

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0307.15
(326 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
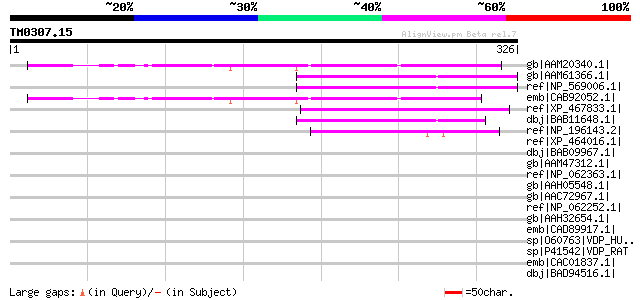
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM20340.1| unknown protein [Arabidopsis thaliana] gi|1738065... 125 2e-27
gb|AAM61366.1| unknown [Arabidopsis thaliana] 119 9e-26
ref|NP_569006.1| expressed protein [Arabidopsis thaliana] 119 9e-26
emb|CAB92052.1| putative protein [Arabidopsis thaliana] gi|11288... 107 5e-22
ref|XP_467833.1| unknown protein [Oryza sativa (japonica cultiva... 102 2e-20
dbj|BAB11648.1| unnamed protein product [Arabidopsis thaliana] 99 2e-19
ref|NP_196143.2| expressed protein [Arabidopsis thaliana] 49 2e-04
ref|XP_464016.1| unknown protein [Oryza sativa (japonica cultiva... 43 0.014
dbj|BAB09967.1| unnamed protein product [Arabidopsis thaliana] 42 0.025
gb|AAM47312.1| At2g40630/T2P4.2 [Arabidopsis thaliana] gi|201968... 42 0.025
ref|NP_062363.1| vesicle docking protein [Mus musculus] gi|16359... 40 0.12
gb|AAH05548.1| Vdp-pending protein [Mus musculus] 40 0.12
gb|AAC72967.1| transcytosis associated protein p115 [Mus musculu... 40 0.12
ref|NP_062252.1| transcytosis associated protein [Rattus norvegi... 39 0.16
gb|AAH32654.1| Vesicle docking protein p115 [Homo sapiens] 39 0.16
emb|CAD89917.1| hypothetical protein [Homo sapiens] 39 0.16
sp|O60763|VDP_HUMAN General vesicular transport factor p115 (Tra... 39 0.16
sp|P41542|VDP_RAT General vesicular transport factor p115 (Trans... 38 0.36
emb|CAC01837.1| putative protein [Arabidopsis thaliana] gi|15237... 38 0.36
dbj|BAD94516.1| putative protein [Arabidopsis thaliana] 38 0.36
>gb|AAM20340.1| unknown protein [Arabidopsis thaliana] gi|17380658|gb|AAL36159.1|
unknown protein [Arabidopsis thaliana]
gi|22326700|ref|NP_196573.2| expressed protein
[Arabidopsis thaliana]
Length = 321
Score = 125 bits (313), Expect = 2e-27
Identities = 113/330 (34%), Positives = 155/330 (46%), Gaps = 54/330 (16%)
Query: 12 VKPQPRPVLSDVTNLRAKRPFSSISADDGDSRFTKKKVQSKLGVQAQQGRSNKDKVVVSQ 71
V Q RPVLSD+TNL KR SSI D D + G++ +V S+
Sbjct: 9 VSEQTRPVLSDLTNLPKKRGISSILGDLLD----------------ESGKTVAHEV--SK 50
Query: 72 QK-GQNPCLELPFCGDDSLGNVSSQESNLQLPSGGLE-EKNLLGGAVVSGPTEVPVAVLG 129
+K + CL + D L V + L G +KN G AV V
Sbjct: 51 EKFSKRLCLVV-----DDL--VKENATPLDTIQGSCSFDKNTSGDAVDKEEYHDAVMEFS 103
Query: 130 EIGQGRPLNEG-----FIDRGIESGQRDVRAVDNLGSPKC-GGRAVELPTMSDSCDSRFP 183
G + L E + + G G R++ A N GG + L ++ +SR
Sbjct: 104 S-GDCKALKESKSIQIYFEPGDRDGARELNAAANADQTDVTGGEGLALSLLTSDTESRNL 162
Query: 184 ----------------GLERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHY 227
+ RCS ++ N + DLL+SC CSFC A++IWSDL+Y
Sbjct: 163 LKTGKELSDCQNLRSFEMSRCSNVK-NKEHVNLNTGDDLLRSCCCSFCLTASYIWSDLNY 221
Query: 228 QDVKGRLSALKKSQKDASNAVQKLSEIKD-SVMPDQQSSSESSKLELALMHQWKSLFDFM 286
QD KGRLSA+KKSQK ASN +Q +K+ S +S S+K E LM QW+SLF M
Sbjct: 222 QDSKGRLSAMKKSQKAASNLIQ--GNVKERSTDFHATGNSVSAKQESKLMAQWRSLFLSM 279
Query: 287 ENTFAEESRQLESSFETLKDLRDNCKNDLD 316
+ +EES L++SF +K LRD+CK DL+
Sbjct: 280 GDILSEESSHLQNSFVRMKKLRDDCKMDLE 309
>gb|AAM61366.1| unknown [Arabidopsis thaliana]
Length = 317
Score = 119 bits (299), Expect = 9e-26
Identities = 64/142 (45%), Positives = 85/142 (59%), Gaps = 1/142 (0%)
Query: 185 LERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDA 244
+ RCS + G A LKSC+CSFC AA+IWSDLHYQD+KGRLS LKKSQK+A
Sbjct: 176 MNRCSNVDGMGIVNHHMEADGELKSCSCSFCLTAAYIWSDLHYQDIKGRLSVLKKSQKEA 235
Query: 245 SNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETL 304
S +Q+ + + ++S+ S+ + M QW SLF ME A ES L +SF +
Sbjct: 236 SGLIQRNGRGTPTDIYGSENSNNSTNTDNP-MEQWTSLFRNMEGILARESNHLHNSFVAM 294
Query: 305 KDLRDNCKNDLDSTDNTHFDNH 326
K+LR+NCK DL+ T N+
Sbjct: 295 KELRENCKIDLERATKTSQQNN 316
>ref|NP_569006.1| expressed protein [Arabidopsis thaliana]
Length = 317
Score = 119 bits (299), Expect = 9e-26
Identities = 64/142 (45%), Positives = 85/142 (59%), Gaps = 1/142 (0%)
Query: 185 LERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDA 244
+ RCS + G A LKSC+CSFC AA+IWSDLHYQD+KGRLS LKKSQK+A
Sbjct: 176 MNRCSNVDGMGIVNHHMEADGELKSCSCSFCLTAAYIWSDLHYQDIKGRLSVLKKSQKEA 235
Query: 245 SNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETL 304
S +Q+ + + ++S+ S+ + M QW SLF ME A ES L +SF +
Sbjct: 236 SGLIQRNDRGTPTDIYGSENSNNSTNTDNP-MEQWTSLFRNMEGILARESNHLHNSFVAM 294
Query: 305 KDLRDNCKNDLDSTDNTHFDNH 326
K+LR+NCK DL+ T N+
Sbjct: 295 KELRENCKIDLERATKTSQQNN 316
>emb|CAB92052.1| putative protein [Arabidopsis thaliana] gi|11288901|pir||T50015
hypothetical protein T31P16.100 - Arabidopsis thaliana
Length = 297
Score = 107 bits (267), Expect = 5e-22
Identities = 105/317 (33%), Positives = 144/317 (45%), Gaps = 54/317 (17%)
Query: 12 VKPQPRPVLSDVTNLRAKRPFSSISADDGDSRFTKKKVQSKLGVQAQQGRSNKDKVVVSQ 71
V Q RPVLSD+TNL KR SSI D D + G++ +V S+
Sbjct: 9 VSEQTRPVLSDLTNLPKKRGISSILGDLLD----------------ESGKTVAHEV--SK 50
Query: 72 QK-GQNPCLELPFCGDDSLGNVSSQESNLQLPSGGLE-EKNLLGGAVVSGPTEVPVAVLG 129
+K + CL + D L V + L G +KN G AV V
Sbjct: 51 EKFSKRLCLVV-----DDL--VKENATPLDTIQGSCSFDKNTSGDAVDKEEYHDAVMEFS 103
Query: 130 EIGQGRPLNEG-----FIDRGIESGQRDVRAVDNLGSPKC-GGRAVELPTMSDSCDSRFP 183
G + L E + + G G R++ A N GG + L ++ +SR
Sbjct: 104 S-GDCKALKESKSIQIYFEPGDRDGARELNAAANADQTDVTGGEGLALSLLTSDTESRNL 162
Query: 184 ----------------GLERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHY 227
+ RCS ++ N + DLL+SC CSFC A++IWSDL+Y
Sbjct: 163 LKTGKELSDCQNLRSFEMSRCSNVK-NKEHVNLNTGDDLLRSCCCSFCLTASYIWSDLNY 221
Query: 228 QDVKGRLSALKKSQKDASNAVQKLSEIKD-SVMPDQQSSSESSKLELALMHQWKSLFDFM 286
QD KGRLSA+KKSQK ASN +Q +K+ S +S S+K E LM QW+SLF M
Sbjct: 222 QDSKGRLSAMKKSQKAASNLIQ--GNVKERSTDFHATGNSVSAKQESKLMAQWRSLFLSM 279
Query: 287 ENTFAEESRQLESSFET 303
+ +EES L S+F +
Sbjct: 280 GDILSEESSHLVSAFRS 296
>ref|XP_467833.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|46390226|dbj|BAD15657.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|46390123|dbj|BAD15558.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 329
Score = 102 bits (253), Expect = 2e-20
Identities = 55/134 (41%), Positives = 78/134 (58%)
Query: 188 CSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDASNA 247
CS + ++ S D KSC CSFC KAA +W+DLHYQD +GRL+ALKKS K A
Sbjct: 192 CSDVHHHSLGNSELENDDTTKSCACSFCLKAAFMWTDLHYQDTRGRLAALKKSIKFARLL 251
Query: 248 VQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETLKDL 307
++ + SV + + ++++E L Q +SLF EN ES QL SS LK+
Sbjct: 252 GKRSQGDEYSVNAGRYNLKRAAEMEFELYQQQRSLFLHTENVLIRESAQLHSSLVKLKEF 311
Query: 308 RDNCKNDLDSTDNT 321
R+NCK DL++ ++
Sbjct: 312 RENCKTDLETASSS 325
>dbj|BAB11648.1| unnamed protein product [Arabidopsis thaliana]
Length = 324
Score = 99.0 bits (245), Expect = 2e-19
Identities = 55/122 (45%), Positives = 72/122 (58%), Gaps = 1/122 (0%)
Query: 185 LERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDA 244
+ RCS + G A LKSC+CSFC AA+IWSDLHYQD+KGRLS LKKSQK+A
Sbjct: 176 MNRCSNVDGMGIVNHHMEADGELKSCSCSFCLTAAYIWSDLHYQDIKGRLSVLKKSQKEA 235
Query: 245 SNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETL 304
S +Q+ + + ++S+ S+ + M QW SLF ME A ES L SS + +
Sbjct: 236 SGLIQRNDRGTPTDIYGSENSNNSTNTDNP-MEQWTSLFRNMEGILARESNHLVSSSDLI 294
Query: 305 KD 306
D
Sbjct: 295 FD 296
>ref|NP_196143.2| expressed protein [Arabidopsis thaliana]
Length = 530
Score = 49.3 bits (116), Expect = 2e-04
Identities = 37/132 (28%), Positives = 64/132 (48%), Gaps = 10/132 (7%)
Query: 194 NAGPTSAAAAADLLKSCTCSF-CSKAAHIWSDLHYQDVKGRLSALKKSQKDASNAVQ-KL 251
N S+ A+DL + + + A W +L +QD+KGR+SAL++S+K V +L
Sbjct: 344 NDSSLSSEDASDLNSASVLTVNAATVASQWLELLHQDIKGRVSALRRSRKRVRAVVTIEL 403
Query: 252 SEIKDSVMPDQQSSSE------SSKLELALMH--QWKSLFDFMENTFAEESRQLESSFET 303
+ P Q + +SK +H +W +LF +E+ +EE QLES
Sbjct: 404 PHLIRKEFPADQENDPTLLLGGASKASTVDIHKSRWMTLFKQLEHKLSEEESQLESWLNQ 463
Query: 304 LKDLRDNCKNDL 315
++ ++ +C L
Sbjct: 464 VRYMQSHCDEGL 475
>ref|XP_464016.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|41052574|dbj|BAD07756.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 534
Score = 42.7 bits (99), Expect = 0.014
Identities = 30/113 (26%), Positives = 52/113 (45%), Gaps = 12/113 (10%)
Query: 215 CSKAAHIWSDLHYQDVKGRLSALKKSQKDASNAVQKL------------SEIKDSVMPDQ 262
CS +A ++ H + ALK+S+K NA+Q E + SV+
Sbjct: 368 CSTSADDFASNHDYMTVLSVKALKRSKKRVRNALQTELPYLISTEFSYNQENEPSVVHSS 427
Query: 263 QSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETLKDLRDNCKNDL 315
S + A + +W+SLF M+ T EE LE+ + +++++ NC+ L
Sbjct: 428 DGGSTGKTVPEAHVARWRSLFLQMDRTLQEEGMHLENRLKEVQEMQMNCEKGL 480
>dbj|BAB09967.1| unnamed protein product [Arabidopsis thaliana]
Length = 570
Score = 42.0 bits (97), Expect = 0.025
Identities = 37/140 (26%), Positives = 64/140 (45%), Gaps = 18/140 (12%)
Query: 194 NAGPTSAAAAADLLKSCTCSF-CSKAAHIWSDLHYQDVKGRLSALKKSQKDASNAVQ-KL 251
N S+ A+DL + + + A W +L +QD+KGR+SAL++S+K V +L
Sbjct: 344 NDSSLSSEDASDLNSASVLTVNAATVASQWLELLHQDIKGRVSALRRSRKRVRAVVTIEL 403
Query: 252 SEIKDSVMPDQQSSSE------SSKLELALMH--QWKSLFDFMENTFAEESRQL------ 297
+ P Q + +SK +H +W +LF +E+ +EE QL
Sbjct: 404 PHLIRKEFPADQENDPTLLLGGASKASTVDIHKSRWMTLFKQLEHKLSEEESQLFVDSAN 463
Query: 298 --ESSFETLKDLRDNCKNDL 315
ES ++ ++ +C L
Sbjct: 464 IQESWLNQVRYMQSHCDEGL 483
>gb|AAM47312.1| At2g40630/T2P4.2 [Arabidopsis thaliana] gi|20196873|gb|AAM14814.1|
expressed protein [Arabidopsis thaliana]
gi|15294202|gb|AAK95278.1| At2g40630/T2P4.2 [Arabidopsis
thaliana] gi|18405458|ref|NP_030608.1| expressed protein
[Arabidopsis thaliana]
Length = 535
Score = 42.0 bits (97), Expect = 0.025
Identities = 26/111 (23%), Positives = 50/111 (44%), Gaps = 19/111 (17%)
Query: 222 WSDLHYQDVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMH---- 277
W + QD+ GRLSA++ S+ N + +E+ + SS++++ LE+ +
Sbjct: 372 WLEFLQQDLSGRLSAVQDSRNRVQNIL--TTELPLLASSRESSSNQANSLEMVTTNTSGD 429
Query: 278 -------------QWKSLFDFMENTFAEESRQLESSFETLKDLRDNCKNDL 315
+W + FD + +E R LE S +K+++ C + L
Sbjct: 430 ASSDKAATETHQKRWTAKFDQINKALYDEQRDLERSLNQVKEMQSRCNHGL 480
>ref|NP_062363.1| vesicle docking protein [Mus musculus] gi|16359203|gb|AAH16069.1|
Vesicle docking protein [Mus musculus]
Length = 959
Score = 39.7 bits (91), Expect = 0.12
Identities = 30/105 (28%), Positives = 51/105 (48%), Gaps = 9/105 (8%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + ++ SS +E KL+L + K
Sbjct: 845 DVEGRLSALLQETKELKNEIKALSEERTAIQKQLDSSNSTIAILQTEKDKLDLEVTDSKK 904
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNCKNDLDSTDNTHFDN 325
D + A++ +++ S LKDL + + +S D D+
Sbjct: 905 EQDDLLV-LLADQDQKILSLKSKLKDLGHPVEEEDESGDQEDDDD 948
>gb|AAH05548.1| Vdp-pending protein [Mus musculus]
Length = 335
Score = 39.7 bits (91), Expect = 0.12
Identities = 30/105 (28%), Positives = 51/105 (48%), Gaps = 9/105 (8%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + ++ SS +E KL+L + K
Sbjct: 221 DVEGRLSALLQETKELKNEIKALSEERTAIQKQLDSSNSTIAILQTEKDKLDLEVTDSKK 280
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNCKNDLDSTDNTHFDN 325
D + A++ +++ S LKDL + + +S D D+
Sbjct: 281 EQDDLLV-LLADQDQKILSLKSKLKDLGHPVEEEDESGDQEDDDD 324
>gb|AAC72967.1| transcytosis associated protein p115 [Mus musculus]
gi|13431959|sp|Q9Z1Z0|VDP_MOUSE General vesicular
transport factor p115 (Transcytosis associated protein)
(TAP) (Vesicle docking protein)
Length = 941
Score = 39.7 bits (91), Expect = 0.12
Identities = 30/105 (28%), Positives = 51/105 (48%), Gaps = 9/105 (8%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + ++ SS +E KL+L + K
Sbjct: 827 DVEGRLSALLQETKELKNEIKALSEERTAIQKQLDSSNSTIAILQTEKDKLDLEVTDSKK 886
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNCKNDLDSTDNTHFDN 325
D + A++ +++ S LKDL + + +S D D+
Sbjct: 887 EQDDLLV-LLADQDQKILSLKSKLKDLGHPVEEEDESGDQEDDDD 930
>ref|NP_062252.1| transcytosis associated protein [Rattus norvegicus]
gi|558475|gb|AAC52151.1| transcytosis associated protein
gi|1083815|pir||A55913 transcytosis-associated protein
p115 - rat
Length = 959
Score = 39.3 bits (90), Expect = 0.16
Identities = 31/105 (29%), Positives = 50/105 (47%), Gaps = 9/105 (8%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + S+ SS +E KL L + K
Sbjct: 845 DVEGRLSALLQETKELKNEIKALSEERTSIQKQLDSSNSTIAILQTEKDKLYLEVTDSKK 904
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNCKNDLDSTDNTHFDN 325
D + A++ +++ S LKDL + + +S D D+
Sbjct: 905 EQDDLLV-LLADQDQKILSLKSKLKDLGHPVEEEDESGDQEDDDD 948
>gb|AAH32654.1| Vesicle docking protein p115 [Homo sapiens]
Length = 961
Score = 39.3 bits (90), Expect = 0.16
Identities = 32/107 (29%), Positives = 53/107 (48%), Gaps = 11/107 (10%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + ++ SS +E KLEL + K
Sbjct: 844 DVEGRLSALLQETKELKNEIKALSEERTAIKEQLDSSNSTIAILQTEKDKLELEITDSKK 903
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNC--KNDLDSTDNTHFDN 325
D + A++ +++ S LKDL +++L+S D D+
Sbjct: 904 EQDDLLV-LLADQDQKILSLKNKLKDLGHPVEEEDELESGDQEDEDD 949
>emb|CAD89917.1| hypothetical protein [Homo sapiens]
Length = 973
Score = 39.3 bits (90), Expect = 0.16
Identities = 32/107 (29%), Positives = 53/107 (48%), Gaps = 11/107 (10%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + ++ SS +E KLEL + K
Sbjct: 856 DVEGRLSALLQETKELKNEIKALSEERTAIKEQLDSSNSTIAILQTEKDKLELEITDSKK 915
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNC--KNDLDSTDNTHFDN 325
D + A++ +++ S LKDL +++L+S D D+
Sbjct: 916 EQDDLLV-LLADQDQKILSLKNKLKDLGHPVEEEDELESGDQEDEDD 961
>sp|O60763|VDP_HUMAN General vesicular transport factor p115 (Transcytosis associated
protein) (TAP) (Vesicle docking protein)
gi|2988344|dbj|BAA25300.1| p115 [Homo sapiens]
gi|4505541|ref|NP_003706.1| vesicle docking protein p115
[Homo sapiens]
Length = 962
Score = 39.3 bits (90), Expect = 0.16
Identities = 32/107 (29%), Positives = 53/107 (48%), Gaps = 11/107 (10%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + ++ SS +E KLEL + K
Sbjct: 845 DVEGRLSALLQETKELKNEIKALSEERTAIKEQLDSSNSTIAILQTEKDKLELEITDSKK 904
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNC--KNDLDSTDNTHFDN 325
D + A++ +++ S LKDL +++L+S D D+
Sbjct: 905 EQDDLLV-LLADQDQKILSLKNKLKDLGHPVEEEDELESGDQEDEDD 950
>sp|P41542|VDP_RAT General vesicular transport factor p115 (Transcytosis associated
protein) (TAP) (Vesicle docking protein)
gi|538153|gb|AAA62632.1| p115
Length = 959
Score = 38.1 bits (87), Expect = 0.36
Identities = 30/105 (28%), Positives = 50/105 (47%), Gaps = 9/105 (8%)
Query: 229 DVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSS--------SESSKLELALMHQWK 280
DV+GRLSAL + K+ N ++ LSE + ++ SS +E KL L + K
Sbjct: 845 DVEGRLSALLQETKELKNEIKALSEERTAIQKQLDSSNSTIAILQTEKDKLYLEVTDSKK 904
Query: 281 SLFDFMENTFAEESRQLESSFETLKDLRDNCKNDLDSTDNTHFDN 325
D + A++ +++ S LKDL + + +S D D+
Sbjct: 905 EQDDLLV-LLADQDQKILSLKSKLKDLGHPVEEEDESGDQEDDDD 948
>emb|CAC01837.1| putative protein [Arabidopsis thaliana]
gi|15237400|ref|NP_197175.1| expressed protein
[Arabidopsis thaliana] gi|11357813|pir||T51505
hypothetical protein F5E19_70 - Arabidopsis thaliana
Length = 853
Score = 38.1 bits (87), Expect = 0.36
Identities = 22/91 (24%), Positives = 47/91 (51%), Gaps = 1/91 (1%)
Query: 233 RLSALKKSQKDASNAVQKLSEIKDSVMPD-QQSSSESSKLELALMHQWKSLFDFMENTFA 291
RLS +K+ K A+ + L + K + + +Q+ E+ ++ L L K+ EN+
Sbjct: 99 RLSQIKEDLKKANERISSLEKDKAKALDELKQAKKEAEQVTLKLDDALKAQKHVEENSEI 158
Query: 292 EESRQLESSFETLKDLRDNCKNDLDSTDNTH 322
E+ + +E+ E +++ + K +L++ N H
Sbjct: 159 EKFQAVEAGIEAVQNNEEELKKELETVKNQH 189
>dbj|BAD94516.1| putative protein [Arabidopsis thaliana]
Length = 454
Score = 38.1 bits (87), Expect = 0.36
Identities = 22/91 (24%), Positives = 47/91 (51%), Gaps = 1/91 (1%)
Query: 233 RLSALKKSQKDASNAVQKLSEIKDSVMPD-QQSSSESSKLELALMHQWKSLFDFMENTFA 291
RLS +K+ K A+ + L + K + + +Q+ E+ ++ L L K+ EN+
Sbjct: 99 RLSQIKEDLKKANERISSLEKDKAKALDELKQAKKEAEQVTLKLDDALKAQKHVEENSEI 158
Query: 292 EESRQLESSFETLKDLRDNCKNDLDSTDNTH 322
E+ + +E+ E +++ + K +L++ N H
Sbjct: 159 EKFQAVEAGIEAVQNNEEELKKELETVKNQH 189
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 549,283,763
Number of Sequences: 2540612
Number of extensions: 23230763
Number of successful extensions: 52638
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 52542
Number of HSP's gapped (non-prelim): 147
length of query: 326
length of database: 863,360,394
effective HSP length: 128
effective length of query: 198
effective length of database: 538,162,058
effective search space: 106556087484
effective search space used: 106556087484
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0307.15


