNLHLEDFIS-------WPTSNW*NCIECRLK 610
RK Y + ++ N + + W T + +
Sbjct: 832 RKI-------------YDQIIEAVYSNSGGVFFVNGFGGTGKTYLWKTLSTYIRSRGEIV 878
Query: 611 WNCINGRQNDTFKIPGGRTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAP 670
N + + GGRTAHSRF+IPL +NET TC +S S A LL ++ LIIWDEAP
Sbjct: 879 LNVASSGM-AALLLDGGRTAHSRFAIPLQVNETSTCSISPDSDLASLLLRAKLIIWDEAP 937
Query: 671 MLNIFCFEALDRTLKDIMSSENRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTAT 730
ML+ C+EALDRTLKDI+ ++N +KPFGGK + GGDF+QIL VI KGSR+ I+ A+
Sbjct: 938 MLHRHCYEALDRTLKDIVQADN----HKPFGGKTITFGGDFRQILPVITKGSREQIIHAS 993
Query: 731 VNSSYLWENCKVLSLTKNMRLLNTNGHQDNDDVKEFSE*ILKLGNGESTPNDDGKMLIDV 790
+ SS LW +CKVL+LTKNMRL T + D++KEFS+ ILKLG+G+ + +DG+ ID+
Sbjct: 994 LTSSRLWNSCKVLTLTKNMRL--TADPTEKDNIKEFSDWILKLGDGKLSEPNDGEAAIDI 1051
Query: 791 PHDLFITDPSEPLLQLIQFVYPDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEG 850
P D+ + D P+ + VYP+++ +L D TF++ERAIL T + V +VN +I+ + G
Sbjct: 1052 PEDMLLLDSLHPIDSIANCVYPNLLQNLNDQTFFRERAILCPTNDDVSEVNNHIMDLLPG 1111
Query: 851 EEK 853
E K
Sbjct: 1112 EVK 1114
>ref|NP_178583.2| hypothetical protein [Arabidopsis thaliana]
Length = 1238
Score = 583 bits (1504), Expect = e-165
Identities = 343/876 (39%), Positives = 490/876 (55%), Gaps = 92/876 (10%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M N DAMA+CK GFP FIT TCN W E+ F NL+ D+RP I+C++FKMK+D
Sbjct: 285 MRNMYLDAMAVCKHFGFPDYFITFTCNPKWPELIRFCGERNLRVDDRPEIICKIFKMKLD 344
Query: 111 NLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPD 160
+L+ DL K NI G+ GLPHAHIL+WL + KLT IDK ISAE+PD
Sbjct: 345 SLMLDLTKRNILGKTSTSMYTIEFQKRGLPHAHILIWLDSKCKLTRAEHIDKAISAEIPD 404
Query: 161 PQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRR 220
P+L++V+ ++HGPCG NP C CM+NGKC+K++PK T ID G+P YRRR
Sbjct: 405 KLKDPELFEVIKEMMVHGPCGVVNPKCPCMENGKCSKFYPKDHVPKTIIDKEGFPIYRRR 464
Query: 221 QSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTA 280
+ V K + D+R V+PYN L +RY AHINVE+CN+S S+KY+FKY++KG DRVT
Sbjct: 465 RIDDFVQKKDFKCDNRYVIPYNRSLSLRYRAHINVEWCNQSGSVKYIFKYIHKGPDRVTV 524
Query: 281 EITTTAES----SGNKRV-----------VDEIKQYYDCRYLSPCEAVWITFKFEIH--- 322
+ ++ S G ++V +E++ +++CRY+S CEA W K+ IH
Sbjct: 525 VVGSSLNSKNKEKGKQKVNADTDGSEPKKKNEVEDFFNCRYVSACEAAWRILKYPIHYRS 584
Query: 323 --------ESENQQSVLFGDHEDITDVLDKKGLYHNVF------------GVDLTYSEFP 362
+Q + F E++ VL+K L ++F LTY P
Sbjct: 585 TSVMKLSFHLPGEQYIYFKGDEEVETVLNKADLDGSMFLSWFELNKVSQIARKLTYPNIP 644
Query: 363 SKFVYNEKFIIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEI 422
++F Y+ K + LRK+G++I R+ Y+P + YY+R+LL V G SFE ++TV G +
Sbjct: 645 TRFTYDPKEKKFNLRKKGFAIGRINYVPRDIEDGYYLRILLNVVPGPRSFEELKTVNGVL 704
Query: 423 HSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTW 482
+ ++ AC ALGLL +D++YI ++ S SG LR LFV I+ +++ P++VW +TW
Sbjct: 705 YKEWKDACEALGLLDNDQEYIDDLKRTSFWSSGWYLRQLFV--IMLDALISPENVWAATW 762
Query: 483 Q-------------------LLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRV 523
Q +L++ + K L +ID+IL+ NG SL + + R
Sbjct: 763 QHLSEDIQNEKKKYFNRPDLILSDEEKKVYALQEIDHILRRNGTSLTYYKTMP--QVPRD 820
Query: 524 EHYQ-NKFGVDEMTYNSEEMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCLRLW 582
+ N +DE Y+ E HA+ + LT EQ+ G N+
Sbjct: 821 PRFDTNVLILDEKGYDRESETKKHADSIKKLTLEQKSVYDNIIGAVNENVGGVFFVYGFG 880
Query: 583 RNEKNLHLEDFISWPTSNW*NCIECRLKWNCINGRQND---TFKIPGGRTAHSRFSIPLN 639
K W T + R K + + + + + GGRTAHSR IPLN
Sbjct: 881 GTGKT------FLWKTLS----AALRSKGDIVLNVASSGIASLLLEGGRTAHSRSGIPLN 930
Query: 640 INETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKP 699
NE TC + GS +A L++++SLIIWDEAPM++ CFE+LDR+L DI + + NKP
Sbjct: 931 PNEFTTCNMKAGSDRANLVKEASLIIWDEAPMMSRHCFESLDRSLSDICGNCD----NKP 986
Query: 700 FGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTN-GHQ 758
FGGKVVV GGDF+Q+L VI DIV A +NSSYLW +CKVL+LTKNM L +
Sbjct: 987 FGGKVVVFGGDFRQVLPVIPGADTADIVMAALNSSYLWSHCKVLTLTKNMCLFSEGLSVS 1046
Query: 759 DNDDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDM--VS 816
+ D+KEFSE IL +G+G +DG+ LID+P + IT +P+ + +Y D+ +
Sbjct: 1047 EAKDIKEFSEWILAVGDGRIGEPNDGEALIDIPSEFLITKAKDPIQAICTEIYGDITKIH 1106
Query: 817 HLTDPTFYQERAILALTLESVEKVNQYILSCIEGEE 852
DP F+QERAIL T E V ++N+ +L ++GEE
Sbjct: 1107 EQKDPVFFQERAILCPTNEDVNQINETMLDNLQGEE 1142
>ref|NP_187933.1| hypothetical protein [Arabidopsis thaliana]
Length = 1419
Score = 583 bits (1503), Expect = e-164
Identities = 343/882 (38%), Positives = 508/882 (56%), Gaps = 96/882 (10%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M DAMAICK GFP LFIT TCN W EI + L D+RP IV R+FK+K+D
Sbjct: 368 MLQTYYDAMAICKHFGFPDLFITFTCNPKWPEITRYCEKRGLTADDRPDIVARIFKIKLD 427
Query: 111 NLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPD 160
+L+ DL + ++ G+ A GLPHAHILL++ SKL + DIDK ISAE+P+
Sbjct: 428 SLMKDLTERHLLGKTVASMYTVEFQKRGLPHAHILLFMAANSKLPTADDIDKIISAEIPN 487
Query: 161 PQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRR 220
P+LY+V+ +MHGPCG AN + CM +G+C+K +PKK Q T + +GYP YRRR
Sbjct: 488 KDKEPELYEVIKNSMMHGPCGSANTSSPCMVDGQCSKLYPKKHQEITKVGADGYPIYRRR 547
Query: 221 QSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRV-- 278
+ + K G++ D+R VVPYN KL +RY AHINVE+CN++ SIKYLFKY+NKG DRV
Sbjct: 548 LTDDYIEKGGVKCDNRYVVPYNKKLSLRYQAHINVEWCNQNGSIKYLFKYINKGPDRVVF 607
Query: 279 --------TAEITT--TAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEI-HESENQ 327
T+ TT ES ++ DEIK ++DCRY+S EA+W FKF I H S
Sbjct: 608 IVEPIKEATSSDTTAPVVESDTTEKKKDEIKDWFDCRYVSASEAIWRIFKFPIQHRSTPV 667
Query: 328 QSVLFGD----------HEDITDVLDKKGLYHNVF------------------GVDLTYS 359
Q + F D + DVL++ + F D Y+
Sbjct: 668 QKLSFHDKGKQPAYFDAKAKMADVLERVSNEDSQFLAWLTLNRKNAVGKNGKRARDCLYA 727
Query: 360 EFPSKFVYNEKFIIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVK 419
E P+ F ++ + ++ R +G+S+ R+ Y+ + YY+R+LL + RG S++ I+TV
Sbjct: 728 EIPAYFTWDGENKQFKKRTRGFSLGRINYVSRKMEDEYYLRVLLNIVRGPQSYDDIKTVN 787
Query: 420 GEIHSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWE 479
G ++ +Y+ AC+A G+L DD+ YI + EAS+ G+ LR F +++++S++RP+HVW
Sbjct: 788 GVVYPSYKLACFARGILDDDQVYINGLIEASQFCFGDYLRNFFSMMLLSDSLARPEHVWS 847
Query: 480 STWQLLA-------------------EGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDL 520
TW LL+ E +++ L +I+ I+ NG +L D F
Sbjct: 848 ETWHLLSEDILIKKRDEFKNQELTLTEAQIQNYTLQEIEKIMLFNGATLEDIEHFPKPSR 907
Query: 521 SRVEHYQNKFGVDEMTYNSE-EMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCL 579
+++ N+ +DE+ YN++ +++ H++ + LT EQR G +QI L
Sbjct: 908 EGIDN-SNRLIIDELRYNNQSDLKKKHSDWIQKLTPEQR-------GIYDQITNAVFNDL 959
Query: 580 R--LWRNEKNLHLEDFISWPTSNW*NCIECRLKWN-CINGRQND--TFKIPGGRTAHSRF 634
+ + FI W T R K C+N + + + GGRTAHSRF
Sbjct: 960 GGVFFVYGFGGTGKTFI-WKTL----AAAVRSKGQICLNVASSGIASLLLEGGRTAHSRF 1014
Query: 635 SIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRA 694
SIPLN +E CK+ S A L++++SLIIWDEAPM++ FCFEALD++ DI+ +
Sbjct: 1015 SIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDEAPMMSKFCFEALDKSFSDIIKRVD-- 1072
Query: 695 NANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNT 754
NK FGGKV+V GGDF+Q+L VI R +IV +++N+SYLW++CKVL LTKNMRLLN
Sbjct: 1073 --NKVFGGKVMVFGGDFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLRLTKNMRLLNN 1130
Query: 755 NGHQDN-DDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVY-- 811
+ D +++EFS+ +L +G+G +DG+++ID+P +L I + P+ + + +Y
Sbjct: 1131 DLSVDEAKEIQEFSDWLLAVGDGRVNEPNDGEVIIDIPEELLIQEADNPIEAISREIYGD 1190
Query: 812 PDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
P + ++DP F+Q RAILA E V +NQY+L ++ EE+
Sbjct: 1191 PTKLHEISDPKFFQRRAILAPKNEDVNTINQYMLEHLDSEER 1232
>gb|AAV44035.1| putative helicase [Oryza sativa (japonica cultivar-group)]
Length = 1634
Score = 579 bits (1493), Expect = e-163
Identities = 328/862 (38%), Positives = 495/862 (57%), Gaps = 89/862 (10%)
Query: 54 NCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINL-KPDERPYIVCRVFKMKVDNL 112
N QDAMAIC+ G+P LF+T TCN+ W EI + + I + KP +RP IV RVF +K+ L
Sbjct: 613 NYQDAMAICRWAGYPDLFVTFTCNAAWPEIQNMLDEIGVQKPSDRPDIVDRVFHIKLREL 672
Query: 113 IADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPDPQ 162
+ D+K + FG+ A GLPHAHIL++L + K +ID+ ISAE+PD +
Sbjct: 673 MTDIKDKQYFGKTLAIIYTIEFQKRGLPHAHILIFLDKKDKCPDASEIDRIISAEIPDKE 732
Query: 163 LYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRRQS 222
+ ++ V +MHGPCG+A CM KC + FPKKF + TT+D++G+P YRRR +
Sbjct: 733 EDREGFEAVENFMMHGPCGEAKSNSPCMIENKCIRNFPKKFHSETTVDEDGFPTYRRRDN 792
Query: 223 GIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTAEI 282
G + K ++LD+R VVPYN LL++Y AHINVE CN+S SIKYLFKY++KG D+ TA I
Sbjct: 793 GRYIEKGNVKLDNRYVVPYNRDLLVKYQAHINVERCNRSKSIKYLFKYMHKGDDQATALI 852
Query: 283 TTTAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH-----------ESENQQSVL 331
+ DEIK+Y +C Y+S +A W F+FE+H EN+Q V+
Sbjct: 853 ESDH---------DEIKKYLECTYISGHDACWRIFQFEMHYRYPSVERLPFHLENEQQVI 903
Query: 332 FGDHEDITDVLDKKGLYHNVF------------GVDLTYSEFPSKFVYNEKFIIWELRKQ 379
F D D+ ++ K+ + F DLTY+EFPSK+V+ K W RK+
Sbjct: 904 FPDSADLRKIVRKERIGVTKFTQWMETNKINDEARDLTYAEFPSKWVWKNKLKQWNKRKK 963
Query: 380 GYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLSDD 439
G I R+ Y P +G+ YY+RMLL +G +FE IRTV G +H +++ AC ALG L DD
Sbjct: 964 GKMIGRIYYAHPASGDKYYLRMLLNTVKGPRTFEEIRTVDGVVHPSFKSACEALGFLDDD 1023
Query: 440 KQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQ---------------- 483
++++ IREAS SGNQLR LF +++ ++ P +WES W+
Sbjct: 1024 REWVECIREASNYASGNQLRHLFTTILCHCEVTDPKRIWESCWEDLSEDIEYKQRKNLNY 1083
Query: 484 ---LLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQNKFGVDEMTYNSE 540
L E + K L++I+ +++ GK+L ++ + + N+ +EM+Y+ +
Sbjct: 1084 PTLRLTEQQKKGHALIEIEKLMRQAGKTLEEYPDIELPKCAELRELGNRLLNEEMSYDKD 1143
Query: 541 EMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLC--------LRLWRN-EKNLHLE 591
+ ++ H ++ L EQ+ +N+ + LWR L E
Sbjct: 1144 KQKEEHDSIFGKLNAEQKVAFDSIIESTNKGLGKLMFVDGYGGTGKTYLWRAITTKLRSE 1203
Query: 592 DFISWPTSNW*NCIECRLKWNCINGRQNDTFKIPGGRTAHSRFSIPLNINETLTCKVSKG 651
I ++ C + ++G GRTAHSRF IPL + E TC + +G
Sbjct: 1204 GKIVLTVAS------CGIAALLLHG----------GRTAHSRFHIPLIVTEESTCDIKQG 1247
Query: 652 SLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPFGGKVVVLGGDF 711
S A+LL+++SLI+WDEAPM N CFEALDR+L+DI+ S+ N+ KPFGG VVLGGDF
Sbjct: 1248 SHLAELLKKTSLILWDEAPMANRICFEALDRSLRDILRSKGEDNSTKPFGGMTVVLGGDF 1307
Query: 712 KQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTNGHQDNDD-VKEFSE*I 770
+QIL V++KG R IV A++ SYLW++ + LT+NMRL + +D +F++ I
Sbjct: 1308 RQILPVVRKGRRTQIVNASIKRSYLWQHFHIFKLTRNMRLSCISRDEDEQKRTADFAQWI 1367
Query: 771 LKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDMVSHLTDPTFYQERAIL 830
L +G+G++T + DG+ I++P DL + +P ++++ +YP++V + F ++RAIL
Sbjct: 1368 LNIGDGKTT-SADGEEWIEIPDDLILKKGGDPKEEIVKSIYPNLVQNYKKRDFLEQRAIL 1426
Query: 831 ALTLESVEKVNQYILSCIEGEE 852
E+ ++N++I++ IEGEE
Sbjct: 1427 CPRNETAREINEFIMNMIEGEE 1448
>gb|AAU44208.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 1525
Score = 579 bits (1493), Expect = e-163
Identities = 328/862 (38%), Positives = 494/862 (57%), Gaps = 89/862 (10%)
Query: 54 NCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINL-KPDERPYIVCRVFKMKVDNL 112
N QDAMAIC+ G+P LF+T TCN+ W EI + + I + KP +RP IV RVF +K+ L
Sbjct: 504 NYQDAMAICRWAGYPDLFVTFTCNAAWPEIQNMLDEIGVQKPSDRPDIVDRVFHIKLREL 563
Query: 113 IADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPDPQ 162
+ D+K + FG+ A GLPHAHIL++L + K +ID+ ISAE+PD +
Sbjct: 564 MTDIKDKQYFGKTLAIIYTIEFQKRGLPHAHILIFLDKKDKCPDASEIDRIISAEIPDKE 623
Query: 163 LYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRRQS 222
+ ++ V +MHGPCG+A CM KC + FPKKF + TT+D++G+P YRRR +
Sbjct: 624 EDREGFEAVENFMMHGPCGEAKSNSPCMIENKCIRNFPKKFHSETTVDEDGFPTYRRRDN 683
Query: 223 GIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTAEI 282
G + K ++LD+R VVPYN LL++Y AHINVE CN+S SIKYLFKY++KG D+ TA I
Sbjct: 684 GRYIEKGNVKLDNRYVVPYNRDLLVKYQAHINVERCNRSKSIKYLFKYMHKGDDQATALI 743
Query: 283 TTTAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH-----------ESENQQSVL 331
+ DEIK+Y +C Y+S +A W F+FE+H EN+Q V+
Sbjct: 744 ESDH---------DEIKKYLECTYISGHDACWRIFQFEMHYRYPSVERLPFHLENEQQVI 794
Query: 332 FGDHEDITDVLDKKGLYHNVF------------GVDLTYSEFPSKFVYNEKFIIWELRKQ 379
F D D+ ++ K+ + F D TY+EFPSK+V+ K W RK+
Sbjct: 795 FPDSADLRKIVRKERIGVTKFTQWMETNKINDEARDFTYAEFPSKWVWKNKLKQWNKRKK 854
Query: 380 GYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLSDD 439
G I R+ Y P +G+ YY+RMLL +G +FE IRTV G +H +++ AC ALG L DD
Sbjct: 855 GKMIGRIYYAHPASGDKYYLRMLLNTVKGPRTFEEIRTVDGVVHPSFKSACEALGFLDDD 914
Query: 440 KQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQ---------------- 483
++++ IREAS SGNQLR LF +++ ++ P +WES W+
Sbjct: 915 REWVECIREASNYASGNQLRHLFTTILCHCEVTDPKRIWESCWEDLGEDIEYKQRKNLNY 974
Query: 484 ---LLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQNKFGVDEMTYNSE 540
L E + K L++I+ +++ GK+L ++ + + N+ +EM+Y+ +
Sbjct: 975 PTLRLTEQQKKGHALIEIEKLMRQAGKTLEEYPDIELPKCAELRELGNRLLNEEMSYDKD 1034
Query: 541 EMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLC--------LRLWRN-EKNLHLE 591
+ ++ H ++ L EQ+ +N+ + LWR L E
Sbjct: 1035 KQKEEHDSIFGKLNAEQKVAFDSIIESTNKGLGKLMFVDGYGGTGKTYLWRAITTKLRSE 1094
Query: 592 DFISWPTSNW*NCIECRLKWNCINGRQNDTFKIPGGRTAHSRFSIPLNINETLTCKVSKG 651
I ++ C + ++G GRTAHSRF IPL + E TC + +G
Sbjct: 1095 GKIVLTVAS------CGIAALLLHG----------GRTAHSRFHIPLIVTEESTCDIKQG 1138
Query: 652 SLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPFGGKVVVLGGDF 711
S A+LL+++SLI+WDEAPM N CFEALDR+L+DI+ S+ N+ KPFGG VVLGGDF
Sbjct: 1139 SHLAELLKKTSLILWDEAPMANRICFEALDRSLRDILRSKGEDNSTKPFGGMTVVLGGDF 1198
Query: 712 KQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTNGHQDNDD-VKEFSE*I 770
+QIL V++KG R IV A++ SYLW++ + LT+NMRL + +D +F++ I
Sbjct: 1199 RQILPVVRKGRRTQIVNASIKRSYLWQHFHIFKLTRNMRLSCISRDEDEQKRTADFAQWI 1258
Query: 771 LKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDMVSHLTDPTFYQERAIL 830
L +G+G++T + DG+ I++P DL + +P ++++ +YP++V + F ++RAIL
Sbjct: 1259 LNIGDGKTT-SADGEEWIEIPDDLILKKGGDPKEEIVKSIYPNLVQNYKKRDFLEQRAIL 1317
Query: 831 ALTLESVEKVNQYILSCIEGEE 852
E+ K+N++I++ IEGEE
Sbjct: 1318 CPRNETARKINEFIMNMIEGEE 1339
>dbj|BAB02793.1| helicase-like protein [Arabidopsis thaliana]
Length = 1428
Score = 575 bits (1483), Expect = e-162
Identities = 343/891 (38%), Positives = 508/891 (56%), Gaps = 105/891 (11%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M DAMAICK GFP LFIT TCN W EI + L D+RP IV R+FK+K+D
Sbjct: 368 MLQTYYDAMAICKHFGFPDLFITFTCNPKWPEITRYCEKRGLTADDRPDIVARIFKIKLD 427
Query: 111 NLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPD 160
+L+ DL + ++ G+ A GLPHAHILL++ SKL + DIDK ISAE+P+
Sbjct: 428 SLMKDLTERHLLGKTVASMYTVEFQKRGLPHAHILLFMAANSKLPTADDIDKIISAEIPN 487
Query: 161 PQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRR 220
P+LY+V+ +MHGPCG AN + CM +G+C+K +PKK Q T + +GYP YRRR
Sbjct: 488 KDKEPELYEVIKNSMMHGPCGSANTSSPCMVDGQCSKLYPKKHQEITKVGADGYPIYRRR 547
Query: 221 QSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRV-- 278
+ + K G++ D+R VVPYN KL +RY AHINVE+CN++ SIKYLFKY+NKG DRV
Sbjct: 548 LTDDYIEKGGVKCDNRYVVPYNKKLSLRYQAHINVEWCNQNGSIKYLFKYINKGPDRVVF 607
Query: 279 --------TAEITT--TAESSGNKRVVDEIKQYYDC---------RYLSPCEAVWITFKF 319
T+ TT ES ++ DEIK ++DC RY+S EA+W FKF
Sbjct: 608 IVEPIKEATSSDTTAPVVESDTTEKKKDEIKDWFDCSSYISFSPARYVSASEAIWRIFKF 667
Query: 320 EI-HESENQQSVLFGD----------HEDITDVLDKKGLYHNVF---------------- 352
I H S Q + F D + DVL++ + F
Sbjct: 668 PIQHRSTPVQKLSFHDKGKQPAYFDAKAKMADVLERVSNEDSQFLAWLTLNRKNAVGKNG 727
Query: 353 --GVDLTYSEFPSKFVYNEKFIIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCT 410
D Y+E P+ F ++ + ++ R +G+S+ R+ Y+ + YY+R+LL + RG
Sbjct: 728 KRARDCLYAEIPAYFTWDGENKQFKKRTRGFSLGRINYVSRKMEDEYYLRVLLNIVRGPQ 787
Query: 411 SFESIRTVKGEIHSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNS 470
S++ I+TV G ++ +Y+ AC+A G+L DD+ YI + EAS+ G+ LR F +++++S
Sbjct: 788 SYDDIKTVNGVVYPSYKLACFARGILDDDQVYINGLIEASQFCFGDYLRNFFSMMLLSDS 847
Query: 471 MSRPDHVWESTWQLLA-------------------EGKMKK*CLLKIDNILQSNGKSLAD 511
++RP+HVW TW LL+ E +++ L +I+ I+ NG +L D
Sbjct: 848 LARPEHVWSETWHLLSEDILIKKRDEFKNQELTLTEAQIQNYTLQEIEKIMLFNGATLED 907
Query: 512 FTCFSNYDLSRVEHYQNKFGVDEMTYNSE-EMQDMHANLVSFLTDEQRKCA**NHGCSNQ 570
F +++ N+ +DE+ YN++ +++ H++ + LT EQR G +Q
Sbjct: 908 IEHFPKPSREGIDN-SNRLIIDELRYNNQSDLKKKHSDWIQKLTPEQR-------GIYDQ 959
Query: 571 IFRYFLLCLR--LWRNEKNLHLEDFISWPTSNW*NCIECRLKWN-CINGRQND--TFKIP 625
I L + + FI W T R K C+N + + +
Sbjct: 960 ITNAVFNDLGGVFFVYGFGGTGKTFI-WKTL----AAAVRSKGQICLNVASSGIASLLLE 1014
Query: 626 GGRTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLK 685
GGRTAHSRFSIPLN +E CK+ S A L++++SLIIWDEAPM++ FCFEALD++
Sbjct: 1015 GGRTAHSRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDEAPMMSKFCFEALDKSFS 1074
Query: 686 DIMSSENRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSL 745
DI+ + NK FGGKV+V GGDF+Q+L VI R +IV +++N+SYLW++CKVL L
Sbjct: 1075 DIIKRVD----NKVFGGKVMVFGGDFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLRL 1130
Query: 746 TKNMRLLNTNGHQDN-DDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLL 804
TKNMRLLN + D +++EFS+ +L +G+G +DG+++ID+P +L I + P+
Sbjct: 1131 TKNMRLLNNDLSVDEAKEIQEFSDWLLAVGDGRVNEPNDGEVIIDIPEELLIQEADNPIE 1190
Query: 805 QLIQFVY--PDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+ + +Y P + ++DP F+Q RAILA E V +NQY+L ++ EE+
Sbjct: 1191 AISREIYGDPTKLHEISDPKFFQRRAILAPKNEDVNTINQYMLEHLDSEER 1241
>gb|AAD25596.1| putative helicase [Arabidopsis thaliana] gi|25353661|pir||H84464
probable helicase [imported] - Arabidopsis thaliana
Length = 1219
Score = 570 bits (1469), Expect = e-161
Identities = 337/870 (38%), Positives = 482/870 (54%), Gaps = 99/870 (11%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M N DAMA+CK GFP FIT TCN W E+ F NL+ D+RP I+C++FKMK+D
Sbjct: 186 MRNMYLDAMAVCKHFGFPDYFITFTCNPKWPELIRFCGERNLRVDDRPEIICKIFKMKLD 245
Query: 111 NLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPD 160
+L+ DL K NI G+ GLPHAHIL+WL + KLT IDK ISAE+PD
Sbjct: 246 SLMLDLTKRNILGKTSTSMYTIEFQKRGLPHAHILIWLDSKCKLTRAEHIDKAISAEIPD 305
Query: 161 PQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRR 220
P+L++V+ ++HGPCG NP C CM+NGKC+K++PK T ID G+P YRRR
Sbjct: 306 KLKDPELFEVIKEMMVHGPCGVVNPKCPCMENGKCSKFYPKDHVPKTIIDKEGFPIYRRR 365
Query: 221 QSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTA 280
+ V K + D+R V+PYN L +RY AHINVE+CN+S S+KY+FKY++KG DRVT
Sbjct: 366 RIDDFVQKKDFKCDNRYVIPYNRSLSLRYRAHINVEWCNQSGSVKYIFKYIHKGPDRVTV 425
Query: 281 EITTTAES----SGNKRV-----------VDEIKQYYDCRYLSPCEAVWITFKFEIH--- 322
+ ++ S G ++V +E++ +++CRY+S CEA W K+ IH
Sbjct: 426 VVGSSLNSKNKEKGKQKVNADTDGSEPKKKNEVEDFFNCRYVSACEAAWRILKYPIHYRS 485
Query: 323 --------ESENQQSVLFGDHEDITDVLDKKGLYHNV-FGVDLTYSEFPSKFVYNEKFII 373
+Q + F E++ VL+K L ++ LTY P++F Y+ K
Sbjct: 486 TSVMKLSFHLPGEQYIYFKGDEEVETVLNKADLDGSIQIARKLTYPNIPTRFTYDPKEKK 545
Query: 374 WELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYAL 433
+ LRK+G++I R+ Y+P + YY+R+LL V G SFE ++TV G ++ ++ AC AL
Sbjct: 546 FNLRKKGFAIGRINYVPRDIEDGYYLRILLNVVPGPRSFEELKTVNGVLYKEWKDACEAL 605
Query: 434 GLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQ---------- 483
GLL +D++YI ++ S SG LR LFV I+ +++ P++VW +TWQ
Sbjct: 606 GLLDNDQEYIDDLKRTSFWSSGWYLRQLFV--IMLDALISPENVWAATWQHLSEDIQNEK 663
Query: 484 ---------------LLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQ- 527
+L++ + K L +ID+IL+ NG SL + + R +
Sbjct: 664 KKYFNRPVTCLFTDLILSDEEKKVYALQEIDHILRRNGTSLTYYKTMP--QVPRDPRFDT 721
Query: 528 NKFGVDEMTYNSEEMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCLRLWRNEKN 587
N +DE Y+ E HA+ + LT EQ+ G N+ K
Sbjct: 722 NVLILDEKGYDRESETKKHADSIKKLTLEQKSVYDNIIGAVNENVGGVFFVYGFGGTGKT 781
Query: 588 LHLEDFISWPTSNW*NCIECRLKWNCINGRQND---TFKIPGGRTAHSRFSIPLNINETL 644
W T + R K + + + + + GGRTAHSR IPLN NE
Sbjct: 782 ------FLWKTLS----AALRSKGDIVLNVASSGIASLLLEGGRTAHSRSGIPLNPNEFT 831
Query: 645 TCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPFGGKV 704
TC + GS +A L++++SLIIWDEAPM++ CFE+LDR+L DI + + NKPFGGKV
Sbjct: 832 TCNMKAGSDRANLVKEASLIIWDEAPMMSRHCFESLDRSLSDICGNCD----NKPFGGKV 887
Query: 705 VVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTNGHQDNDDVK 764
VV GGDF+Q+L VI DIV A +NSSYLW +CKVL+LTKNM L +
Sbjct: 888 VVFGGDFRQVLPVIPGADTADIVMAALNSSYLWSHCKVLTLTKNMCLFS----------- 936
Query: 765 EFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDM--VSHLTDPT 822
E IL +G+G +DG+ LID+P + IT +P+ + +Y D+ + DP
Sbjct: 937 --EEWILAVGDGRIGEPNDGEALIDIPSEFLITKAKDPIQAICTEIYGDITKIHEQKDPV 994
Query: 823 FYQERAILALTLESVEKVNQYILSCIEGEE 852
F+QERAIL T E V ++N+ +L ++GEE
Sbjct: 995 FFQERAILCPTNEDVNQINETMLDNLQGEE 1024
>ref|NP_189677.1| hypothetical protein [Arabidopsis thaliana]
Length = 1473
Score = 560 bits (1443), Expect = e-158
Identities = 324/861 (37%), Positives = 477/861 (54%), Gaps = 107/861 (12%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M N DAM CK GFP LFIT TCNS W +I FV L +++P I+CR++KMK++
Sbjct: 475 MQQNYLDAMGTCKHFGFPDLFITFTCNSKWPKITRFVKERKLNAEDKPDIICRIYKMKLE 534
Query: 111 NLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPD 160
NL+ DL K +IFG+ GLPHAHI++W+ K +DK I AE+PD
Sbjct: 535 NLMDDLTKNHIFGKTKLATYTIEFQKRGLPHAHIIVWMDPRYKFPIADHVDKIIFAEIPD 594
Query: 161 PQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRR 220
+ PKLY VS ++HGPC NP CM+NGKC+KY+PK +T +D+ GYP YRRR
Sbjct: 595 KENDPKLYQAVSECMIHGPCRLVNPNSPCMENGKCSKYYPKNHVENTFLDNEGYPIYRRR 654
Query: 221 QSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVT- 279
+G + KN D+R VVPYN LL +Y AHINVE+CN+S S+KYLFKYVNKG DRVT
Sbjct: 655 DTGRFIKKNKYPCDNRYVVPYNDFLLRKYRAHINVEWCNQSVSVKYLFKYVNKGPDRVTV 714
Query: 280 ----------AEITTTAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH------- 322
+E E++ + + ++++ Y+DCRY+S CEA+W + IH
Sbjct: 715 SLEPHRKEVVSEENNVGETNNDPQEQNQVEDYFDCRYVSACEAMWRIKGYPIHYRQTLVT 774
Query: 323 ---------------ESENQQSVLFGDHEDITDV-----LDKKGLYHNVFGVDLTYSEFP 362
E E +SVL+ ++D T L+K+ + L Y + P
Sbjct: 775 KLTFHEKGKQPIYVKEGETAESVLYRVNDDETQFTAWFELNKR----DPEAAKLLYEQIP 830
Query: 363 SKFVYNEKFIIWELRKQ-GYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGE 421
+ + +N K + RK G+ + R+ ++PP + Y++R+L+ RG F+ I+T++G
Sbjct: 831 NFYTWNGKDKNFRRRKMPGFVVGRINHVPPKIDDAYHLRILINNIRGPKGFDDIKTIEGV 890
Query: 422 IHSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWEST 481
+H TY+ ACYALGLL DDK YI I EA+ +R LFV ++I S+S P VWE T
Sbjct: 891 VHKTYRDACYALGLLDDDKDYINGIEEANFWCFHKYVRKLFVIMLIFESLSSPAVVWEHT 950
Query: 482 WQLLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQ-------NKFGVDE 534
W++L+E +K + + L+ + T + + + + N+ +DE
Sbjct: 951 WKILSEDFQRK-----VRDKLKCPAAKNIETTWNRSLEEKMLPQLKPGDEPAFNQLILDE 1005
Query: 535 MTYNSEEMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCLRLWRNEKNLHLEDFI 594
YN E ++ +H + + LT EQ+K ++I L N K +
Sbjct: 1006 RNYNRETLKTIHDDWLKMLTTEQKK-------VYDKIMDAVL-------NNKGGDI---- 1047
Query: 595 SWPTSNW*NCIECRLKWNCINGRQND--TFKIPGGRTAHSRFSIPLNINETLTCKVSKGS 652
C+N + + + GGRTAHSRF IPL +ET TC + +GS
Sbjct: 1048 ------------------CLNVASSGIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGS 1089
Query: 653 LKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPFGGKVVVLGGDFK 712
A+L+ + LIIWDEAPM++ +CFE+LD++LKDI+S+ + PFGGK+++ GGDF+
Sbjct: 1090 DLAELVTAAKLIIWDEAPMMSKYCFESLDKSLKDILSTPE----DMPFGGKLIIFGGDFR 1145
Query: 713 QILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTNGHQDNDDVKEFSE*ILK 772
QIL VI R+ IV +++NSS+LW+ CKV LTKNMRLL + ++++FS+ IL
Sbjct: 1146 QILPVILAAGRELIVKSSLNSSHLWQYCKVFKLTKNMRLLQDIDINEAREIEDFSKWILA 1205
Query: 773 LGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDMVSHLTDPTFYQERAILAL 832
+G G+ +DG I + D+ I + P+ +I+ VY DP F+Q+RAIL
Sbjct: 1206 VGEGKLNQPNDGVTQIQIRDDILIPEGDNPIESIIKAVYGTSFDEERDPKFFQDRAILCP 1265
Query: 833 TLESVEKVNQYILSCIEGEEK 853
T + V +N ++LS + GEEK
Sbjct: 1266 TNDDVNSINDHMLSKLTGEEK 1286
>gb|AAP54108.1| putative helicase [Oryza sativa (japonica cultivar-group)]
gi|37535038|ref|NP_921821.1| putative helicase [Oryza
sativa (japonica cultivar-group)]
gi|14140296|gb|AAK54302.1| putative helicase [Oryza
sativa (japonica cultivar-group)]
Length = 1573
Score = 555 bits (1430), Expect = e-156
Identities = 314/858 (36%), Positives = 480/858 (55%), Gaps = 76/858 (8%)
Query: 54 NCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSI-NLKPDERPYIVCRVFKMKVDNL 112
N QDAMAIC+ G P LF+T TCN W EI + + + KP ERP IV RVF +K+ L
Sbjct: 546 NYQDAMAICRWAGNPDLFVTFTCNPKWPEIQCMLEKVGHQKPSERPDIVVRVFMIKLREL 605
Query: 113 IADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPDPQ 162
++D+K+ FG+ A GLPHAHIL++L + K P IDK I AE+PD
Sbjct: 606 MSDIKRNQHFGKTKAIIFTIEFQKKGLPHAHILIFLDKKEKCLKPSQIDKMICAEIPDSN 665
Query: 163 LYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRRQS 222
P+ ++ V +MHGPCG+ NP CM + KC +YFPK F + T ID+ +P Y+RR
Sbjct: 666 KDPETFEAVKNFMMHGPCGETNPKSPCMVDHKCDRYFPKGFSDETIIDEVNFPIYKRRDD 725
Query: 223 GIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTAEI 282
G ++ K I L++ VVPYN LL+++ AHINVE+ N+S SI+YLFK + G D+ TA +
Sbjct: 726 GRQIKKGRINLNNGFVVPYNKDLLVKFQAHINVEWFNRSKSIRYLFKSIYNGDDQATAVV 785
Query: 283 TTTAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH-----------ESENQQSVL 331
T + N DEIK+Y C Y++ EA W F F +H EN+Q V+
Sbjct: 786 EETDTAKDN----DEIKRYLGCSYMTATEACWRIFTFPLHYQEPSVQRLFFHVENEQQVI 841
Query: 332 FGDHEDITDVLDKKGLYHNVF------------GVDLTYSEFPSKFVYNEKFIIWELRKQ 379
F D D+ +++ +F +LTYSEFP+K+ + K W RK
Sbjct: 842 FPDSTDLQEIIRHPRSGVTMFTEWMETNKKHEDARELTYSEFPTKWTWVNKVKKWVRRKG 901
Query: 380 GYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLSDD 439
I R+ P +GE YY+R++L +GCT+FE IRTV G +HS+Y+ AC+ALG L+DD
Sbjct: 902 RKKIGRIYNAHPASGERYYLRVILNTAKGCTTFEDIRTVNGFVHSSYKSACHALGFLNDD 961
Query: 440 KQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAE------------ 487
++I I+EAS SG +L+ LF +++ ++ P +WES W+ L++
Sbjct: 962 NEWIECIKEASCWASGIELQQLFATILCHCEVTDPKSLWESIWEELSKDIQHTQSWLLNF 1021
Query: 488 -------GKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQNKFGVDEMTYNSE 540
+K L++I+ ++ GKSL ++T +++++ +N +EM Y+ E
Sbjct: 1022 PASCLTPSHKRKCALIEIEKNMRQAGKSLKEYTGIEPPNVAKLSEIENSLINEEMNYDKE 1081
Query: 541 EMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCLRLWRNEKNLHLEDFISWPTSN 600
++ H +++ L EQ+K ++Q K + ++ + +
Sbjct: 1082 RLKHQHLQILNTLNIEQKKAFDAIIESAHQSL------------GKLIFVDGYGGTGKTY 1129
Query: 601 W*NCIECRLKWN-----CINGRQNDTFKIPGGRTAHSRFSIPLNINETLTCKVSKGSLKA 655
I RL+ + + GGRTAHS F+IP+N+ + TC + +GS A
Sbjct: 1130 LWKAITTRLRSEGKIVIAVASSGIAALLLQGGRTAHSAFNIPINLTDESTCFIKQGSRIA 1189
Query: 656 KLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPFGGKVVVLGGDFKQIL 715
LL ++SLI+WDEAPM N CFEALD++L+D+ N + KPFGG VVLGGDF+QIL
Sbjct: 1190 DLLMKTSLILWDEAPMANRNCFEALDKSLRDVQRFRNENSYQKPFGGMTVVLGGDFRQIL 1249
Query: 716 HVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTNGHQ-DNDDVKEFSE*ILKLG 774
++ KG R+ V A++ SYLW++ +V +LTKNMRL + + Q ++ EF+E IL++G
Sbjct: 1250 PIVPKGRREHTVNASIKFSYLWQHFEVFNLTKNMRLNSVSKDQAEHQKTAEFAEWILRIG 1309
Query: 775 NGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDMVSHLTDPTFYQERAILALTL 834
NG++ D+ K + +P DL + +P Q++ YP + + P + +ERAIL T
Sbjct: 1310 NGDTILLDE-KGWVSMPSDLLLQKGDDPKAQIVDSTYPGLQYNCCKPKYLEERAILCPTN 1368
Query: 835 ESVEKVNQYILSCIEGEE 852
+ V ++N+YI+ I+G++
Sbjct: 1369 DDVNELNEYIMDQIQGDK 1386
>gb|AAX95983.1| hypothetical protein LOC_Os11g13920 [Oryza sativa (japonica
cultivar-group)]
Length = 2157
Score = 509 bits (1310), Expect = e-142
Identities = 323/878 (36%), Positives = 473/878 (53%), Gaps = 95/878 (10%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFV-CSINLKPDERPYIVCRVFKMKV 109
M N QDAMAIC+ G P LF+T TCNS W+EIAD + +P +R I+ RVF MKV
Sbjct: 523 MVMNYQDAMAICRVYGPPDLFVTYTCNSKWQEIADAIRFEPGQQPSDRADIIVRVFNMKV 582
Query: 110 DNLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELP 159
+ I D+++ FG+V A GLPH H L+WL + S ID I AE+P
Sbjct: 583 NEFITDIREGRTFGKVLAVLYTVEFQKRGLPHIHCLVWLAAATADVSASIIDGFICAEIP 642
Query: 160 DPQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRR 219
D Y++VS +MHGPCG AN C CMKN KC+K++PK FQ+ T +D++G+ YRR
Sbjct: 643 DYDTDRLGYELVSEFMMHGPCGDANKKCPCMKNDKCSKHYPKDFQDETIVDESGFTIYRR 702
Query: 220 RQSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVT 279
R G ++KNGI LD+R+VVPYN LL +Y AHIN+E+CNKSN IKYLFKY+ KG DR
Sbjct: 703 RNDGRSIMKNGILLDNRSVVPYNMALLKKYEAHINIEWCNKSNLIKYLFKYITKGHDRAR 762
Query: 280 AEITTTAE---SSGNKRVV--DEIKQYYDCRYLSPCEAVWITFKFEIH-----------E 323
TT + +S N + +EI +Y D R+LS CEA+ F+F+IH
Sbjct: 763 IYFETTGKTQNASPNHDLAPRNEILEYMDARFLSTCEALHRLFEFDIHYRVPPVERLVVH 822
Query: 324 SENQQSVLFGDHEDITDVLDKKGLYHNVF------------GVDLTYSEFPSKFVYNEKF 371
+ V + D+ VL+ G ++ LTY EFP ++ +
Sbjct: 823 LPGKNFVRYEKGADLRAVLESPGAKRSMLTEWFETNKKNSKAHSLTYCEFPKEWTWEPSS 882
Query: 372 IIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACY 431
W R I R+ Y+ P GELYY+RMLL + +G S+ +RT G ++ TY++AC
Sbjct: 883 KTWHERTPAPKIGRIYYVHPTAGELYYLRMLLMIVKGAQSYADVRTYDGVVYGTYREACE 942
Query: 432 ALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAE---G 488
A GLL D ++ EA S QLR LFV++++ S+ +++ W + +
Sbjct: 943 ARGLLEGDNEWHLLFDEAIVTASSAQLRQLFVTVLLYCSVGDVRSLFDKYWLYMTDDIHN 1002
Query: 489 KMKK-----*CLLKIDNILQ-----------SNGKSLADFTCFSNYDLSRVEHY--QNKF 530
++KK C++ D++L ++G ++ DF S V H N+
Sbjct: 1003 RLKKALDNPHCVIPHDHLLNMLLHELIAVFANSGGNIKDFNL---PHPSSVPHVLGTNRL 1059
Query: 531 GVDEMTYNSEEMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCLRLWRNEKNLHL 590
+DE M MHA+ SF+ ++QI + +C R NE
Sbjct: 1060 -IDEEITIDPLMLAMHAD--SFVQQLN----------NDQITVFNTICSRAIANEPGFF- 1105
Query: 591 EDFIS-----WPTSNW*NCIECRLKWN-----CINGRQNDTFKIPGGRTAHSRFSIPLNI 640
F+S T W N I +L+ + + +P GRTAHSRF IP++I
Sbjct: 1106 --FVSGHGGTGKTFLW-NTIIAKLRSQNKIVLAVASSGVASLLLPRGRTAHSRFKIPIDI 1162
Query: 641 NETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPF 700
+ET C + +G++ A+LL +++LIIWDEAPM + CFEALDRTL+DI+S +N+ PF
Sbjct: 1163 DETSICNIKRGTMLAELLAETALIIWDEAPMTHRRCFEALDRTLRDILSETCPSNSIIPF 1222
Query: 701 GGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTN-GHQD 759
GGK +VLGGDFKQIL VI KGSRQ I+ A++ +S LW++ +LSL NMRLLN
Sbjct: 1223 GGKPIVLGGDFKQILPVIPKGSRQAIINASITNSELWKHVALLSLNINMRLLNPMLPDNQ 1282
Query: 760 NDDVKEFSE*ILKLGNG----ESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDMV 815
++ +FS+ +L +GNG + ++ I +P DL + + + ++ VY D +
Sbjct: 1283 KKELHDFSQWVLAIGNGTLPMTAKEGENYPAWITIPDDLLVMTSGDKIAAIVHEVYSDFL 1342
Query: 816 SHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+ D + RAI+ T +V+++N YI+ + G+ +
Sbjct: 1343 TCYRDIEYLASRAIVCPTNTTVDEINDYIIGLVPGDSR 1380
>ref|NP_913627.1| putative helicase [Oryza sativa (japonica cultivar-group)]
Length = 1453
Score = 504 bits (1297), Expect = e-141
Identities = 311/887 (35%), Positives = 463/887 (52%), Gaps = 113/887 (12%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIAD-FVCSINLKPDERPYIVCRVFKMKV 109
M N QDAMA+C+ G P LF+T TCNS W+EI D V P +R ++ RVF MKV
Sbjct: 396 MVQNYQDAMAVCRVFGSPDLFVTFTCNSKWQEIYDALVFEPGQVPSDRSDMIVRVFSMKV 455
Query: 110 DNLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELP 159
D I+D+K+ FG V A GLPH H ++W S +D I AE+P
Sbjct: 456 DEFISDIKEGRTFGPVLAVLYTVEFQKRGLPHIHCIVWRAAADAEFSATAVDSLICAEIP 515
Query: 160 DPQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRR 219
D P Y +V ++HGPCG N +C CMKNG C+K+FPK FQ TT+D+ G+ YRR
Sbjct: 516 DVFSDPLGYALVDEFMIHGPCGDKNKSCVCMKNGHCSKHFPKGFQEETTMDEFGFTVYRR 575
Query: 220 RQSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVT 279
R G V+KNGI+LD+R VVPYN KLL +Y AHINVE CNKSN IKYLFKY+ KG DR
Sbjct: 576 RNDGRYVVKNGIKLDNRWVVPYNMKLLKKYQAHINVESCNKSNMIKYLFKYITKGGDRTK 635
Query: 280 AEITTTAES-----SGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH-----------E 323
TT + G +EI +Y + R+LS CEA W F+F+IH
Sbjct: 636 LYFETTGNTPNKTVDGTVLPPNEIDEYINARFLSTCEAFWRAFEFDIHYRVPAVERLPIH 695
Query: 324 SENQQSVLFGDHEDITDVLD----KKGLYHNVFGVD--------LTYSEFPSKFVYNEKF 371
N V + D+ +LD KK + F + LTY +FP ++ ++
Sbjct: 696 LPNMNFVQYKKGTDLKKLLDSPAAKKTMLTEWFECNKKHPNARTLTYCDFPKQWTWDNSA 755
Query: 372 IIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACY 431
W R I R+ Y+ P GELYY+RMLL +G S+ +RT +G ++ T+++AC
Sbjct: 756 RCWRPRTPVEKIGRIYYVSPAAGELYYLRMLLMTVKGAKSYADVRTFEGTVYPTFRQACE 815
Query: 432 ALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTW--------- 482
+ GLL +D + EA S QLR LFV++++ S+ +++ W
Sbjct: 816 SRGLLENDNDWHLLFDEAIVSASSLQLRQLFVTVVMFCSVGNVRSLFDKYWLYFTDDIQH 875
Query: 483 ----------QLLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRV-----EHYQ 527
++ ++ + ++ + ++G ++ D YDL R + +
Sbjct: 876 RLRTALSNPAYVVPHDRLLSLLIKELHSAFANSGGNIDD------YDLPRSTIHSDDEFG 929
Query: 528 NKFGVDEMTYNSEEMQDMHANLVSFLTDEQRK----------------CA**NHGCSNQI 571
N+ +E+ ++ + + ++ L EQ+ HG + +
Sbjct: 930 NRMVNEELALDTAALAAHASLMIPRLNSEQQNFFDTIVSRVSESRPGFFFVYGHGGTGKT 989
Query: 572 FRYFLLCLRLWRNEKNLHLEDFISWPTSNW*NCIECRLKWNCINGRQNDTFKIPGGRTAH 631
F + +L ++ R+E N+ L S S +P GRTAH
Sbjct: 990 FLWNVLISKI-RSEGNIVLAVASSGVAS----------------------LLLPRGRTAH 1026
Query: 632 SRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSE 691
SRF IP++I+E C + +G++ A+L+Q++SLIIWDEAPM + CFEALDRTL+D++S
Sbjct: 1027 SRFKIPIDIDENSICSIKRGTMLAELIQKTSLIIWDEAPMTHRRCFEALDRTLRDLLSEH 1086
Query: 692 NRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRL 751
N +N+ PFGGK VVLGGDF+QIL VI+KG+R IV A++ +S LW++ +L LT NMRL
Sbjct: 1087 NPSNSVLPFGGKFVVLGGDFRQILPVIKKGTRNSIVDASITNSPLWQHVVLLKLTVNMRL 1146
Query: 752 LNTN-GHQDNDDVKEFSE*ILKLGNG----ESTPNDDGKMLIDVPHDLFITDPSEPLLQL 806
+ D+++F+ +L LG+G ++ ID+P DL I + + +
Sbjct: 1147 FQSGLSEGRRHDLEQFARWVLALGDGMLPVSKRIDESEATWIDIPDDLLIRASDDKIYSI 1206
Query: 807 IQFVYPDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+ V+P V TD ++ RAI+ +V+++N Y+++ I GE K
Sbjct: 1207 VNEVFPCYVHRYTDSSYLASRAIVCPNNSTVDEINDYMVAMIPGEMK 1253
>dbj|BAD81603.1| helicase-like protein [Oryza sativa (japonica cultivar-group)]
Length = 1652
Score = 504 bits (1297), Expect = e-141
Identities = 311/887 (35%), Positives = 463/887 (52%), Gaps = 113/887 (12%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIAD-FVCSINLKPDERPYIVCRVFKMKV 109
M N QDAMA+C+ G P LF+T TCNS W+EI D V P +R ++ RVF MKV
Sbjct: 595 MVQNYQDAMAVCRVFGSPDLFVTFTCNSKWQEIYDALVFEPGQVPSDRSDMIVRVFSMKV 654
Query: 110 DNLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELP 159
D I+D+K+ FG V A GLPH H ++W S +D I AE+P
Sbjct: 655 DEFISDIKEGRTFGPVLAVLYTVEFQKRGLPHIHCIVWRAAADAEFSATAVDSLICAEIP 714
Query: 160 DPQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRR 219
D P Y +V ++HGPCG N +C CMKNG C+K+FPK FQ TT+D+ G+ YRR
Sbjct: 715 DVFSDPLGYALVDEFMIHGPCGDKNKSCVCMKNGHCSKHFPKGFQEETTMDEFGFTVYRR 774
Query: 220 RQSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVT 279
R G V+KNGI+LD+R VVPYN KLL +Y AHINVE CNKSN IKYLFKY+ KG DR
Sbjct: 775 RNDGRYVVKNGIKLDNRWVVPYNMKLLKKYQAHINVESCNKSNMIKYLFKYITKGGDRTK 834
Query: 280 AEITTTAES-----SGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH-----------E 323
TT + G +EI +Y + R+LS CEA W F+F+IH
Sbjct: 835 LYFETTGNTPNKTVDGTVLPPNEIDEYINARFLSTCEAFWRAFEFDIHYRVPAVERLPIH 894
Query: 324 SENQQSVLFGDHEDITDVLD----KKGLYHNVFGVD--------LTYSEFPSKFVYNEKF 371
N V + D+ +LD KK + F + LTY +FP ++ ++
Sbjct: 895 LPNMNFVQYKKGTDLKKLLDSPAAKKTMLTEWFECNKKHPNARTLTYCDFPKQWTWDNSA 954
Query: 372 IIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACY 431
W R I R+ Y+ P GELYY+RMLL +G S+ +RT +G ++ T+++AC
Sbjct: 955 RCWRPRTPVEKIGRIYYVSPAAGELYYLRMLLMTVKGAKSYADVRTFEGTVYPTFRQACE 1014
Query: 432 ALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTW--------- 482
+ GLL +D + EA S QLR LFV++++ S+ +++ W
Sbjct: 1015 SRGLLENDNDWHLLFDEAIVSASSLQLRQLFVTVVMFCSVGNVRSLFDKYWLYFTDDIQH 1074
Query: 483 ----------QLLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRV-----EHYQ 527
++ ++ + ++ + ++G ++ D YDL R + +
Sbjct: 1075 RLRTALSNPAYVVPHDRLLSLLIKELHSAFANSGGNIDD------YDLPRSTIHSDDEFG 1128
Query: 528 NKFGVDEMTYNSEEMQDMHANLVSFLTDEQRK----------------CA**NHGCSNQI 571
N+ +E+ ++ + + ++ L EQ+ HG + +
Sbjct: 1129 NRMVNEELALDTAALAAHASLMIPRLNSEQQNFFDTIVSRVSESRPGFFFVYGHGGTGKT 1188
Query: 572 FRYFLLCLRLWRNEKNLHLEDFISWPTSNW*NCIECRLKWNCINGRQNDTFKIPGGRTAH 631
F + +L ++ R+E N+ L S S +P GRTAH
Sbjct: 1189 FLWNVLISKI-RSEGNIVLAVASSGVAS----------------------LLLPRGRTAH 1225
Query: 632 SRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSE 691
SRF IP++I+E C + +G++ A+L+Q++SLIIWDEAPM + CFEALDRTL+D++S
Sbjct: 1226 SRFKIPIDIDENSICSIKRGTMLAELIQKTSLIIWDEAPMTHRRCFEALDRTLRDLLSEH 1285
Query: 692 NRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRL 751
N +N+ PFGGK VVLGGDF+QIL VI+KG+R IV A++ +S LW++ +L LT NMRL
Sbjct: 1286 NPSNSVLPFGGKFVVLGGDFRQILPVIKKGTRNSIVDASITNSPLWQHVVLLKLTVNMRL 1345
Query: 752 LNTN-GHQDNDDVKEFSE*ILKLGNG----ESTPNDDGKMLIDVPHDLFITDPSEPLLQL 806
+ D+++F+ +L LG+G ++ ID+P DL I + + +
Sbjct: 1346 FQSGLSEGRRHDLEQFARWVLALGDGMLPVSKRIDESEATWIDIPDDLLIRASDDKIYSI 1405
Query: 807 IQFVYPDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+ V+P V TD ++ RAI+ +V+++N Y+++ I GE K
Sbjct: 1406 VNEVFPCYVHRYTDSSYLASRAIVCPNNSTVDEINDYMVAMIPGEMK 1452
>ref|NP_175845.1| hypothetical protein [Arabidopsis thaliana]
Length = 1639
Score = 499 bits (1285), Expect = e-139
Identities = 314/869 (36%), Positives = 477/869 (54%), Gaps = 112/869 (12%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSI-NLKPDERPYIVCRVFKMKV 109
M DAMAIC+ G P LFIT+T N W+EI++ + + N + RP I CRVFK+K+
Sbjct: 620 MVEKYHDAMAICRWYGNPDLFITMTTNPKWEEISEHLKTYGNDAANVRPDIECRVFKIKL 679
Query: 110 DNLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELP 159
D L+AD K F + A GLPHAHILLWL G+ K +P+DIDK ISAE+P
Sbjct: 680 DELLADFNKGLFFPKPIAIVYTIEFQKRGLPHAHILLWLQGDLKKPTPNDIDKYISAEIP 739
Query: 160 DPQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRR 219
P+ + +V +MHGPCGK P+ CM+ G C+K FP++F N T ++++G+ YRR
Sbjct: 740 VKDKDPEGHTLVEQHMMHGPCGKDRPSSPCMEKGICSKKFPREFVNHTKMNESGFILYRR 799
Query: 220 RQSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVT 279
R VLK LD+R VVP+N ++L +Y AHINVE+CNKS++IKYLFKY+ KG D+ T
Sbjct: 800 RNDQRYVLKGQTRLDNRFVVPHNLEILKKYKAHINVEWCNKSSAIKYLFKYITKGVDKAT 859
Query: 280 AEI----TTTAESSGN---KRVVDEIKQYYDCRYLSPCEAVWITFKFEIHESE------- 325
I + + SGN ++ +EI +Y DCRYLS CEA+W F F IH
Sbjct: 860 FIIQKGNSVNGQGSGNGFEEKPRNEINEYLDCRYLSACEAMWRIFMFNIHHHNPPVQRLP 919
Query: 326 ----NQQSVLFGDHEDITDVLDKKG----LYHNVFGVD--------LTYSEFPSKFVYNE 369
+QS +F + E++ +V + G + F ++ L Y + P+ FV++
Sbjct: 920 LHLPGEQSTIFEEEENLENVEYRYGHERTMLTEYFELNKICEDARKLKYVQVPTMFVWDS 979
Query: 370 KFIIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKA 429
++ RKQ +I R+ I P G+LYY+R+LL +G TSF+ ++TV G +H +++ A
Sbjct: 980 TNKMYTRRKQRENIGRIVNILPTAGDLYYLRILLNKVKGATSFDYLKTVGGVVHESFKAA 1039
Query: 430 CYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAEG- 488
C+ GLL DK++ A+ EA++ + LR LFV ++I +S P +W W+ +A+
Sbjct: 1040 CHTRGLLDGDKEWHDAMDEAAQWSTSYLLRSLFVLILIYCEVSEPLKLWSHCWESMADDV 1099
Query: 489 ------------------KMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQNKF 530
+++K L++I+ +L+ + KSL+D+ + +NK
Sbjct: 1100 FRKQQKVLNFPQLELKAEELEKYTLIEIETLLRQHEKSLSDY--------PEMPQPENKL 1151
Query: 531 GVDEMTYNSEEMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCLRLWRNEKNLHL 590
N ++ L S + E + G + + F Y + L N KN+
Sbjct: 1152 -------NEQQRIIYDDVLKSVINKEGKLFFLYGAGGTGKTFLYKTIISALRSNGKNV-- 1202
Query: 591 EDFISWPTSNW*NCIECRLKWNCINGRQNDTFKIPGGRTAHSRFSIPLNINETLTCKVSK 650
P ++ + I L +PGGRTAHSRF IP+N++E C +
Sbjct: 1203 -----MPVAS--SAIAALL--------------LPGGRTAHSRFKIPINVHEDSICDIKI 1241
Query: 651 GSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPFGGKVVVLGGD 710
GS+ A +L + LIIWDEAPM + FEA+DRTL+DI+S + K FGGK V+LGGD
Sbjct: 1242 GSMLANVLSKVDLIIWDEAPMAHRHTFEAVDRTLRDILSVGDEKALTKTFGGKTVLLGGD 1301
Query: 711 FKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTNGHQDNDDVKEFSE*I 770
F+QIL VI +G+RQ+ V+A +N SYLWE+C L++NMR+ +++K F+E I
Sbjct: 1302 FRQILPVIPQGTRQETVSAAINRSYLWESCHKYLLSQNMRV-------QPEEIK-FAEWI 1353
Query: 771 LKLGNGES------TPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDMVSHLTDPTFY 824
L++G+GE+ +D + I + +L + + PL L + V+PD + D
Sbjct: 1354 LQVGDGEAPRKTHGIDDDQEEDNIIIDKNLLLPETENPLEVLCRSVFPDFTNTFQDLENL 1413
Query: 825 QERAILALTLESVEKVNQYILSCIEGEEK 853
+ A+L E+V+++N Y+LS + G K
Sbjct: 1414 KGTAVLTPRNETVDEINDYLLSKVPGLAK 1442
>gb|AAD20107.1| putative helicase [Arabidopsis thaliana] gi|25353665|pir||H84515
probable helicase [imported] - Arabidopsis thaliana
Length = 1230
Score = 494 bits (1273), Expect = e-138
Identities = 303/817 (37%), Positives = 439/817 (53%), Gaps = 77/817 (9%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M N DA+ CK GFP LFIT TCN W EI FV L ++RP I+CR++KMK+D
Sbjct: 332 MQQNYLDAIGTCKHFGFPDLFITFTCNPKWPEITRFVKERKLNAEDRPDIICRIYKMKLD 391
Query: 111 NLIADLKKENIFGEVDA-----GLPHAHILLWLHGESKLTSPHDIDKKISAELPDPQLYP 165
NL+ DL K +IF GLPHAHI++W+ K + +DK I AE+PD + +P
Sbjct: 392 NLMDDLTKNHIFAMYTVEFQKRGLPHAHIIVWMDPRYKFHTADHVDKIIFAEIPDKEKHP 451
Query: 166 KLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRRQSGIK 225
+LY VS ++HGPC NP CM+NGKC+KY+PK +T++D GYP YRRR SG
Sbjct: 452 ELYQAVSECMIHGPCRLVNPNSPCMENGKCSKYYPKNHVENTSLDYKGYPIYRRRDSGRF 511
Query: 226 VLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTAEITTT 285
+ KN + D+ VVPYN LL +Y AHINVE+CN+S SIKYLFKYVNKG DRVT
Sbjct: 512 IEKNKYQCDNWYVVPYNDVLLRKYRAHINVEWCNQSVSIKYLFKYVNKGPDRVTQ--NNV 569
Query: 286 AESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIHESENQQSVLFGDHEDITDVLDKK 345
E + + + ++++ Y+DCR K HE + +QSV + E VL
Sbjct: 570 GEINNDPQERNQVQDYFDCRGYPIHYRQTSVTKLTFHE-KGKQSVYVKEGETAESVL--- 625
Query: 346 GLYHNVFGVDLTYSEFPSKFVYNEK-----FIIWELRKQGYSIRRLTYIPPGTGELYYMR 400
+ V+ ++F + F N++ +++E Y+I ++PP + Y++R
Sbjct: 626 ------YRVNNDETQFIAWFELNKRDPEAAKLLYEQIPNFYTI---NHVPPKIDDAYHLR 676
Query: 401 MLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRG 460
+L+ R F+ I+TV+G +H TY+ ACYALGLL DDK+YI I EA+ S +R
Sbjct: 677 ILINNIRAPKGFDDIKTVEGVVHKTYRDACYALGLLDDDKEYIHGIEEANFWCSPKYVRK 736
Query: 461 LFVSLIITNSMSRPDHVWESTWQLLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDL 520
FV ++I+ S+S P VWE TW++L E + K+ + L+ D +
Sbjct: 737 SFVIMLISESLSSPVVVWEHTWKILFEDFQR-----KVRDKLER-----PDLWRYKMLLE 786
Query: 521 SRVEHYQNKFGVDEMTYNSEEMQDMHANLVSFLTDEQRKCA**NHGCSNQIF--RYFLLC 578
E N +DE YN E ++ H N + LT E +K + + + + +
Sbjct: 787 PGDEPAFNPLIIDERNYNRESLKKKHDNWLKTLTPEHKKVY--HDEIMDDVLNDKGGVFF 844
Query: 579 LRLWRNEKNLHLEDFISWPTSNW*NCIECRLKWNCINGRQND--TFKIPGGRTAHSRFSI 636
L + L +S I C+ C+N + + + GGRTAHSRF I
Sbjct: 845 LYAFGGTGKTFLWKVLS-------AAIRCKGD-TCLNVASSSIASLLLEGGRTAHSRFGI 896
Query: 637 PLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANA 696
PL +ET TC + +GS A+L+ + LIIWDE
Sbjct: 897 PLTPHETSTCNMERGSDLAELVTAAKLIIWDE---------------------------- 928
Query: 697 NKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTNG 756
+ PFG KV++ GGDF+QILHVI R+ IV +++NSSYLW++CKVL LTKNMRLL
Sbjct: 929 DMPFGRKVILFGGDFRQILHVIPAAGRELIVKSSLNSSYLWQHCKVLKLTKNMRLLQDID 988
Query: 757 HQDNDDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVYPDMVS 816
+ ++++F + IL +G G+ DG I +P D+ I + P+ +I+ VY +
Sbjct: 989 INEAREIEDFLKWILTVGEGKLNEPSDGVTHIQIPDDILIPEGDNPIESIIKAVYGTTFA 1048
Query: 817 HLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
DP F+Q +AIL T + V +N ++LS + GEE+
Sbjct: 1049 QKRDPKFFQHKAILCPTNDDVNSINDHMLSKLTGEER 1085
>ref|NP_174828.1| AT hook motif-containing protein-related [Arabidopsis thaliana]
gi|25511694|pir||D86481 189.6K hypothetical protein
F10O5.11 - Arabidopsis thaliana
gi|12322087|gb|AAG51081.1| hypothetical protein
[Arabidopsis thaliana]
Length = 1678
Score = 475 bits (1223), Expect = e-132
Identities = 312/873 (35%), Positives = 453/873 (51%), Gaps = 162/873 (18%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M + DAMAICK GFP LFIT TCN W EI + L P++RP I+ R+FK+K+D
Sbjct: 710 MVQSYYDAMAICKHYGFPDLFITFTCNPKWPEITRYYDKRGLNPEDRPNIIARIFKIKLD 769
Query: 111 NLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPD 160
+L+ DL + + G+ A GLPHAHILL++H +SKL + DIDK ISAE+PD
Sbjct: 770 SLMNDLTVKKMLGKTVASMYTVEFQKRGLPHAHILLFMHAKSKLPTADDIDKLISAEIPD 829
Query: 161 PQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRR 220
+ P LY+V+ ++HGPCG AN CM +G+C+K +PKK Q+ T I +GYP YRRR
Sbjct: 830 KEKEPDLYEVIKNSMIHGPCGSANVNSPCMVDGECSKLYPKKHQDITKIGSDGYPIYRRR 889
Query: 221 QSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTA 280
++ V K GI+ D+R VVPYN KL +RY+AHINVE+CN++ SIKYLFKY+NKG D+V
Sbjct: 890 KTDDYVEKGGIKCDNRYVVPYNKKLSLRYNAHINVEWCNQNGSIKYLFKYINKGPDKVVF 949
Query: 281 EITTTAESSGN------------KRVVDEIKQYYDCRYLSPCEAVWITFKFEIHES---- 324
+ T +++ ++ +EIK ++DCRY+S EAVW FK+ I
Sbjct: 950 IVEPTQQTTAGDSETPQQEQGSAEKKKNEIKDWFDCRYVSASEAVWRIFKYPIQHISTPV 1009
Query: 325 -------ENQQSVLFGDHEDITDVLDKKGLYHNVFGVDLT------------------YS 359
E +Q F +I DVL++ + F LT Y+
Sbjct: 1010 QKLSFHVEGKQPAYFDPKSNIEDVLERVANVDSQFMAWLTLNRRNAVGKNGKRARECLYA 1069
Query: 360 EFPSKFVYNEKFIIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVK 419
E P+ F ++ + ++ R +G+SI R+ Y+ + Y++R+LL + RG TS+ I+T
Sbjct: 1070 EIPAYFTWDGENKSFKKRTRGFSIGRIHYVSRKMEDEYFLRVLLNIVRGPTSYAEIKTYD 1129
Query: 420 GEIHSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWE 479
G ++ T+++AC+A G+L DD+ +I + EA+ HV
Sbjct: 1130 GVVYKTFKEACFARGILDDDQVFIDGLVEAT-------------------------HVRS 1164
Query: 480 STWQLLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQNKFG-----VDE 534
TW LLAE +K K D N T NY L +E G +DE
Sbjct: 1165 QTWHLLAEDILKT----KRDEF--KNPDLTLTETEIKNYTLQEIEKIMLSNGATLEDIDE 1218
Query: 535 MTYNS-EEMQDMHANLVSFLTDEQRKCA**NHGCSNQIFRYFLLCLRLWRNEKNLHL--- 590
+ +E + M LT EQR G N I ++ N +
Sbjct: 1219 FPKPTRDEWKQM-------LTPEQR-------GVYNAITE------AVFNNLGGVFFVYG 1258
Query: 591 -----EDFISWPTSNW*NCIECRLKWNCINGRQND--TFKIPGGRTAHSRFSIPLNINET 643
+ FI W T + I CR + +N + + + GGRTAHSRF IPLN +E
Sbjct: 1259 FGGTGKTFI-WKTLS--AAIRCRGQI-VLNVASSGIASLLLEGGRTAHSRFGIPLNHDEF 1314
Query: 644 LTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLKDIMSSENRANANKPFGGK 703
+LD++ DI+ + N NK FGGK
Sbjct: 1315 SV---------------------------------SLDKSFSDIIKNTN----NKVFGGK 1337
Query: 704 VVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSLTKNMRLLNTN-GHQDNDD 762
VVV GGDF+Q+L VI R +IV +++N+SYLW++CKVL LTKNMRLL N + +
Sbjct: 1338 VVVFGGDFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLANNLSATEAKE 1397
Query: 763 VKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQLIQFVY--PDMVSHLTD 820
++EFS+ +L + +G +DG ID+P DL IT+ +P+ + +Y P ++ +TD
Sbjct: 1398 IQEFSDWLLAVSDGRINEPNDGVATIDIPEDLLITNADKPIETITNEIYGDPKILHEITD 1457
Query: 821 PTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
P F+Q RAILA E V +N+Y+L ++ EE+
Sbjct: 1458 PKFFQGRAILAPKNEDVNTINEYLLEQLDAEER 1490
>emb|CAB91581.1| putative protein [Arabidopsis thaliana] gi|15228453|ref|NP_189796.1|
AT hook motif-containing protein-related [Arabidopsis
thaliana] gi|11357786|pir||T48965 hypothetical protein
F4M19.60 - Arabidopsis thaliana
Length = 1752
Score = 392 bits (1006), Expect = e-107
Identities = 228/616 (37%), Positives = 332/616 (53%), Gaps = 72/616 (11%)
Query: 14 CLNFQEKTTAVVIKETNIPMFGFSFIPYSQLINLSIDMFNNCQDAMAICKKMGFPYLFIT 73
C N+ A T++ G + S L M N DAMAICK GFP LFIT
Sbjct: 663 CENYNSLKKASEAGTTSMNEEGNQVLIPSSLTGGPRYMVQNYYDAMAICKHYGFPDLFIT 722
Query: 74 ITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVDNLIADLKKENIFGEVDA------ 127
TCN W EI + L D+RP IV R+FK+K+D+L+ DL + G+ A
Sbjct: 723 FTCNPKWPEITRHCQARGLSVDDRPDIVARIFKIKLDSLMKDLTDGKMLGKTVASMHTVE 782
Query: 128 ----GLPHAHILLWLHGESKLTSPHDIDKKISAELPDPQLYPKLYDVVST*IMHGPCGKA 183
GLPHAHILL++ +SKL + DIDK ISAE+PD P+LY+V+ ++HGPCG A
Sbjct: 783 FQKRGLPHAHILLFMDAKSKLPTADDIDKIISAEIPDKDKEPELYEVIKNSMIHGPCGAA 842
Query: 184 NPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRRQSGIKVLKNGIELDSRNVVPYNP 243
N CM GKC+K +PKK Q+ T + +GYP YRRR + + K G + D+ VVPYN
Sbjct: 843 NMNSPCMVEGKCSKQYPKKHQDITKVGKDGYPIYRRRMTEDYIEKGGFKCDNGYVVPYNK 902
Query: 244 KLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRV-----------TAEITTTAE-SSGN 291
KL +RY AHINVE+CN+S SIKYLFKY+NKG+DRV T E T+ E +
Sbjct: 903 KLSLRYQAHINVEWCNQSGSIKYLFKYINKGADRVVFIVEPVNQDKTTENATSGEPPNST 962
Query: 292 KRVVDEIKQYYDCRYLSPCEAVWITFKFEIHE-----------SENQQSVLFGDHEDITD 340
++ DEIK ++DCRY+S EAVW +KF + + E +Q V DI D
Sbjct: 963 EKKKDEIKDWFDCRYVSASEAVWRIYKFPLQDRSTAVQRLSFHDEGKQPVYAKPDADIED 1022
Query: 341 VLDKKGLYHNVF------------------GVDLTYSEFPSKFVYNEKFIIWELRKQGYS 382
VL++ ++F +L YS+ P+ F ++ K W R +G+S
Sbjct: 1023 VLERISNEDSMFMAWLTLNKNNDVGKNGKRARELLYSQIPAYFTWDGKNKQWVKRIRGFS 1082
Query: 383 IRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLSDDKQY 442
+ R+ Y+ YY+R+LL + +G S++ I+T G ++ ++++AC+A G+L DD+ Y
Sbjct: 1083 LGRINYVCRKMEVEYYLRVLLNIVKGPMSYDDIKTFNGVVYPSFKEACFARGILDDDQVY 1142
Query: 443 IGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAE--------------- 487
I + EAS+ G+ LR F L++++S++RP+HVW TW LLAE
Sbjct: 1143 IDGLHEASQFCFGDYLRNFFAMLLLSDSLARPEHVWSETWHLLAEDIENKKREDFKNPDL 1202
Query: 488 ----GKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQNKFGVDEMTYN-SEEM 542
+++ L +I+ I+ NG +L + F +++ N+ VDE+ YN +
Sbjct: 1203 KLTLAEIRNYTLQEIEKIMLRNGATLKEIQDFPQPSREGIDN-SNRLVVDELRYNIDSNL 1261
Query: 543 QDMHANLVSFLTDEQR 558
++ H L EQR
Sbjct: 1262 KEKHDEWFQMLNTEQR 1277
Score = 216 bits (549), Expect = 4e-54
Identities = 108/233 (46%), Positives = 164/233 (70%), Gaps = 7/233 (3%)
Query: 624 IPGGRTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRT 683
+ GGRTAHSRF+IPLN +E CK++ S A L++++SLIIWDEAPM++ FCFE+LD++
Sbjct: 1336 LEGGRTAHSRFAIPLNPDEFSVCKITPKSDLANLIKEASLIIWDEAPMMSKFCFESLDKS 1395
Query: 684 LKDIMSSENRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVL 743
DI+++++ NK FGGKVVV GGDF+Q+L VI R +IV +++N+SYLW++CKVL
Sbjct: 1396 FYDILNNKD----NKVFGGKVVVFGGDFRQVLPVINGAGRVEIVMSSLNASYLWDHCKVL 1451
Query: 744 SLTKNMRLLNTN-GHQDNDDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEP 802
LTKNMRLL+ ++ ++++FS+ +L +G+G +DG+ LID+P +L I + P
Sbjct: 1452 KLTKNMRLLSGGLSSEEAKEIQQFSDWLLAVGDGRINEPNDGEALIDIPEELLIKEAGNP 1511
Query: 803 LLQLIQFVY--PDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+ + + +Y P + + DP F+Q RAILA T E V +NQY+L ++ EE+
Sbjct: 1512 IEAISKEIYGDPSELHMINDPKFFQRRAILAPTNEDVNTINQYMLEHLKSEER 1564
>dbj|BAB01023.1| helicase-like protein [Arabidopsis thaliana]
Length = 1669
Score = 385 bits (988), Expect = e-105
Identities = 214/565 (37%), Positives = 314/565 (54%), Gaps = 66/565 (11%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVD 110
M N DAM CK GFP LFIT TCNS W +I FV L +++P I+CR++KMK++
Sbjct: 690 MQQNYLDAMGTCKHFGFPDLFITFTCNSKWPKITRFVKERKLNAEDKPDIICRIYKMKLE 749
Query: 111 NLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPD 160
NL+ DL K +IFG+ GLPHAHI++W+ K +DK I AE+PD
Sbjct: 750 NLMDDLTKNHIFGKTKLATYTIEFQKRGLPHAHIIVWMDPRYKFPIADHVDKIIFAEIPD 809
Query: 161 PQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRR 220
+ PKLY VS ++HGPC NP CM+NGKC+KY+PK +T +D+ GYP YRRR
Sbjct: 810 KENDPKLYQAVSECMIHGPCRLVNPNSPCMENGKCSKYYPKNHVENTFLDNEGYPIYRRR 869
Query: 221 QSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVT- 279
+G + KN D+R VVPYN LL +Y AHINVE+CN+S S+KYLFKYVNKG DRVT
Sbjct: 870 DTGRFIKKNKYPCDNRYVVPYNDFLLRKYRAHINVEWCNQSVSVKYLFKYVNKGPDRVTV 929
Query: 280 ----------AEITTTAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH------- 322
+E E++ + + ++++ Y+DCRY+S CEA+W + IH
Sbjct: 930 SLEPHRKEVVSEENNVGETNNDPQEQNQVEDYFDCRYVSACEAMWRIKGYPIHYRQTLVT 989
Query: 323 ---------------ESENQQSVLFGDHEDITDV-----LDKKGLYHNVFGVDLTYSEFP 362
E E +SVL+ ++D T L+K+ + L Y + P
Sbjct: 990 KLTFHEKGKQPIYVKEGETAESVLYRVNDDETQFTAWFELNKR----DPEAAKLLYEQIP 1045
Query: 363 SKFVYNEKFIIWELRKQ-GYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGE 421
+ + +N K + RK G+ + R+ ++PP + Y++R+L+ RG F+ I+T++G
Sbjct: 1046 NFYTWNGKDKNFRRRKMPGFVVGRINHVPPKIDDAYHLRILINNIRGPKGFDDIKTIEGV 1105
Query: 422 IHSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWEST 481
+H TY+ ACYALGLL DDK YI I EA+ +R LFV ++I S+S P VWE T
Sbjct: 1106 VHKTYRDACYALGLLDDDKDYINGIEEANFWCFHKYVRKLFVIMLIFESLSSPAVVWEHT 1165
Query: 482 WQLLAEGKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQ-------NKFGVDE 534
W++L+E +K D+++ + + T + + + + N+ +DE
Sbjct: 1166 WKILSEDFQRK------DSVMFTRAAKNIETTWNRSLEEKMLPQLKPGDEPAFNQLILDE 1219
Query: 535 MTYNSEEMQDMHANLVSFLTDEQRK 559
YN E ++ +H + + LT EQ+K
Sbjct: 1220 RNYNRETLKTIHDDWLKMLTTEQKK 1244
Score = 205 bits (522), Expect = 6e-51
Identities = 104/228 (45%), Positives = 151/228 (65%), Gaps = 4/228 (1%)
Query: 626 GGRTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRTLK 685
GGRTAHSRF IPL +ET TC + +GS A+L+ + LIIWDEAPM++ +CFE+LD++LK
Sbjct: 1259 GGRTAHSRFGIPLTPHETSTCNMERGSDLAELVTAAKLIIWDEAPMMSKYCFESLDKSLK 1318
Query: 686 DIMSSENRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVLSL 745
DI+S+ + PFGGK+++ GGDF+QIL VI R+ IV +++NSS+LW+ CKV L
Sbjct: 1319 DILSTPE----DMPFGGKLIIFGGDFRQILPVILAAGRELIVKSSLNSSHLWQYCKVFKL 1374
Query: 746 TKNMRLLNTNGHQDNDDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPLLQ 805
TKNMRLL + ++++FS+ IL +G G+ +DG I + D+ I + P+
Sbjct: 1375 TKNMRLLQDIDINEAREIEDFSKWILAVGEGKLNQPNDGVTQIQIRDDILIPEGDNPIES 1434
Query: 806 LIQFVYPDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+I+ VY DP F+Q+RAIL T + V +N ++LS + GEEK
Sbjct: 1435 IIKAVYGTSFDEERDPKFFQDRAILCPTNDDVNSINDHMLSKLTGEEK 1482
>gb|AAK54292.1| putative helicase [Oryza sativa (japonica cultivar-group)]
Length = 1501
Score = 363 bits (932), Expect = 2e-98
Identities = 196/528 (37%), Positives = 296/528 (55%), Gaps = 58/528 (10%)
Query: 54 NCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSINL-KPDERPYIVCRVFKMKVDNL 112
N QDAMAIC+ G P LF+T TCN W EI + + KP ERP I+ RVF +K+ L
Sbjct: 531 NYQDAMAICRWAGHPDLFVTFTCNPKWPEIQCMLDKVGYQKPSERPDILVRVFMIKLKEL 590
Query: 113 IADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELPDPQ 162
++D+K+ FG+ A GLPHAHIL++L K P ID+ I AE+PD
Sbjct: 591 MSDIKRNQHFGKTKAIVFTIEFQKRGLPHAHILIFLDKRGKSLEPSQIDELICAEIPDRD 650
Query: 163 LYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRRQS 222
P+ ++ V +MHGPCG+ANP CM + KC ++FP+ F + T ID+ +P YRRR
Sbjct: 651 KDPETFEAVKNFMMHGPCGEANPKSPCMVDHKCNRFFPRGFSDETIIDEVNFPIYRRRDD 710
Query: 223 GIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTAEI 282
G ++ K + L++ VVPYN LL ++ H+NVE+ N+S SIKYLFK + D+ TA +
Sbjct: 711 GRQIKKGRVNLNNGFVVPYNKDLLAKFQVHMNVEWFNRSRSIKYLFKSICNEDDQATAVV 770
Query: 283 TTTAESSGNKRVVDEIKQYYDCRYLSPCEAVWITFKFEIH-----------ESENQQSVL 331
T E + DEIK+Y C+Y + EA W FKF +H EN+Q V+
Sbjct: 771 EETDEKNN-----DEIKRYLGCKYTTATEACWRIFKFPLHYQEPPVERLSFHEENEQHVI 825
Query: 332 FGDHEDITDVLDKKGLYHNVF------------GVDLTYSEFPSKFVYNEKFIIWELRKQ 379
F D D+ +++ + +F +LTYSEFP+K+ +++ W RK
Sbjct: 826 FPDSTDLQEIVRRPRSGVTMFTEWMETNKRHEDARELTYSEFPTKWTWDKNVKKWVRRKG 885
Query: 380 GYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLSDD 439
G I R+ P +GE YY+R++L +GCT+FE IRTV G +HS+Y+ AC+ALG L+DD
Sbjct: 886 GMKIGRIYNAHPASGERYYLRVILNTAKGCTTFEDIRTVNGTVHSSYKSACHALGFLNDD 945
Query: 440 KQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAE------------ 487
++I I+EAS SG +LR LF +++ ++ P +WES+W+ L++
Sbjct: 946 SEWIECIKEASCWASGMKLRQLFATVLCHCEVTDPKRLWESSWEKLSKDIQHTQSWALNF 1005
Query: 488 -------GKMKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQN 528
+K L++I+ ++ GKSL ++ +++++ +N
Sbjct: 1006 PTSCLTPSHRRKCALIEIEKNMRQAGKSLKEYAGIEPPNMAKLSEIEN 1053
Score = 204 bits (520), Expect = 1e-50
Identities = 102/230 (44%), Positives = 153/230 (66%), Gaps = 2/230 (0%)
Query: 624 IPGGRTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRT 683
+ GGRTAHS F+IP+N+ + TC + +GS A LL ++SLI+WDEAPM N CFEALD++
Sbjct: 1086 LQGGRTAHSAFNIPINLTDEYTCFIKQGSHIADLLMKTSLILWDEAPMANRNCFEALDKS 1145
Query: 684 LKDIMSSENRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVL 743
L+D+ N + KPFGG VVLGGDF+QIL ++ KG R+ V AT+ SYLW++ +V
Sbjct: 1146 LRDVQRCRNENSCQKPFGGMTVVLGGDFRQILPIVPKGRREHTVNATIKCSYLWQHFEVF 1205
Query: 744 SLTKNMRLLNTNGHQ-DNDDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEP 802
+LTKNMRL + Q ++ EF+E IL++GNG++ D+ K + +P DL + +P
Sbjct: 1206 NLTKNMRLNYVSKDQTEHQKSAEFAEWILQIGNGDTISLDE-KGWVRMPSDLLLQKGDDP 1264
Query: 803 LLQLIQFVYPDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEE 852
Q+I+ YPD+ + + +ERAIL E+V ++N+YI+ I+G++
Sbjct: 1265 KAQIIESTYPDLQDNCCKQNYLEERAILCPVNENVNELNEYIMDQIQGDK 1314
>ref|NP_175704.1| hypothetical protein [Arabidopsis thaliana]
gi|12324645|gb|AAG52281.1| hypothetical protein;
100703-104276 [Arabidopsis thaliana]
Length = 996
Score = 355 bits (911), Expect = 4e-96
Identities = 187/490 (38%), Positives = 282/490 (57%), Gaps = 60/490 (12%)
Query: 128 GLPHAHILLWLHGESKLTSPHDIDKKISAELPDPQLYPKLYDVVST*IMHGPCGKANPTC 187
GLPHAHILL++H SKL++ D DK I+AE+PD + P LY VV ++HGPCG +P
Sbjct: 63 GLPHAHILLFMHPTSKLSTAEDTDKIITAEIPDKKKKPGLYAVVKDCMIHGPCGVGHPNS 122
Query: 188 ACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRRRQSGIKVLKNGIELDSRNVVPYNPKLLM 247
CM+NGKC KYFPK + ++T +D++G+P YRRR +GI V KNG + D+R V+PYN K+ +
Sbjct: 123 PCMENGKCKKYFPKSYSDTTKVDNDGFPVYRRRDTGIYVEKNGFQCDNRYVIPYNEKVSL 182
Query: 248 RYHAHINVEYCNKSNSIKYLFKYVNKGSDRVTAEITTTAESSGNKRVVDEIKQYYDCRYL 307
RY AHINVE CN+S SIKYLFKYV+KG DRVT + + + K+ DE+K Y+DCRY+
Sbjct: 183 RYQAHINVELCNQSGSIKYLFKYVHKGHDRVTVTVEPNDQDTA-KKEKDEVKDYFDCRYV 241
Query: 308 SPCEAVWITFKFEIH-----------ESENQQSVLFGDHEDITDVLDKKGLYHNVF---- 352
S CEA+W FKF IH E +Q V + E V+D+ F
Sbjct: 242 SACEAMWRIFKFPIHYRTTPVVKLFFHEEGKQPVYYKPGETTESVMDRLSSEATQFLAWF 301
Query: 353 ------------------------GVDLTYSEFPSKFVYNEKFIIWELRKQGYSIRRLTY 388
L + E P+ F +N K + +R++G++I R+ +
Sbjct: 302 QLNKKPPSRTIRANAKKLPKAAPDPTKLLFEEIPNHFTWNSKEKKFMIRERGFAIGRINF 361
Query: 389 IPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKACYALGLLSDDKQYIGAIRE 448
+P + YY+R+LL ++RG TS++ ++TVKG +H +++ A +ALGLL DDK+YI I++
Sbjct: 362 VPRTIEDAYYLRILLNIKRGVTSYKDLKTVKGVVHKSFRDAVFALGLLDDDKEYINGIKD 421
Query: 449 ASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAEG-------------------K 489
A S +R LFV ++++ S+++P+ VW+ TW++L+E +
Sbjct: 422 AKFWCSAKYVRRLFVIMLLSESLTKPEMVWDETWRILSEDIERRKRKEWKRPDLQLSDEE 481
Query: 490 MKK*CLLKIDNILQSNGKSLADFTCFSNYDLSRVEHYQNKFGVDEMTYNSEEMQDMHANL 549
++ CL +I +L NG SL+ + VE N F +DE Y+ + + H
Sbjct: 482 RQQYCLQEIARLLTKNGVSLSKWKQMPQISDEHVEKC-NHFILDERKYDRAYLIEKHEEW 540
Query: 550 VSFLTDEQRK 559
++ +T EQ+K
Sbjct: 541 LTMVTSEQKK 550
Score = 226 bits (577), Expect = 2e-57
Identities = 109/230 (47%), Positives = 161/230 (69%), Gaps = 4/230 (1%)
Query: 624 IPGGRTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRT 683
+ GGRT HSRF IP+N NE+ TC +S+GS +L+++++LIIWDE PM++ CFE+LDRT
Sbjct: 608 LDGGRTTHSRFGIPINPNESSTCNISRGSDLGELVKEANLIIWDETPMMSKHCFESLDRT 667
Query: 684 LKDIMSSENRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVL 743
L+DIM++ +KPFGGK +V GGDF+Q+L VI R++IV A +NSSY+WE+CKVL
Sbjct: 668 LRDIMNNPG----DKPFGGKGIVFGGDFRQVLPVINGAGREEIVFAALNSSYIWEHCKVL 723
Query: 744 SLTKNMRLLNTNGHQDNDDVKEFSE*ILKLGNGESTPNDDGKMLIDVPHDLFITDPSEPL 803
LTKNMRLL + D++ FS+ IL +G+G+ + +DG LID+P + I ++P+
Sbjct: 724 ELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQPNDGIALIDIPEEFLINGDNDPV 783
Query: 804 LQLIQFVYPDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+I+ VY + DP F+Q RAIL T E V +N++++S ++GEE+
Sbjct: 784 ESIIEAVYGNTFMEEKDPKFFQGRAILCPTNEDVNSINEHMMSMLDGEER 833
>gb|AAD25621.1| Hypothetical protein [Arabidopsis thaliana]
gi|25353659|pir||A96586 hypothetical protein F20D21.24
[imported] - Arabidopsis thaliana
Length = 1250
Score = 346 bits (887), Expect = 3e-93
Identities = 200/522 (38%), Positives = 301/522 (57%), Gaps = 60/522 (11%)
Query: 51 MFNNCQDAMAICKKMGFPYLFITITCNSNWKEIADFVCSI-NLKPDERPYIVCRVFKMKV 109
M DAMAIC+ G P LFIT+T N W+EI++ + + N + RP I CRVFK+K+
Sbjct: 278 MVEKYHDAMAICRWYGNPDLFITMTTNPKWEEISEHLKTYGNDAANVRPDIECRVFKIKL 337
Query: 110 DNLIADLKKENIFGEVDA----------GLPHAHILLWLHGESKLTSPHDIDKKISAELP 159
D L+AD K F + A GLPHAHILLWL G+ K +P+DIDK ISAE+P
Sbjct: 338 DELLADFNKGLFFPKPIAIVYTIEFQKRGLPHAHILLWLQGDLKKPTPNDIDKYISAEIP 397
Query: 160 DPQLYPKLYDVVST*IMHGPCGKANPTCACMKNGKCTKYFPKKFQNSTTIDDNGYPHYRR 219
P+ + +V +MHGPCGK P+ CM+ G C+K FP++F N T ++++G+ YRR
Sbjct: 398 VKDKDPEGHTLVEQHMMHGPCGKDRPSSPCMEKGICSKKFPREFVNHTKMNESGFILYRR 457
Query: 220 RQSGIKVLKNGIELDSRNVVPYNPKLLMRYHAHINVEYCNKSNSIKYLFKYVNKGSDRVT 279
R VLK LD+R VVP+N ++L +Y AHINVE+CNKS++IKYLFKY+ KG D+ T
Sbjct: 458 RNDQRYVLKGQTRLDNRFVVPHNLEILKKYKAHINVEWCNKSSAIKYLFKYITKGVDKAT 517
Query: 280 AEI----TTTAESSGN---KRVVDEIKQYYDCRYLSPCEAVWITFKFEIHESE------- 325
I + + SGN ++ +EI +Y DCRYLS CEA+W F F IH
Sbjct: 518 FIIQKGNSVNGQGSGNGFEEKPRNEINEYLDCRYLSACEAMWRIFMFNIHHHNPPVQRLP 577
Query: 326 ----NQQSVLFGDHEDITDVLDKKG----LYHNVFGVD--------LTYSEFPSKFVYNE 369
+QS +F + E++ +V + G + F ++ L Y + P+ FV++
Sbjct: 578 LHLPGEQSTIFEEEENLENVEYRYGHERTMLTEYFELNKICEDARKLKYVQVPTMFVWDS 637
Query: 370 KFIIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSFESIRTVKGEIHSTYQKA 429
++ RKQ +I R+ I P G+LYY+R+LL +G TSF+ ++TV G +H +++ A
Sbjct: 638 TNKMYTRRKQRENIGRIVNILPTAGDLYYLRILLNKVKGATSFDYLKTVGGVVHESFKAA 697
Query: 430 CYALGLLSDDKQYIGAIREASELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAEG- 488
C+ GLL DK++ A+ EA++ + LR LFV ++I +S P +W W+ +A+
Sbjct: 698 CHTRGLLDGDKEWHDAMDEAAQWSTSYLLRSLFVLILIYCEVSEPLKLWSHCWESMADDV 757
Query: 489 ------------------KMKK*CLLKIDNILQSNGKSLADF 512
+++K L++I+ +L+ + KSL+D+
Sbjct: 758 FRKQQKVLNFPQLELKAEELEKYTLIEIETLLRQHEKSLSDY 799
Score = 178 bits (452), Expect = 7e-43
Identities = 98/236 (41%), Positives = 147/236 (61%), Gaps = 14/236 (5%)
Query: 624 IPGGRTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAPMLNIFCFEALDRT 683
+PGGRTAHSRF IP+N++E C + GS+ A +L + LIIWDEAPM + FEA+DRT
Sbjct: 826 LPGGRTAHSRFKIPINVHEDSICDIKIGSMLANVLSKVDLIIWDEAPMAHRHTFEAVDRT 885
Query: 684 LKDIMSSENRANANKPFGGKVVVLGGDFKQILHVIQKGSRQDIVTATVNSSYLWENCKVL 743
L+DI+S + K FGGK V+LGGDF+QIL VI +G+RQ+ V+A +N SYLWE+C
Sbjct: 886 LRDILSVGDEKALTKTFGGKTVLLGGDFRQILPVIPQGTRQETVSAAINRSYLWESCHKY 945
Query: 744 SLTKNMRLLNTNGHQDNDDVKEFSE*ILKLGNGES------TPNDDGKMLIDVPHDLFIT 797
L++NMR+ +++K F+E IL++G+GE+ +D + I + +L +
Sbjct: 946 LLSQNMRV-------QPEEIK-FAEWILQVGDGEAPRKTHGIDDDQEEDNIIIDKNLLLP 997
Query: 798 DPSEPLLQLIQFVYPDMVSHLTDPTFYQERAILALTLESVEKVNQYILSCIEGEEK 853
+ PL L + V+PD + D + A+L E+V+++N Y+LS + G K
Sbjct: 998 ETENPLEVLCRSVFPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLLSKVPGLAK 1053
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.333 0.145 0.469
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,731,836,189
Number of Sequences: 2540612
Number of extensions: 71801828
Number of successful extensions: 221637
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 126
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 220813
Number of HSP's gapped (non-prelim): 365
length of query: 1046
length of database: 863,360,394
effective HSP length: 138
effective length of query: 908
effective length of database: 512,755,938
effective search space: 465582391704
effective search space used: 465582391704
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 81 (35.8 bits)
Lotus: description of TM0310b.7


