

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.5
(165 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
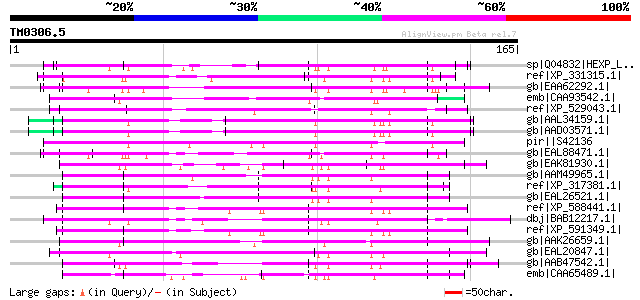
Score E
Sequences producing significant alignments: (bits) Value
sp|Q04832|HEXP_LEIMA DNA-binding protein HEXBP (Hexamer-binding ... 95 8e-19
ref|XP_331315.1| hypothetical protein [Neurospora crassa] gi|289... 93 2e-18
gb|EAA62292.1| hypothetical protein AN5111.2 [Aspergillus nidula... 91 2e-17
emb|CAA93542.1| byr3 [Schizosaccharomyces pombe] gi|254734|gb|AA... 90 3e-17
ref|XP_529043.1| PREDICTED: similar to zinc finger, CCHC domain ... 89 6e-17
gb|AAL34159.1| putative glycine-rich, zinc-finger DNA-binding pr... 88 7e-17
gb|AAD03571.1| putative glycine-rich, zinc-finger DNA-binding pr... 88 7e-17
pir||S42136 cnjB protein - Tetrahymena thermophila gi|161752|gb|... 88 7e-17
gb|EAL88471.1| zinc knuckle domain protein (Byr3), putative [Asp... 86 3e-16
gb|EAK81930.1| hypothetical protein UM00856.1 [Ustilago maydis 5... 86 3e-16
gb|AAM49965.1| LD48005p [Drosophila melanogaster] gi|23240122|gb... 84 1e-15
ref|XP_317381.1| ENSANGP00000011651 [Anopheles gambiae str. PEST... 84 1e-15
gb|EAL26521.1| GA17695-PA [Drosophila pseudoobscura] 84 1e-15
ref|XP_588441.1| PREDICTED: similar to cellular nucleic acid bin... 84 1e-15
dbj|BAB12217.1| vasa homolog [Ciona savignyi] 84 2e-15
ref|XP_591349.1| PREDICTED: similar to cellular nucleic acid bin... 83 2e-15
gb|AAK26659.1| putative DNA binding protein [Schizophyllum commune] 83 3e-15
gb|EAL20847.1| hypothetical protein CNBE2080 [Cryptococcus neofo... 82 4e-15
gb|AAB47542.1| nucleic acid binding protein [Trypanosoma equiper... 82 4e-15
emb|CAA65489.1| unnamed protein product [Saccharomyces cerevisia... 82 5e-15
>sp|Q04832|HEXP_LEIMA DNA-binding protein HEXBP (Hexamer-binding protein)
gi|159342|gb|AAA29245.1| HEXBP DNA binding protein
Length = 271
Score = 94.7 bits (234), Expect = 8e-19
Identities = 53/182 (29%), Positives = 75/182 (41%), Gaps = 47/182 (25%)
Query: 16 VTCFCCGKFGQFANNC-------SVIGPRCFKCNREGHLAVNCKTLQGGP---------- 58
+TCF CG+ G + +C + G C+KC +EGHL+ +C + QGG
Sbjct: 70 MTCFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQEGHLSRDCPSSQGGSRGGYGQKRGR 129
Query: 59 ----------------SDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDG--- 99
D+ D G G + TCY+C GH + CP+G
Sbjct: 130 SGAQGGYSGDRTCYKCGDAGHISRDCPNGQGGYSGAGDRTCYKCGDAGHISRDCPNGQGG 189
Query: 100 -----GPLCFNCGEIGHYASKCKIKG------RLCFNCNQPGHFSRDCKAPKGESSGNTG 148
C+ CGE GH + +C G R C+ C +PGH SR+C G G+ G
Sbjct: 190 YSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRACYKCGKPGHISRECPEAGGSYGGSRG 249
Query: 149 KG 150
G
Sbjct: 250 GG 251
Score = 79.3 bits (194), Expect = 3e-14
Identities = 46/147 (31%), Positives = 65/147 (43%), Gaps = 42/147 (28%)
Query: 15 DVTCFCCGKFGQFANNC-------SVIGPR-CFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
D TC+ CG G + +C S G R C+KC GH++ +C QGG S
Sbjct: 139 DRTCYKCGDAGHISRDCPNGQGGYSGAGDRTCYKCGDAGHISRDCPNGQGGYS------- 191
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPL------CFNCGEIGHYASKCKIKG 120
G G++ CY+C + GH + +CP G C+ CG+ GH + +C G
Sbjct: 192 ----GAGDRK------CYKCGESGHMSRECPSAGSTGSGDRACYKCGKPGHISRECPEAG 241
Query: 121 -----------RLCFNCNQPGHFSRDC 136
R C+ C + GH SRDC
Sbjct: 242 GSYGGSRGGGDRTCYKCGEAGHISRDC 268
Score = 75.9 bits (185), Expect = 4e-13
Identities = 44/126 (34%), Positives = 58/126 (45%), Gaps = 32/126 (25%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
C C +EGH A C +DSK G++ S TC+RC + GH + +CP
Sbjct: 18 CRNCGKEGHYARECPE-----ADSK----------GDERS---TTCFRCGEEGHMSRECP 59
Query: 98 D-------GGPLCFNCGEIGHYASKCK-------IKGRLCFNCNQPGHFSRDCKAPKGES 143
+ G CF CGE GH + C KG C+ C Q GH SRDC + +G S
Sbjct: 60 NEARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQEGHLSRDCPSSQGGS 119
Query: 144 SGNTGK 149
G G+
Sbjct: 120 RGGYGQ 125
Score = 58.5 bits (140), Expect = 6e-08
Identities = 29/80 (36%), Positives = 37/80 (46%), Gaps = 16/80 (20%)
Query: 82 TCYRCKKVGHYTNKCPDG-------GPLCFNCGEIGHYASKCKIKGR-------LCFNCN 127
+C C K GHY +CP+ CF CGE GH + +C + R CF C
Sbjct: 17 SCRNCGKEGHYARECPEADSKGDERSTTCFRCGEEGHMSRECPNEARSGAAGAMTCFRCG 76
Query: 128 QPGHFSRDC--KAPKGESSG 145
+ GH SRDC A G + G
Sbjct: 77 EAGHMSRDCPNSAKPGAAKG 96
Score = 52.8 bits (125), Expect = 3e-06
Identities = 31/106 (29%), Positives = 43/106 (40%), Gaps = 20/106 (18%)
Query: 12 GDKDVTCFCCGKFGQFANNC-------SVIGPR-CFKCNREGHLAVNCKTLQ-------- 55
G D TC+ CG G + +C S G R C+KC GH++ C +
Sbjct: 164 GAGDRTCYKCGDAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRA 223
Query: 56 ----GGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
G P + +A G G G TCY+C + GH + CP
Sbjct: 224 CYKCGKPGHISRECPEAGGSYGGSRGGGDRTCYKCGEAGHISRDCP 269
>ref|XP_331315.1| hypothetical protein [Neurospora crassa] gi|28922111|gb|EAA31369.1|
hypothetical protein [Neurospora crassa]
Length = 183
Score = 93.2 bits (230), Expect = 2e-18
Identities = 52/139 (37%), Positives = 68/139 (48%), Gaps = 18/139 (12%)
Query: 10 PIGDKDV--TCFCCGKFGQFANNCSVIG------PRCFKCNREGHLAVNCKTLQGGPSDS 61
P G KD TC+ CG+ G + +CS G C+KC GH+A NC +GG S
Sbjct: 48 PEGPKDNARTCYRCGQTGHISRDCSQSGGGQSSGAECYKCGEVGHIARNCS--KGGASYG 105
Query: 62 KMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKC-KIKG 120
G G G G TCY C +GH + C +G C+NCGE GH++ C K G
Sbjct: 106 ---GGYQNSGYGGGFGGPQKTCYSCGGIGHMSRDCVNGSK-CYNCGESGHFSRDCPKDSG 161
Query: 121 ---RLCFNCNQPGHFSRDC 136
++C+ C QPGH C
Sbjct: 162 SGEKICYKCQQPGHVQSQC 180
Score = 87.8 bits (216), Expect = 1e-16
Identities = 51/159 (32%), Positives = 70/159 (43%), Gaps = 39/159 (24%)
Query: 18 CFCCGKFGQFANNCSVIGP-RCFKCNREGHLAVNCKTLQGGPSDSKMK----------GN 66
CF CG+ A +C G +C+ C EGH++ +C GP D+
Sbjct: 13 CFTCGQTTHQARDCPNKGAAKCYNCGNEGHMSRDCPE---GPKDNARTCYRCGQTGHISR 69
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGP--------------------LCFNC 106
D + G ++SG CY+C +VGH C GG C++C
Sbjct: 70 DCSQSGGGQSSG--AECYKCGEVGHIARNCSKGGASYGGGYQNSGYGGGFGGPQKTCYSC 127
Query: 107 GEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSG 145
G IGH + C + G C+NC + GHFSRDC PK SG
Sbjct: 128 GGIGHMSRDC-VNGSKCYNCGESGHFSRDC--PKDSGSG 163
Score = 52.4 bits (124), Expect = 5e-06
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 22/86 (25%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGG 71
G TC+ CG G + +C V G +C+ C GH + +C P DS G
Sbjct: 118 GGPQKTCYSCGGIGHMSRDC-VNGSKCYNCGESGHFSRDC------PKDS---------G 161
Query: 72 TGNKTSGKIVTCYRCKKVGHYTNKCP 97
+G K CY+C++ GH ++CP
Sbjct: 162 SGEKI------CYKCQQPGHVQSQCP 181
Score = 48.1 bits (113), Expect = 9e-05
Identities = 22/46 (47%), Positives = 25/46 (53%), Gaps = 2/46 (4%)
Query: 97 PDGGPLCFNCGEIGHYASKCKIKGRL-CFNCNQPGHFSRDC-KAPK 140
P G CF CG+ H A C KG C+NC GH SRDC + PK
Sbjct: 7 PQGTRACFTCGQTTHQARDCPNKGAAKCYNCGNEGHMSRDCPEGPK 52
>gb|EAA62292.1| hypothetical protein AN5111.2 [Aspergillus nidulans FGSC A4]
gi|67537882|ref|XP_662715.1| hypothetical protein
AN5111_2 [Aspergillus nidulans FGSC A4]
gi|49095574|ref|XP_409248.1| hypothetical protein
AN5111.2 [Aspergillus nidulans FGSC A4]
Length = 176
Score = 90.5 bits (223), Expect = 2e-17
Identities = 47/137 (34%), Positives = 61/137 (44%), Gaps = 23/137 (16%)
Query: 11 IGDKDVTCFCCGKFGQFANNCSVIGPR--------CFKCNREGHLAVNCKTLQGGPSDSK 62
+ K+ +C+ CG G + C G C+KC R GH+A NC
Sbjct: 49 VAPKEKSCYRCGAVGHISRECPQAGENERPAGGQECYKCGRVGHIARNCS---------- 98
Query: 63 MKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKC--KIKG 120
+G GG G G+ TCY C GH C G C+NCGE GH + C + KG
Sbjct: 99 -QGGSYGGGFGGGYGGRQQTCYSCGGFGHMARDCTQGQK-CYNCGETGHVSRDCPTEAKG 156
Query: 121 -RLCFNCNQPGHFSRDC 136
R+C+ C QPGH C
Sbjct: 157 ERVCYQCKQPGHIQSAC 173
Score = 73.9 bits (180), Expect = 1e-12
Identities = 44/148 (29%), Positives = 62/148 (41%), Gaps = 40/148 (27%)
Query: 17 TCFCCGKF-----GQFANNCSVIGPR--CFKCNREGHLAVNCKTLQGGPSDSKMKGNDAT 69
TC+ CG G + C+V C++C GH++ C Q G ++ G +
Sbjct: 28 TCYNCGVLDRLGQGHVSRECTVAPKEKSCYRCGAVGHISRECP--QAGENERPAGGQE-- 83
Query: 70 GGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPL--------------CFNCGEIGHYASK 115
CY+C +VGH C GG C++CG GH A
Sbjct: 84 -------------CYKCGRVGHIARNCSQGGSYGGGFGGGYGGRQQTCYSCGGFGHMARD 130
Query: 116 CKIKGRLCFNCNQPGHFSRDCKA-PKGE 142
C +G+ C+NC + GH SRDC KGE
Sbjct: 131 CT-QGQKCYNCGETGHVSRDCPTEAKGE 157
Score = 65.9 bits (159), Expect = 4e-10
Identities = 44/160 (27%), Positives = 62/160 (38%), Gaps = 52/160 (32%)
Query: 18 CFCCGKFGQFANNCSVIG-PRCFKCN-----REGHLAVNCKTLQGGPSDSKMKGNDATGG 71
CF CG+ A +C G P C+ C +GH++ C
Sbjct: 8 CFNCGEATHQARDCPKKGTPTCYNCGVLDRLGQGHVSRECTV------------------ 49
Query: 72 TGNKTSGKIVTCYRCKKVGHYTNKCPD--------GGPLCFNCGEIGHYASKCKIKG--- 120
+ K +CYRC VGH + +CP GG C+ CG +GH A C G
Sbjct: 50 -----APKEKSCYRCGAVGHISRECPQAGENERPAGGQECYKCGRVGHIARNCSQGGSYG 104
Query: 121 -----------RLCFNCNQPGHFSRDC-KAPKGESSGNTG 148
+ C++C GH +RDC + K + G TG
Sbjct: 105 GGFGGGYGGRQQTCYSCGGFGHMARDCTQGQKCYNCGETG 144
Score = 52.8 bits (125), Expect = 3e-06
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 23/87 (26%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGG 71
G + TC+ CG FG A +C+ G +C+ C GH++ +C T G
Sbjct: 112 GGRQQTCYSCGGFGHMARDCTQ-GQKCYNCGETGHVSRDCPTEAKGER------------ 158
Query: 72 TGNKTSGKIVTCYRCKKVGHYTNKCPD 98
CY+CK+ GH + CP+
Sbjct: 159 ----------VCYQCKQPGHIQSACPN 175
Score = 46.6 bits (109), Expect = 2e-04
Identities = 27/65 (41%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query: 99 GGPLCFNCGEIGHYASKCKIKGR-LCFNCN-----QPGHFSRDCK-APKGESSGNTGKGK 151
GG +CFNCGE H A C KG C+NC GH SR+C APK +S G
Sbjct: 4 GGRVCFNCGEATHQARDCPKKGTPTCYNCGVLDRLGQGHVSRECTVAPKEKSCYRCGAVG 63
Query: 152 QLAAE 156
++ E
Sbjct: 64 HISRE 68
>emb|CAA93542.1| byr3 [Schizosaccharomyces pombe] gi|254734|gb|AAB23116.1| human
cellular nucleic acid binding protein (CNBP) homolog
[Schizosaccharomyces pombe] gi|19114592|ref|NP_593680.1|
hypothetical protein SPAC13D6.02c [Schizosaccharomyces
pombe 972h-] gi|543908|sp|P36627|BYR3_SCHPO Cellular
nucleic acid binding protein homolog
Length = 179
Score = 89.7 bits (221), Expect = 3e-17
Identities = 45/133 (33%), Positives = 66/133 (48%), Gaps = 20/133 (15%)
Query: 14 KDVTCFCCGKFGQFANNCSVI-----GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDA 68
++ TC+ CG G +C G C+KC R GH+A +C+T G +
Sbjct: 56 QEKTCYACGTAGHLVRDCPSSPNPRQGAECYKCGRVGHIARDCRT----------NGQQS 105
Query: 69 TGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCK--IKGRLCFNC 126
G G S + CY C GH C G C++CG+IGH + +C+ G+LC+ C
Sbjct: 106 GGRFGGHRSN--MNCYACGSYGHQARDCTMGVK-CYSCGKIGHRSFECQQASDGQLCYKC 162
Query: 127 NQPGHFSRDCKAP 139
NQPGH + +C +P
Sbjct: 163 NQPGHIAVNCTSP 175
Score = 65.9 bits (159), Expect = 4e-10
Identities = 36/121 (29%), Positives = 47/121 (38%), Gaps = 33/121 (27%)
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
GPRC+ C GH A C T G I CY C + GH +
Sbjct: 16 GPRCYNCGENGHQAREC------------------------TKGSI--CYNCNQTGHKAS 49
Query: 95 KC--PDGGPLCFNCGEIGHYASKCKI-----KGRLCFNCNQPGHFSRDCKAPKGESSGNT 147
+C P C+ CG GH C +G C+ C + GH +RDC+ +S G
Sbjct: 50 ECTEPQQEKTCYACGTAGHLVRDCPSSPNPRQGAECYKCGRVGHIARDCRTNGQQSGGRF 109
Query: 148 G 148
G
Sbjct: 110 G 110
>ref|XP_529043.1| PREDICTED: similar to zinc finger, CCHC domain containing 13;
cellular nucleic acid binding protein-like;
4930513O09RIK [Pan troglodytes]
Length = 346
Score = 88.6 bits (218), Expect = 6e-17
Identities = 46/131 (35%), Positives = 66/131 (50%), Gaps = 13/131 (9%)
Query: 17 TCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKT 76
TC+CCG+ G+ A N ++G C+ C R GH+A +CK P + + G G+
Sbjct: 46 TCYCCGESGRNAKNRVLLGNICYNCGRSGHIAKDCKE----PKRERRQHCYTCGRLGHLA 101
Query: 77 SG----KIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKC-KIKGRLCFNCNQPGH 131
K CY C K+GH C C+ CGEIGH A C K + C+ C + GH
Sbjct: 102 RDCDRQKEQKCYSCGKLGHIQKDCAQ--VKCYRCGEIGHVAINCSKTRPGQCYRCGKSGH 159
Query: 132 FSRDCKAPKGE 142
+++C PKG+
Sbjct: 160 LAKEC--PKGD 168
Score = 63.5 bits (153), Expect = 2e-09
Identities = 37/115 (32%), Positives = 48/115 (41%), Gaps = 8/115 (6%)
Query: 39 FKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD 98
F C GH A C G + + G G G T+ TCY C + G
Sbjct: 7 FACGHSGHWARGCPR---GGAGGRRGGGHGRGSQGGSTTLSY-TCYCCGESGRNAKNRVL 62
Query: 99 GGPLCFNCGEIGHYASKCKIKGRL----CFNCNQPGHFSRDCKAPKGESSGNTGK 149
G +C+NCG GH A CK R C+ C + GH +RDC K + + GK
Sbjct: 63 LGNICYNCGRSGHIAKDCKEPKRERRQHCYTCGRLGHLARDCDRQKEQKCYSCGK 117
Score = 55.1 bits (131), Expect = 7e-07
Identities = 25/86 (29%), Positives = 37/86 (42%), Gaps = 26/86 (30%)
Query: 14 KDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTG 73
K+ C+ CGK G +C+ + +C++C GH+A+NC + G
Sbjct: 108 KEQKCYSCGKLGHIQKDCAQV--KCYRCGEIGHVAINCSKTRPG---------------- 149
Query: 74 NKTSGKIVTCYRCKKVGHYTNKCPDG 99
CYRC K GH +CP G
Sbjct: 150 --------QCYRCGKSGHLAKECPKG 167
>gb|AAL34159.1| putative glycine-rich, zinc-finger DNA-binding protein [Arabidopsis
thaliana] gi|14334920|gb|AAK59638.1| putative
glycine-rich, zinc-finger DNA-binding protein
[Arabidopsis thaliana] gi|18398546|ref|NP_565427.1|
cold-shock DNA-binding family protein [Arabidopsis
thaliana]
Length = 301
Score = 88.2 bits (217), Expect = 7e-17
Identities = 53/162 (32%), Positives = 67/162 (40%), Gaps = 38/162 (23%)
Query: 18 CFCCGKFGQFANNCSVIGPR-----------CFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
C+ CG G FA +C G C+ C GHLA +C+ GG ++ G
Sbjct: 132 CYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCR---GGSGGNRYGGG 188
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD--------GGPLCFNCGEIGHYASKCKI 118
GG G+ G CY C VGH+ C GG C+ CG +GH A C
Sbjct: 189 ---GGRGSGGDG----CYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTS 241
Query: 119 K--------GRLCFNCNQPGHFSRDC-KAPKGESSGNTGKGK 151
K GR C+ C GH +RDC + G S G G K
Sbjct: 242 KIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNK 283
Score = 79.7 bits (195), Expect = 3e-14
Identities = 51/162 (31%), Positives = 65/162 (39%), Gaps = 38/162 (23%)
Query: 15 DVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGN 74
+VT G + N+ G CF C GH+A +C GG S GG G
Sbjct: 73 EVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKS---------FGGGGG 123
Query: 75 KTSGKIVTCYRCKKVGHYTNKCPD-----------GGPLCFNCGEIGHYASKCK------ 117
+ SG CY C VGH+ C GG C++CGE+GH A C+
Sbjct: 124 RRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGGN 183
Query: 118 ---------IKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKG 150
G C+ C GHF+RDC+ G GN G G
Sbjct: 184 RYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGG---GNVGGG 222
Score = 57.8 bits (138), Expect = 1e-07
Identities = 40/148 (27%), Positives = 49/148 (33%), Gaps = 55/148 (37%)
Query: 7 GG*PIGDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
GG G C+ CG G FA +C + G
Sbjct: 187 GGGGRGSGGDGCYMCGGVGHFARDC------------------------------RQNGG 216
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD--------GGPLCFNCGEIGHYASKCKI 118
GG G+ TCY C VGH C GG C+ CG GH A C
Sbjct: 217 GNVGGGGS-------TCYTCGGVGHIAKVCTSKIPSGGGGGGRACYECGGTGHLARDCDR 269
Query: 119 KG----------RLCFNCNQPGHFSRDC 136
+G CF C + GHF+R+C
Sbjct: 270 RGSGSSGGGGGSNKCFICGKEGHFAREC 297
Score = 51.2 bits (121), Expect = 1e-05
Identities = 32/96 (33%), Positives = 39/96 (40%), Gaps = 16/96 (16%)
Query: 71 GTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIK----------G 120
G+ KT VT + N G CFNCGE+GH A C G
Sbjct: 64 GSDGKTKAIEVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGG 123
Query: 121 RL------CFNCNQPGHFSRDCKAPKGESSGNTGKG 150
R C+ C GHF+RDC+ G +SG G G
Sbjct: 124 RRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGG 159
Score = 35.8 bits (81), Expect = 0.44
Identities = 15/50 (30%), Positives = 23/50 (46%)
Query: 107 GEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAE 156
G + + + G CFNC + GH ++DC G S G G++ E
Sbjct: 80 GSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGE 129
Score = 32.3 bits (72), Expect = 4.9
Identities = 15/47 (31%), Positives = 21/47 (43%), Gaps = 10/47 (21%)
Query: 18 CFCCGKFGQFANNCSVIGP----------RCFKCNREGHLAVNCKTL 54
C+ CG G A +C G +CF C +EGH A C ++
Sbjct: 254 CYECGGTGHLARDCDRRGSGSSGGGGGSNKCFICGKEGHFARECTSV 300
>gb|AAD03571.1| putative glycine-rich, zinc-finger DNA-binding protein [Arabidopsis
thaliana] gi|7442089|pir||T00837 glycine-rich protein
T13L16.11 - Arabidopsis thaliana
Length = 299
Score = 88.2 bits (217), Expect = 7e-17
Identities = 53/162 (32%), Positives = 67/162 (40%), Gaps = 38/162 (23%)
Query: 18 CFCCGKFGQFANNCSVIGPR-----------CFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
C+ CG G FA +C G C+ C GHLA +C+ GG ++ G
Sbjct: 130 CYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCR---GGSGGNRYGGG 186
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD--------GGPLCFNCGEIGHYASKCKI 118
GG G+ G CY C VGH+ C GG C+ CG +GH A C
Sbjct: 187 ---GGRGSGGDG----CYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTS 239
Query: 119 K--------GRLCFNCNQPGHFSRDC-KAPKGESSGNTGKGK 151
K GR C+ C GH +RDC + G S G G K
Sbjct: 240 KIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNK 281
Score = 79.7 bits (195), Expect = 3e-14
Identities = 51/162 (31%), Positives = 65/162 (39%), Gaps = 38/162 (23%)
Query: 15 DVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGN 74
+VT G + N+ G CF C GH+A +C GG S GG G
Sbjct: 71 EVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKS---------FGGGGG 121
Query: 75 KTSGKIVTCYRCKKVGHYTNKCPD-----------GGPLCFNCGEIGHYASKCK------ 117
+ SG CY C VGH+ C GG C++CGE+GH A C+
Sbjct: 122 RRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGGN 181
Query: 118 ---------IKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKG 150
G C+ C GHF+RDC+ G GN G G
Sbjct: 182 RYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGG---GNVGGG 220
Score = 57.8 bits (138), Expect = 1e-07
Identities = 40/148 (27%), Positives = 49/148 (33%), Gaps = 55/148 (37%)
Query: 7 GG*PIGDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGN 66
GG G C+ CG G FA +C + G
Sbjct: 185 GGGGRGSGGDGCYMCGGVGHFARDC------------------------------RQNGG 214
Query: 67 DATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD--------GGPLCFNCGEIGHYASKCKI 118
GG G+ TCY C VGH C GG C+ CG GH A C
Sbjct: 215 GNVGGGGS-------TCYTCGGVGHIAKVCTSKIPSGGGGGGRACYECGGTGHLARDCDR 267
Query: 119 KG----------RLCFNCNQPGHFSRDC 136
+G CF C + GHF+R+C
Sbjct: 268 RGSGSSGGGGGSNKCFICGKEGHFAREC 295
Score = 51.2 bits (121), Expect = 1e-05
Identities = 32/96 (33%), Positives = 39/96 (40%), Gaps = 16/96 (16%)
Query: 71 GTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIK----------G 120
G+ KT VT + N G CFNCGE+GH A C G
Sbjct: 62 GSDGKTKAIEVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGG 121
Query: 121 RL------CFNCNQPGHFSRDCKAPKGESSGNTGKG 150
R C+ C GHF+RDC+ G +SG G G
Sbjct: 122 RRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGG 157
Score = 35.8 bits (81), Expect = 0.44
Identities = 15/50 (30%), Positives = 23/50 (46%)
Query: 107 GEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAE 156
G + + + G CFNC + GH ++DC G S G G++ E
Sbjct: 78 GSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGE 127
Score = 32.3 bits (72), Expect = 4.9
Identities = 15/47 (31%), Positives = 21/47 (43%), Gaps = 10/47 (21%)
Query: 18 CFCCGKFGQFANNCSVIGP----------RCFKCNREGHLAVNCKTL 54
C+ CG G A +C G +CF C +EGH A C ++
Sbjct: 252 CYECGGTGHLARDCDRRGSGSSGGGGGSNKCFICGKEGHFARECTSV 298
>pir||S42136 cnjB protein - Tetrahymena thermophila gi|161752|gb|AAC37171.1| cnjB
[Tetrahymena thermophila] gi|737494|prf||1922371A cnjB
gene
Length = 1748
Score = 88.2 bits (217), Expect = 7e-17
Identities = 59/191 (30%), Positives = 82/191 (42%), Gaps = 56/191 (29%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPR-------CFKCNREGHLAVNCKTLQGG------- 57
G+K CF CGK G A +C+ + CFKCN+EGH++ +C Q
Sbjct: 1445 GNKGKGCFKCGKVGHMAKDCTEPQQQGRKQSGACFKCNQEGHMSKDCPNQQQKKSGCFKC 1504
Query: 58 ----------PSDSKMKGNDATGGTGNKTS--GKIV-------------TCYRCKKVGHY 92
P+ K + GG K G I TC++CK+ GH
Sbjct: 1505 GEEGHFSKDCPNPQKQQQQKPRGGACFKCGEEGHISKDCPNPQKQQQKNTCFKCKQEGHI 1564
Query: 93 TNKCPD----GGPLCFNCGEIGHYASKC-----KIKGRLCFNCNQPGHFSRDC------K 137
+ CP+ GG CFNC + GH + C K KG CFNC + GH SR+C +
Sbjct: 1565 SKDCPNSQNSGGNKCFNCNQEGHMSKDCPNPSQKKKG--CFNCGEEGHQSRECTKERKER 1622
Query: 138 APKGESSGNTG 148
P+ ++ N G
Sbjct: 1623 PPRNNNNNNNG 1633
Score = 35.8 bits (81), Expect = 0.44
Identities = 21/73 (28%), Positives = 31/73 (41%), Gaps = 11/73 (15%)
Query: 18 CFCCGKFGQFANNCSVIGPR---CFKCNREGHLAVNC-KTLQGGP-------SDSKMKGN 66
CF C + G + +C + CF C EGH + C K + P ++ +GN
Sbjct: 1579 CFNCNQEGHMSKDCPNPSQKKKGCFNCGEEGHQSRECTKERKERPPRNNNNNNNGNFRGN 1638
Query: 67 DATGGTGNKTSGK 79
GG GN G+
Sbjct: 1639 KQFGGGGNSNGGQ 1651
>gb|EAL88471.1| zinc knuckle domain protein (Byr3), putative [Aspergillus fumigatus
Af293]
Length = 190
Score = 86.3 bits (212), Expect = 3e-16
Identities = 49/141 (34%), Positives = 65/141 (45%), Gaps = 26/141 (18%)
Query: 11 IGDKDVTCFCCGKFGQFANNCSVIGP------------RCFKCNREGHLAVNCKTLQGGP 58
+ K+ +C+ CG G + CS G C+KC + GH+A NC QGG
Sbjct: 59 VAPKEKSCYRCGVAGHISRECSQAGSGDNYNGAPSGGQECYKCGQVGHIARNCS--QGG- 115
Query: 59 SDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKC-- 116
G GG G G+ TCY C GH C G C+NCG++GH + C
Sbjct: 116 ---NYGGGFGHGGYG----GRQQTCYSCGGFGHMARDCTHGQK-CYNCGDVGHVSRDCPT 167
Query: 117 KIKG-RLCFNCNQPGHFSRDC 136
+ KG R+C+ C QPGH C
Sbjct: 168 EAKGERVCYKCKQPGHVQAAC 188
Score = 72.4 bits (176), Expect = 4e-12
Identities = 46/147 (31%), Positives = 62/147 (41%), Gaps = 35/147 (23%)
Query: 17 TCFCCGKF---GQFANNCSVIGPR--CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGG 71
TC+ CG G + C+V C++C GH++ C Q G D+
Sbjct: 40 TCYNCGATIGQGHVSRECTVAPKEKSCYRCGVAGHISRECS--QAGSGDNY--------- 88
Query: 72 TGNKTSGKIVTCYRCKKVGHYTNKCPDGG---------------PLCFNCGEIGHYASKC 116
G + G+ CY+C +VGH C GG C++CG GH A C
Sbjct: 89 NGAPSGGQ--ECYKCGQVGHIARNCSQGGNYGGGFGHGGYGGRQQTCYSCGGFGHMARDC 146
Query: 117 KIKGRLCFNCNQPGHFSRDCKA-PKGE 142
G+ C+NC GH SRDC KGE
Sbjct: 147 T-HGQKCYNCGDVGHVSRDCPTEAKGE 172
Score = 53.9 bits (128), Expect = 2e-06
Identities = 35/140 (25%), Positives = 51/140 (36%), Gaps = 54/140 (38%)
Query: 28 ANNCSVIG-PRCFKCNR---EGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTC 83
A +C G P C+ C +GH++ C + K +C
Sbjct: 30 ARDCPKKGTPTCYNCGATIGQGHVSRECTV-----------------------APKEKSC 66
Query: 84 YRCKKVGHYTNKC------------PDGGPLCFNCGEIGHYASKCKIKG----------- 120
YRC GH + +C P GG C+ CG++GH A C G
Sbjct: 67 YRCGVAGHISRECSQAGSGDNYNGAPSGGQECYKCGQVGHIARNCSQGGNYGGGFGHGGY 126
Query: 121 ----RLCFNCNQPGHFSRDC 136
+ C++C GH +RDC
Sbjct: 127 GGRQQTCYSCGGFGHMARDC 146
Score = 50.4 bits (119), Expect = 2e-05
Identities = 25/87 (28%), Positives = 37/87 (41%), Gaps = 23/87 (26%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGG 71
G + TC+ CG FG A +C+ G +C+ C GH++ +C T G
Sbjct: 127 GGRQQTCYSCGGFGHMARDCT-HGQKCYNCGDVGHVSRDCPTEAKGER------------ 173
Query: 72 TGNKTSGKIVTCYRCKKVGHYTNKCPD 98
CY+CK+ GH CP+
Sbjct: 174 ----------VCYKCKQPGHVQAACPN 190
>gb|EAK81930.1| hypothetical protein UM00856.1 [Ustilago maydis 521]
gi|49068364|ref|XP_398471.1| hypothetical protein
UM00856.1 [Ustilago maydis 521]
Length = 189
Score = 86.3 bits (212), Expect = 3e-16
Identities = 48/161 (29%), Positives = 73/161 (44%), Gaps = 39/161 (24%)
Query: 17 TCFCCGKFGQFANNCSVIGP--RCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGN 74
+C+ CG+ G ++ C + C+KC+ GH++ C T P+ A GG G
Sbjct: 39 SCYNCGQQGHISSQCGMEAQPKTCYKCSETGHISRECPT---NPAP-------AAGGPGG 88
Query: 75 KTSGKIVTCYRCKKVGHYTNKCP--------------DGGPLCFNCGEIGHYASKCKIK- 119
+ CY+C + GH CP GG C+NCG +GH + +C
Sbjct: 89 E-------CYKCGQHGHIARACPTAGGSSRGGFGGARSGGRSCYNCGGVGHLSRECTSPA 141
Query: 120 -----GRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAA 155
G+ C+NCN+ GH SR+C P+ +S G L+A
Sbjct: 142 GAAAGGQRCYNCNESGHISRECPKPQTKSCYRCGDEGHLSA 182
Score = 57.0 bits (136), Expect = 2e-07
Identities = 23/62 (37%), Positives = 36/62 (57%), Gaps = 3/62 (4%)
Query: 90 GHYTNKCPDGG-PLCFNCGEIGHYASKCKIKG--RLCFNCNQPGHFSRDCKAPKGESSGN 146
GH CP G P C+NCG+ GH +S+C ++ + C+ C++ GH SR+C ++G
Sbjct: 26 GHNAAACPTAGNPSCYNCGQQGHISSQCGMEAQPKTCYKCSETGHISRECPTNPAPAAGG 85
Query: 147 TG 148
G
Sbjct: 86 PG 87
Score = 55.8 bits (133), Expect = 4e-07
Identities = 34/124 (27%), Positives = 53/124 (42%), Gaps = 24/124 (19%)
Query: 17 TCFCCGKFGQFANNCSVI--------GPRCFKCNREGHLAVNCKTL----QGGPSDSKMK 64
TC+ C + G + C G C+KC + GH+A C T +GG ++
Sbjct: 61 TCYKCSETGHISRECPTNPAPAAGGPGGECYKCGQHGHIARACPTAGGSSRGGFGGARSG 120
Query: 65 GNDA--TGGTGNKT------SGKIV---TCYRCKKVGHYTNKCPDGGPL-CFNCGEIGHY 112
G GG G+ + +G CY C + GH + +CP C+ CG+ GH
Sbjct: 121 GRSCYNCGGVGHLSRECTSPAGAAAGGQRCYNCNESGHISRECPKPQTKSCYRCGDEGHL 180
Query: 113 ASKC 116
++ C
Sbjct: 181 SAAC 184
Score = 42.4 bits (98), Expect = 0.005
Identities = 30/110 (27%), Positives = 41/110 (37%), Gaps = 20/110 (18%)
Query: 4 AATGG*PIGDKDVTCFCCGKFGQFANNCSVIGPR--------------CFKCNREGHLAV 49
A G P G+ C+ CG+ G A C G C+ C GHL+
Sbjct: 80 APAAGGPGGE----CYKCGQHGHIARACPTAGGSSRGGFGGARSGGRSCYNCGGVGHLSR 135
Query: 50 NCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVT--CYRCKKVGHYTNKCP 97
C + G + + N G ++ K T CYRC GH + CP
Sbjct: 136 ECTSPAGAAAGGQRCYNCNESGHISRECPKPQTKSCYRCGDEGHLSAACP 185
>gb|AAM49965.1| LD48005p [Drosophila melanogaster] gi|23240122|gb|AAN16117.1|
CG3800-PA [Drosophila melanogaster]
gi|24658883|ref|NP_611739.1| CG3800-PA [Drosophila
melanogaster] gi|18447272|gb|AAL68216.1| GM14667p
[Drosophila melanogaster]
Length = 165
Score = 84.3 bits (207), Expect = 1e-15
Identities = 39/133 (29%), Positives = 55/133 (41%), Gaps = 31/133 (23%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C+ C +FG FA C RC++CN GH++ +C T
Sbjct: 57 CYKCNQFGHFARACPEEAERCYRCNGIGHISKDC------------------------TQ 92
Query: 78 GKIVTCYRCKKVGHYTNKCPDG----GPL---CFNCGEIGHYASKCKIKGRLCFNCNQPG 130
TCYRC K GH+ CP+ GP C+ C GH + C + C+ C + G
Sbjct: 93 ADNPTCYRCNKTGHWVRNCPEAVNERGPTNVSCYKCNRTGHISKNCPETSKTCYGCGKSG 152
Query: 131 HFSRDCKAPKGES 143
H R+C G +
Sbjct: 153 HLRRECDEKGGRN 165
Score = 79.3 bits (194), Expect = 3e-14
Identities = 37/110 (33%), Positives = 52/110 (46%), Gaps = 16/110 (14%)
Query: 38 CFKCNREGHLAVNCKTLQGGP----------SDSKMKGNDATGGTGNKTSGKIVTCYRCK 87
C+KCNR GH A +C GG M+GND G N+ CY+C
Sbjct: 7 CYKCNRPGHFARDCSLGGGGGPGGVGGGGGGGGGGMRGNDGGGMRRNREK-----CYKCN 61
Query: 88 KVGHYTNKCPDGGPLCFNCGEIGHYASKC-KIKGRLCFNCNQPGHFSRDC 136
+ GH+ CP+ C+ C IGH + C + C+ CN+ GH+ R+C
Sbjct: 62 QFGHFARACPEEAERCYRCNGIGHISKDCTQADNPTCYRCNKTGHWVRNC 111
Score = 52.8 bits (125), Expect = 3e-06
Identities = 24/85 (28%), Positives = 34/85 (39%), Gaps = 30/85 (35%)
Query: 82 TCYRCKKVGHYTNKCP------------------------DGGPL------CFNCGEIGH 111
TCY+C + GH+ C DGG + C+ C + GH
Sbjct: 6 TCYKCNRPGHFARDCSLGGGGGPGGVGGGGGGGGGGMRGNDGGGMRRNREKCYKCNQFGH 65
Query: 112 YASKCKIKGRLCFNCNQPGHFSRDC 136
+A C + C+ CN GH S+DC
Sbjct: 66 FARACPEEAERCYRCNGIGHISKDC 90
Score = 39.3 bits (90), Expect = 0.040
Identities = 14/28 (50%), Positives = 17/28 (60%)
Query: 123 CFNCNQPGHFSRDCKAPKGESSGNTGKG 150
C+ CN+PGHF+RDC G G G G
Sbjct: 7 CYKCNRPGHFARDCSLGGGGGPGGVGGG 34
Score = 38.5 bits (88), Expect = 0.068
Identities = 13/40 (32%), Positives = 20/40 (49%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNC 51
G +V+C+ C + G + NC C+ C + GHL C
Sbjct: 119 GPTNVSCYKCNRTGHISKNCPETSKTCYGCGKSGHLRREC 158
>ref|XP_317381.1| ENSANGP00000011651 [Anopheles gambiae str. PEST]
gi|30175401|gb|EAA12317.2| ENSANGP00000011651 [Anopheles
gambiae str. PEST]
Length = 153
Score = 84.3 bits (207), Expect = 1e-15
Identities = 41/129 (31%), Positives = 56/129 (42%), Gaps = 28/129 (21%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C+ C + G FA +C RC++CN GH+A +C P DS
Sbjct: 46 CYKCNQMGHFARDCKEDLDRCYRCNGSGHIARDCSL---SPDDS---------------- 86
Query: 78 GKIVTCYRCKKVGHYTNKCP-----DGGPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHF 132
CY C + GH CP D C+NC + GH + C + C++C + GH
Sbjct: 87 ----CCYNCNQSGHLARNCPEKSDRDMNVSCYNCNKSGHISRNCPSGDKSCYSCGKIGHL 142
Query: 133 SRDCKAPKG 141
SRDC KG
Sbjct: 143 SRDCTENKG 151
Score = 80.9 bits (198), Expect = 1e-14
Identities = 35/101 (34%), Positives = 51/101 (49%), Gaps = 8/101 (7%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
CFKC+R GH A +C+ + GG G G + G+ CY+C ++GH+ C
Sbjct: 7 CFKCDRPGHYARDCQNVGGGG------GRGVGGPRDRRDFGRREKCYKCNQMGHFARDCK 60
Query: 98 DGGPLCFNCGEIGHYASKCKIK--GRLCFNCNQPGHFSRDC 136
+ C+ C GH A C + C+NCNQ GH +R+C
Sbjct: 61 EDLDRCYRCNGSGHIARDCSLSPDDSCCYNCNQSGHLARNC 101
Score = 57.8 bits (138), Expect = 1e-07
Identities = 26/81 (32%), Positives = 37/81 (45%), Gaps = 19/81 (23%)
Query: 82 TCYRCKKVGHYTNKCPD---------GGPL----------CFNCGEIGHYASKCKIKGRL 122
TC++C + GHY C + GGP C+ C ++GH+A CK
Sbjct: 6 TCFKCDRPGHYARDCQNVGGGGGRGVGGPRDRRDFGRREKCYKCNQMGHFARDCKEDLDR 65
Query: 123 CFNCNQPGHFSRDCKAPKGES 143
C+ CN GH +RDC +S
Sbjct: 66 CYRCNGSGHIARDCSLSPDDS 86
Score = 45.8 bits (107), Expect = 4e-04
Identities = 25/89 (28%), Positives = 35/89 (39%), Gaps = 30/89 (33%)
Query: 15 DVTCFCCGKFGQFANNCSVIGPR-----CFKCNREGHLAVNCKTLQGGPSDSKMKGNDAT 69
D C+ C + G A NC R C+ CN+ GH++ NC PS K
Sbjct: 85 DSCCYNCNQSGHLARNCPEKSDRDMNVSCYNCNKSGHISRNC------PSGDK------- 131
Query: 70 GGTGNKTSGKIVTCYRCKKVGHYTNKCPD 98
+CY C K+GH + C +
Sbjct: 132 ------------SCYSCGKIGHLSRDCTE 148
Score = 40.0 bits (92), Expect = 0.023
Identities = 15/44 (34%), Positives = 24/44 (54%)
Query: 13 DKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQG 56
D +V+C+ C K G + NC C+ C + GHL+ +C +G
Sbjct: 108 DMNVSCYNCNKSGHISRNCPSGDKSCYSCGKIGHLSRDCTENKG 151
Score = 33.1 bits (74), Expect = 2.9
Identities = 11/26 (42%), Positives = 16/26 (61%)
Query: 123 CFNCNQPGHFSRDCKAPKGESSGNTG 148
CF C++PGH++RDC+ G G
Sbjct: 7 CFKCDRPGHYARDCQNVGGGGGRGVG 32
>gb|EAL26521.1| GA17695-PA [Drosophila pseudoobscura]
Length = 159
Score = 84.3 bits (207), Expect = 1e-15
Identities = 39/133 (29%), Positives = 55/133 (41%), Gaps = 31/133 (23%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C+ C +FG FA C RC++CN GH++ +C T
Sbjct: 51 CYKCNQFGHFARACPEEAERCYRCNGIGHISKDC------------------------TQ 86
Query: 78 GKIVTCYRCKKVGHYTNKCPDG----GPL---CFNCGEIGHYASKCKIKGRLCFNCNQPG 130
TCYRC K GH+ CP+ GP C+ C GH + C + C+ C + G
Sbjct: 87 ADNPTCYRCNKTGHWVRNCPEAVNERGPANVSCYKCNRTGHISKNCPETSKTCYGCGKSG 146
Query: 131 HFSRDCKAPKGES 143
H R+C G +
Sbjct: 147 HLRRECDEKGGRN 159
Score = 83.6 bits (205), Expect = 2e-15
Identities = 36/100 (36%), Positives = 49/100 (49%), Gaps = 2/100 (2%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
C+KCNR GH A +C GGP G GG G CY+C + GH+ CP
Sbjct: 7 CYKCNRPGHFARDCSLGGGGPGGGG-PGGGMRGGDGGGMRRNREKCYKCNQFGHFARACP 65
Query: 98 DGGPLCFNCGEIGHYASKC-KIKGRLCFNCNQPGHFSRDC 136
+ C+ C IGH + C + C+ CN+ GH+ R+C
Sbjct: 66 EEAERCYRCNGIGHISKDCTQADNPTCYRCNKTGHWVRNC 105
Score = 55.1 bits (131), Expect = 7e-07
Identities = 24/79 (30%), Positives = 34/79 (42%), Gaps = 24/79 (30%)
Query: 82 TCYRCKKVGHYTNKCP------------------DGGPL------CFNCGEIGHYASKCK 117
TCY+C + GH+ C DGG + C+ C + GH+A C
Sbjct: 6 TCYKCNRPGHFARDCSLGGGGPGGGGPGGGMRGGDGGGMRRNREKCYKCNQFGHFARACP 65
Query: 118 IKGRLCFNCNQPGHFSRDC 136
+ C+ CN GH S+DC
Sbjct: 66 EEAERCYRCNGIGHISKDC 84
Score = 38.5 bits (88), Expect = 0.068
Identities = 13/40 (32%), Positives = 20/40 (49%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNC 51
G +V+C+ C + G + NC C+ C + GHL C
Sbjct: 113 GPANVSCYKCNRTGHISKNCPETSKTCYGCGKSGHLRREC 152
Score = 35.4 bits (80), Expect = 0.57
Identities = 12/23 (52%), Positives = 15/23 (65%)
Query: 123 CFNCNQPGHFSRDCKAPKGESSG 145
C+ CN+PGHF+RDC G G
Sbjct: 7 CYKCNRPGHFARDCSLGGGGPGG 29
>ref|XP_588441.1| PREDICTED: similar to cellular nucleic acid binding protein [Bos
taurus]
Length = 171
Score = 84.0 bits (206), Expect = 1e-15
Identities = 48/119 (40%), Positives = 61/119 (50%), Gaps = 13/119 (10%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATG-GTGNKTSGKI--VTCYRCKKVGHYTN 94
CFKC R GH A C +GG + +G A G G G + S V CYRC K GHY
Sbjct: 6 CFKCGRVGHWAPACS--KGG----RARGRGARGRGHGAQCSSTTLPVICYRCGKFGHYAK 59
Query: 95 KCPDGGPLCFNCGEIGHYASKC---KIKG-RLCFNCNQPGHFSRDCKAPKGESSGNTGK 149
C +C+NCG+ GH A C K +G R C+ C +PGH +RDC + + GK
Sbjct: 60 DCDLLDDICYNCGKSGHIAKDCAEPKREGERCCYTCGRPGHLARDCDRQEERKCYSCGK 118
Score = 81.6 bits (200), Expect = 7e-15
Identities = 41/131 (31%), Positives = 61/131 (46%), Gaps = 21/131 (16%)
Query: 16 VTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNK 75
V C+ CGKFG +A +C ++ C+ C + GH+A +C ++ K +G G
Sbjct: 46 VICYRCGKFGHYAKDCDLLDDICYNCGKSGHIAKDC-------AEPKREGERCCYTCGR- 97
Query: 76 TSGKIV---------TCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIKGRL-CFN 125
G + CY C K GH C C+ CGEIGH A C+ + C+
Sbjct: 98 -PGHLARDCDRQEERKCYSCGKSGHIQKYCTQ--VKCYRCGEIGHVAINCRKMNEVNCYR 154
Query: 126 CNQPGHFSRDC 136
C + GH +R+C
Sbjct: 155 CGESGHLTREC 165
Score = 51.2 bits (121), Expect = 1e-05
Identities = 23/80 (28%), Positives = 35/80 (43%), Gaps = 26/80 (32%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C+ CGK G C+ + +C++C GH+A+NC+ +
Sbjct: 113 CYSCGKSGHIQKYCTQV--KCYRCGEIGHVAINCRKMNE--------------------- 149
Query: 78 GKIVTCYRCKKVGHYTNKCP 97
V CYRC + GH T +CP
Sbjct: 150 ---VNCYRCGESGHLTRECP 166
>dbj|BAB12217.1| vasa homolog [Ciona savignyi]
Length = 770
Score = 83.6 bits (205), Expect = 2e-15
Identities = 53/166 (31%), Positives = 70/166 (41%), Gaps = 41/166 (24%)
Query: 18 CFCCGKFGQFANNC-----SVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGT 72
CF CG+ G + C G CFKC EGH++ C KG GG
Sbjct: 108 CFKCGEEGHMSRECPQGGGGSRGKGCFKCGEEGHMSRECP-----------KGGGGGGGG 156
Query: 73 GNKTSGKIVTCYRCKKVGHYTNKCPDGGPL----------CFNCGEIGHYASKCKI---- 118
G C++C + GH + +CP GG CF CGE GH + +C
Sbjct: 157 GRG-------CFKCGEEGHMSRECPKGGDSGFEGRSRSKGCFKCGEEGHMSRECPQGGGG 209
Query: 119 -KGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAEERIHTKE 163
+G CF C + GH SR+C P+G G G G EE ++E
Sbjct: 210 GRGSGCFKCGEEGHMSREC--PQG-GGGGRGSGCFKCGEEGHMSRE 252
Score = 81.6 bits (200), Expect = 7e-15
Identities = 47/142 (33%), Positives = 65/142 (45%), Gaps = 32/142 (22%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPR-------CFKCNREGHLAVNCKTLQGGPSDSKMK 64
G + CF CG+ G + C G CFKC EGH++ C +GG DS +
Sbjct: 127 GSRGKGCFKCGEEGHMSRECPKGGGGGGGGGRGCFKCGEEGHMSRECP--KGG--DSGFE 182
Query: 65 GNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGG-----PLCFNCGEIGHYASKCKI- 118
G + G C++C + GH + +CP GG CF CGE GH + +C
Sbjct: 183 GRSRSKG-----------CFKCGEEGHMSRECPQGGGGGRGSGCFKCGEEGHMSRECPQG 231
Query: 119 ----KGRLCFNCNQPGHFSRDC 136
+G CF C + GH SR+C
Sbjct: 232 GGGGRGSGCFKCGEEGHMSREC 253
Score = 49.7 bits (117), Expect = 3e-05
Identities = 30/99 (30%), Positives = 40/99 (40%), Gaps = 23/99 (23%)
Query: 18 CFCCGKFGQFANNCSVIGPR----------CFKCNREGHLAVNCKTLQGGPSDS------ 61
CF CG+ G + C G CFKC EGH++ C GG S
Sbjct: 160 CFKCGEEGHMSRECPKGGDSGFEGRSRSKGCFKCGEEGHMSRECPQGGGGGRGSGCFKCG 219
Query: 62 ---KMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
M GG G + SG C++C + GH + +CP
Sbjct: 220 EEGHMSRECPQGGGGGRGSG----CFKCGEEGHMSRECP 254
Score = 37.0 bits (84), Expect = 0.20
Identities = 18/56 (32%), Positives = 23/56 (40%), Gaps = 5/56 (8%)
Query: 12 GDKDVTCFCCGKFGQFANNCSV-----IGPRCFKCNREGHLAVNCKTLQGGPSDSK 62
G + CF CG+ G + C G CFKC EGH++ C G K
Sbjct: 209 GGRGSGCFKCGEEGHMSRECPQGGGGGRGSGCFKCGEEGHMSRECPRNTSGEGGEK 264
>ref|XP_591349.1| PREDICTED: similar to cellular nucleic acid binding protein [Bos
taurus]
Length = 171
Score = 83.2 bits (204), Expect = 2e-15
Identities = 48/119 (40%), Positives = 61/119 (50%), Gaps = 13/119 (10%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATG-GTGNKTSGKI--VTCYRCKKVGHYTN 94
CFKC R GH A C +GG + +G A G G G + S V CYRC K GHY
Sbjct: 6 CFKCGRVGHWAPACS--KGGGA----RGRGARGRGHGAQCSSTTLPVICYRCGKFGHYAK 59
Query: 95 KCPDGGPLCFNCGEIGHYASKC---KIKG-RLCFNCNQPGHFSRDCKAPKGESSGNTGK 149
C +C+NCG+ GH A C K +G R C+ C +PGH +RDC + + GK
Sbjct: 60 DCDLLDDICYNCGKSGHIAKDCAEPKREGERCCYTCGRPGHLARDCDRQEERKCYSCGK 118
Score = 81.6 bits (200), Expect = 7e-15
Identities = 41/131 (31%), Positives = 61/131 (46%), Gaps = 21/131 (16%)
Query: 16 VTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNK 75
V C+ CGKFG +A +C ++ C+ C + GH+A +C ++ K +G G
Sbjct: 46 VICYRCGKFGHYAKDCDLLDDICYNCGKSGHIAKDC-------AEPKREGERCCYTCGR- 97
Query: 76 TSGKIV---------TCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCKIKGRL-CFN 125
G + CY C K GH C C+ CGEIGH A C+ + C+
Sbjct: 98 -PGHLARDCDRQEERKCYSCGKSGHIQKYCTQ--VKCYRCGEIGHVAINCRKMNEVNCYR 154
Query: 126 CNQPGHFSRDC 136
C + GH +R+C
Sbjct: 155 CGESGHLTREC 165
Score = 51.2 bits (121), Expect = 1e-05
Identities = 23/80 (28%), Positives = 35/80 (43%), Gaps = 26/80 (32%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTS 77
C+ CGK G C+ + +C++C GH+A+NC+ +
Sbjct: 113 CYSCGKSGHIQKYCTQV--KCYRCGEIGHVAINCRKMNE--------------------- 149
Query: 78 GKIVTCYRCKKVGHYTNKCP 97
V CYRC + GH T +CP
Sbjct: 150 ---VNCYRCGESGHLTRECP 166
>gb|AAK26659.1| putative DNA binding protein [Schizophyllum commune]
Length = 146
Score = 82.8 bits (203), Expect = 3e-15
Identities = 39/141 (27%), Positives = 63/141 (44%), Gaps = 30/141 (21%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
C+KC EGH++ +C + G + G G G + CYRC K GH CP
Sbjct: 6 CYKCGGEGHISRDCSSADAGGAGGYSGGGFGGGARGGE-------CYRCGKAGHMARACP 58
Query: 98 DGGP----------------------LCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRD 135
+ P C+ CG +GH + C ++G+ C+NC++ GH SRD
Sbjct: 59 EPAPGGNASYGGGGSYGYGGGFQSQKSCYTCGGVGHLSKDC-VQGQRCYNCSETGHISRD 117
Query: 136 CKAPKGESSGNTGKGKQLAAE 156
C P+ ++ + G ++ +
Sbjct: 118 CPNPQKKACYSCGSESHISRD 138
Score = 72.8 bits (177), Expect = 3e-12
Identities = 43/143 (30%), Positives = 60/143 (41%), Gaps = 31/143 (21%)
Query: 17 TCFCCGKFGQFANNCSVI------------------GPRCFKCNREGHLAVNCKTLQGGP 58
TC+ CG G + +CS G C++C + GH+A C G
Sbjct: 5 TCYKCGGEGHISRDCSSADAGGAGGYSGGGFGGGARGGECYRCGKAGHMARACPEPAPG- 63
Query: 59 SDSKMKGNDATGGTGNKTSG----KIVTCYRCKKVGHYTNKCPDGGPLCFNCGEIGHYAS 114
GN + GG G+ G +CY C VGH + C G C+NC E GH +
Sbjct: 64 ------GNASYGGGGSYGYGGGFQSQKSCYTCGGVGHLSKDCVQ-GQRCYNCSETGHISR 116
Query: 115 KC-KIKGRLCFNCNQPGHFSRDC 136
C + + C++C H SRDC
Sbjct: 117 DCPNPQKKACYSCGSESHISRDC 139
>gb|EAL20847.1| hypothetical protein CNBE2080 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57227106|gb|AAW43565.1|
DNA-binding protein hexbp, putative [Cryptococcus
neoformans var. neoformans JEC21]
gi|58267432|ref|XP_570872.1| DNA-binding protein hexbp,
putative [Cryptococcus neoformans var. neoformans JEC21]
Length = 204
Score = 82.4 bits (202), Expect = 4e-15
Identities = 50/186 (26%), Positives = 69/186 (36%), Gaps = 49/186 (26%)
Query: 17 TCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTL---------QGGPSDSKMKGND 67
+CF CG+ G A C P C+ C GHL+ C Q G S
Sbjct: 9 SCFKCGQQGHVAAACPAEAPTCYNCGLSGHLSRECPQPKNKACYTCGQEGHLSSACPQGS 68
Query: 68 ATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDGGPL------------------------C 103
GG G + G CYRC K GH CP+ G C
Sbjct: 69 GAGGFGGASGGG--ECYRCGKPGHIARMCPESGDAAAGGFGGAGGYGGFGGGAGFGNKSC 126
Query: 104 FNCGEIGHYASKCKIKG--------------RLCFNCNQPGHFSRDCKAPKGESSGNTGK 149
+ CG +GH + +C R C+NC Q GH SR+C +G++ + G+
Sbjct: 127 YTCGGVGHISRECPSGASRGFGGGGGGFGGPRKCYNCGQDGHISRECPQEQGKTCYSCGQ 186
Query: 150 GKQLAA 155
+A+
Sbjct: 187 PGHIAS 192
Score = 82.0 bits (201), Expect = 5e-15
Identities = 45/156 (28%), Positives = 70/156 (44%), Gaps = 27/156 (17%)
Query: 14 KDVTCFCCGKFGQFANNC-----------SVIGPRCFKCNREGHLAVNCKTLQGGPSDSK 62
K+ C+ CG+ G ++ C + G C++C + GH+A C G +
Sbjct: 47 KNKACYTCGQEGHLSSACPQGSGAGGFGGASGGGECYRCGKPGHIARMCPE-SGDAAAGG 105
Query: 63 MKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPDG-------------GPL-CFNCGE 108
G GG G +CY C VGH + +CP G GP C+NCG+
Sbjct: 106 FGGAGGYGGFGGGAGFGNKSCYTCGGVGHISRECPSGASRGFGGGGGGFGGPRKCYNCGQ 165
Query: 109 IGHYASKC-KIKGRLCFNCNQPGHFSRDCKAPKGES 143
GH + +C + +G+ C++C QPGH + C E+
Sbjct: 166 DGHISRECPQEQGKTCYSCGQPGHIASACPGAGAEA 201
Score = 53.9 bits (128), Expect = 2e-06
Identities = 21/56 (37%), Positives = 31/56 (54%)
Query: 100 GPLCFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAA 155
G CF CG+ GH A+ C + C+NC GH SR+C PK ++ G+ L++
Sbjct: 7 GSSCFKCGQQGHVAAACPAEAPTCYNCGLSGHLSRECPQPKNKACYTCGQEGHLSS 62
>gb|AAB47542.1| nucleic acid binding protein [Trypanosoma equiperdum]
Length = 270
Score = 82.4 bits (202), Expect = 4e-15
Identities = 50/172 (29%), Positives = 76/172 (44%), Gaps = 47/172 (27%)
Query: 18 CFCCGKFGQFANNCSVIGP------RCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGG 71
C CG+ G FA C + P C+ C + HL+ +C + +G + M G A
Sbjct: 19 CHRCGQPGHFARECPNVPPGAMGDRACYTCGQPDHLSRDCPSNRG---TAPMGGGRA--- 72
Query: 72 TGNKTSGKIVTCYRCKKVGHYTNKCPD--GGPL----------CFNCGEIGHYASKCKIK 119
CY C + GH++ +CP+ GGP+ C+NC + GH++ +C
Sbjct: 73 -----------CYNCGQPGHFSRECPNMRGGPMGGAPMGGGRACYNCVQPGHFSRECPNM 121
Query: 120 ------------GRLCFNCNQPGHFSRDCKAPKGESSGNTGKGKQLAAEERI 159
GR C++C QPGHFSR+C +G + G + Q E I
Sbjct: 122 RGGPMGGAPMGGGRACYHCGQPGHFSRECPNMRGANMGGGRECYQCRQEGHI 173
Score = 81.3 bits (199), Expect = 9e-15
Identities = 46/151 (30%), Positives = 64/151 (41%), Gaps = 45/151 (29%)
Query: 18 CFCCGKFGQFANNCSVI------------GPRCFKCNREGHLAVNCKTLQGGPSDSKMKG 65
C+ CG+ G F+ C + G C+ C + GH + C ++GGP M G
Sbjct: 73 CYNCGQPGHFSRECPNMRGGPMGGAPMGGGRACYNCVQPGHFSRECPNMRGGP----MGG 128
Query: 66 NDATGGTGNKTSGKIVTCYRCKKVGHYTNKCPD-------GGPLCFNCGEIGHYASKCKI 118
GG CY C + GH++ +CP+ GG C+ C + GH AS+C
Sbjct: 129 APMGGGRA---------CYHCGQPGHFSRECPNMRGANMGGGRECYQCRQEGHIASECPN 179
Query: 119 K-------------GRLCFNCNQPGHFSRDC 136
GR C+ C QPGH SR C
Sbjct: 180 APDDAAAGGTAAGGGRACYKCGQPGHLSRAC 210
Score = 73.2 bits (178), Expect = 2e-12
Identities = 41/136 (30%), Positives = 55/136 (40%), Gaps = 39/136 (28%)
Query: 35 GPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTN 94
G C +C + GH A C N G G++ CY C + H +
Sbjct: 16 GNNCHRCGQPGHFARECP-------------NVPPGAMGDRA------CYTCGQPDHLSR 56
Query: 95 KCPD--------GGPLCFNCGEIGHYASKCKIK------------GRLCFNCNQPGHFSR 134
CP GG C+NCG+ GH++ +C GR C+NC QPGHFSR
Sbjct: 57 DCPSNRGTAPMGGGRACYNCGQPGHFSRECPNMRGGPMGGAPMGGGRACYNCVQPGHFSR 116
Query: 135 DCKAPKGESSGNTGKG 150
+C +G G G
Sbjct: 117 ECPNMRGGPMGGAPMG 132
Score = 49.7 bits (117), Expect = 3e-05
Identities = 21/58 (36%), Positives = 31/58 (53%), Gaps = 6/58 (10%)
Query: 98 DGGPLCFNCGEIGHYASKC------KIKGRLCFNCNQPGHFSRDCKAPKGESSGNTGK 149
+GG C CG+ GH+A +C + R C+ C QP H SRDC + +G + G+
Sbjct: 14 EGGNNCHRCGQPGHFARECPNVPPGAMGDRACYTCGQPDHLSRDCPSNRGTAPMGGGR 71
>emb|CAA65489.1| unnamed protein product [Saccharomyces cerevisiae]
gi|6324074|ref|NP_014144.1| Putative zinc finger protein
with similarity to human CNBP, proposed to be involved
in the RAS/cAMP signaling pathway; Gis2p [Saccharomyces
cerevisiae] gi|1302303|emb|CAA96162.1| unnamed protein
product [Saccharomyces cerevisiae]
gi|1730843|sp|P53849|GIS2_YEAST Zinc-finger protein GIS2
gi|45269894|gb|AAS56328.1| YNL255C [Saccharomyces
cerevisiae]
Length = 153
Score = 82.0 bits (201), Expect = 5e-15
Identities = 44/151 (29%), Positives = 67/151 (44%), Gaps = 38/151 (25%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGN-KT 76
C+ CGK G A +C C+ CN+ GH+ +C P + K G TG+ ++
Sbjct: 6 CYVCGKIGHLAEDCDS-ERLCYNCNKPGHVQTDCTM----PRTVEFKQCYNCGETGHVRS 60
Query: 77 SGKIVTCYRCKKVGHYTNKCPD---------------GGP----------------LCFN 105
+ C+ C + GH + +CP+ GGP C+
Sbjct: 61 ECTVQRCFNCNQTGHISRECPEPKKTSRFSKVSCYKCGGPNHMAKDCMKEDGISGLKCYT 120
Query: 106 CGEIGHYASKCKIKGRLCFNCNQPGHFSRDC 136
CG+ GH + C+ RLC+NCN+ GH S+DC
Sbjct: 121 CGQAGHMSRDCQ-NDRLCYNCNETGHISKDC 150
Score = 70.1 bits (170), Expect = 2e-11
Identities = 39/110 (35%), Positives = 49/110 (44%), Gaps = 32/110 (29%)
Query: 38 CFKCNREGHLAVNCKTLQGGPSDSKMKGNDATGGTGNKTSGKIVTCYRCKKVGHYTNKCP 97
C+ C + GHLA +C DS+ CY C K GH C
Sbjct: 6 CYVCGKIGHLAEDC--------DSER------------------LCYNCNKPGHVQTDCT 39
Query: 98 DGGPL----CFNCGEIGHYASKCKIKGRLCFNCNQPGHFSRDCKAPKGES 143
+ C+NCGE GH S+C ++ CFNCNQ GH SR+C PK S
Sbjct: 40 MPRTVEFKQCYNCGETGHVRSECTVQR--CFNCNQTGHISRECPEPKKTS 87
Score = 54.3 bits (129), Expect = 1e-06
Identities = 23/70 (32%), Positives = 35/70 (49%), Gaps = 5/70 (7%)
Query: 83 CYRCKKVGHYTNKCPDGGPLCFNCGEIGHYASKCK----IKGRLCFNCNQPGHFSRDCKA 138
CY C K+GH C D LC+NC + GH + C ++ + C+NC + GH +C
Sbjct: 6 CYVCGKIGHLAEDC-DSERLCYNCNKPGHVQTDCTMPRTVEFKQCYNCGETGHVRSECTV 64
Query: 139 PKGESSGNTG 148
+ + TG
Sbjct: 65 QRCFNCNQTG 74
Score = 45.1 bits (105), Expect = 7e-04
Identities = 31/105 (29%), Positives = 44/105 (41%), Gaps = 27/105 (25%)
Query: 18 CFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNC----KTLQ---------GGPSDSKMK 64
C+ CG+ G + C+V RCF CN+ GH++ C KT + GGP+
Sbjct: 49 CYNCGETGHVRSECTV--QRCFNCNQTGHISRECPEPKKTSRFSKVSCYKCGGPNHMAKD 106
Query: 65 GNDATGGTGNK--TSGKIV----------TCYRCKKVGHYTNKCP 97
G +G K T G+ CY C + GH + CP
Sbjct: 107 CMKEDGISGLKCYTCGQAGHMSRDCQNDRLCYNCNETGHISKDCP 151
Score = 33.5 bits (75), Expect = 2.2
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Query: 12 GDKDVTCFCCGKFGQFANNCSVIGPRCFKCNREGHLAVNC 51
G + C+ CG+ G + +C C+ CN GH++ +C
Sbjct: 112 GISGLKCYTCGQAGHMSRDCQ-NDRLCYNCNETGHISKDC 150
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.140 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 341,414,153
Number of Sequences: 2540612
Number of extensions: 16154960
Number of successful extensions: 77329
Number of sequences better than 10.0: 5360
Number of HSP's better than 10.0 without gapping: 4446
Number of HSP's successfully gapped in prelim test: 930
Number of HSP's that attempted gapping in prelim test: 36143
Number of HSP's gapped (non-prelim): 24171
length of query: 165
length of database: 863,360,394
effective HSP length: 118
effective length of query: 47
effective length of database: 563,568,178
effective search space: 26487704366
effective search space used: 26487704366
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0306.5


