

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0304.6
(243 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
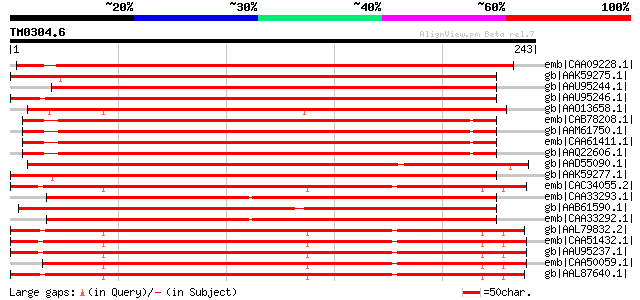
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA09228.1| thaumatin-like protein PR-5b [Cicer arietinum] 454 e-126
gb|AAK59275.1| thaumatin-like protein [Sambucus nigra] 369 e-101
gb|AAU95244.1| putative thaumatin-like protein [Solanum tuberosum] 363 2e-99
gb|AAU95246.1| putative thaumatin-like protein [Solanum tuberosum] 362 4e-99
gb|AAO13658.1| osmotin-like protein linusitin [Linum usitatissimum] 362 4e-99
emb|CAB78208.1| osmotin precursor [Arabidopsis thaliana] gi|4539... 361 9e-99
gb|AAM61750.1| osmotin precursor [Arabidopsis thaliana] 361 9e-99
emb|CAA61411.1| osmotin [Arabidopsis thaliana] 360 2e-98
gb|AAQ22606.1| At4g11650 [Arabidopsis thaliana] 359 4e-98
gb|AAD55090.1| thaumatin [Vitis riparia] 356 4e-97
gb|AAK59277.1| thaumatin-like protein [Sambucus nigra] 351 9e-96
emb|CAC34055.2| osmotin-like protein [Capsicum annuum] gi|154198... 349 4e-95
emb|CAA33293.1| thaumatin-like protein [Nicotiana tabacum] gi|19... 348 1e-94
gb|AAB61590.1| VVTL1 [Vitis vinifera] 345 6e-94
emb|CAA33292.1| thaumatin-like protein [Nicotiana tabacum] gi|20... 344 1e-93
gb|AAL79832.2| osmotin-like protein [Solanum nigrum] 343 2e-93
emb|CAA51432.1| osmotin-like protein [Solanum commersonii] gi|21... 343 3e-93
gb|AAU95237.1| osmotin-like protein [Solanum phureja] 343 3e-93
emb|CAA50059.1| pathogenesis-related protein PR P23 [Lycopersico... 342 7e-93
gb|AAL87640.1| osmotin-like protein precursor [Solanum nigrum] 341 9e-93
>emb|CAA09228.1| thaumatin-like protein PR-5b [Cicer arietinum]
Length = 239
Score = 454 bits (1168), Expect = e-126
Identities = 202/230 (87%), Positives = 215/230 (92%), Gaps = 5/230 (2%)
Query: 4 LTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTA 63
+TL SLL +LT S +AANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNL VNAGT+
Sbjct: 7 ITLCSLLFLLTPS-----QAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLWVNAGTS 61
Query: 64 MARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLV 123
MARIWGRTGCNFDG+GRGRC+TGDCTGGL C GWGVPPNTLAEFALNQYGN DFYDISLV
Sbjct: 62 MARIWGRTGCNFDGSGRGRCETGDCTGGLQCTGWGVPPNTLAEFALNQYGNLDFYDISLV 121
Query: 124 DGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTC 183
DGFNIPMDFFP+NGGCHKISCTADINGQCPNELRA GGCNNPCTVYKTNE+CCTNGQG+C
Sbjct: 122 DGFNIPMDFFPINGGCHKISCTADINGQCPNELRAQGGCNNPCTVYKTNEYCCTNGQGSC 181
Query: 184 GPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGSPHVTL 233
GPTNFS FFKDRCHD+YSYPQDDPTSTFTCPAGSNYKVVFCPLG+PH+ +
Sbjct: 182 GPTNFSTFFKDRCHDAYSYPQDDPTSTFTCPAGSNYKVVFCPLGAPHIEM 231
>gb|AAK59275.1| thaumatin-like protein [Sambucus nigra]
Length = 226
Score = 369 bits (947), Expect = e-101
Identities = 164/226 (72%), Positives = 187/226 (82%), Gaps = 1/226 (0%)
Query: 1 MGYLTLSSLLLMLTASLVTLTR-AANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVN 59
MG+L + + +L++ + AANF I NNCP+TVWAAA PGGGR+L+RGQ W+L V
Sbjct: 1 MGFLKSLPISIFFVIALISSSAYAANFNIRNNCPFTVWAAAVPGGGRQLNRGQVWSLDVP 60
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
AGT ARIW RT CNFDGAGRGRCQTGDC G L+CQ +G PPNTLAE+ALNQ+ N DF+D
Sbjct: 61 AGTRGARIWARTNCNFDGAGRGRCQTGDCNGLLSCQAYGAPPNTLAEYALNQFNNLDFFD 120
Query: 120 ISLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNG 179
ISLVDGFN+ MDF P G I CTADINGQCPNELRAPGGCNNPCTVY+TNE+CCTNG
Sbjct: 121 ISLVDGFNVAMDFSPTGGCARGIQCTADINGQCPNELRAPGGCNNPCTVYRTNEYCCTNG 180
Query: 180 QGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
QGTCGPTNFSRFFK+RC D+YSYPQDDPTSTFTCP G+NY+VVFCP
Sbjct: 181 QGTCGPTNFSRFFKERCRDAYSYPQDDPTSTFTCPGGTNYRVVFCP 226
>gb|AAU95244.1| putative thaumatin-like protein [Solanum tuberosum]
Length = 227
Score = 363 bits (932), Expect = 2e-99
Identities = 159/206 (77%), Positives = 173/206 (83%)
Query: 20 LTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDGAG 79
+T AA F+I N C Y VWAAASPGGGRRLD GQTWNL VN GT ARIWGRT CNFDG+G
Sbjct: 22 VTHAATFDITNRCTYPVWAAASPGGGRRLDSGQTWNLNVNPGTIQARIWGRTNCNFDGSG 81
Query: 80 RGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNGGC 139
RG+C+TGDC G L CQG+G PPNTLAEFALNQ N DF DISLVDGFNIPM+F P+NGGC
Sbjct: 82 RGKCETGDCNGLLECQGYGSPPNTLAEFALNQPNNLDFVDISLVDGFNIPMEFSPINGGC 141
Query: 140 HKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRFFKDRCHDS 199
+ C A IN QCPNELR PGGCNNPCTV+KTNEFCCTNG G+CGPT+ SRFFK RC D+
Sbjct: 142 RNLLCNAPINDQCPNELRTPGGCNNPCTVFKTNEFCCTNGPGSCGPTDLSRFFKQRCPDA 201
Query: 200 YSYPQDDPTSTFTCPAGSNYKVVFCP 225
YSYPQDDPTS FTCPAG+NYKVVFCP
Sbjct: 202 YSYPQDDPTSLFTCPAGTNYKVVFCP 227
>gb|AAU95246.1| putative thaumatin-like protein [Solanum tuberosum]
Length = 246
Score = 362 bits (930), Expect = 4e-99
Identities = 162/225 (72%), Positives = 183/225 (81%), Gaps = 2/225 (0%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNA 60
M +L S L + L +++T AA F+I N C Y VWAAASPGGGRRLD GQTWN+ VN
Sbjct: 1 MHFLKFSPLFVFLY--FLSVTHAATFDITNRCTYPVWAAASPGGGRRLDLGQTWNINVNP 58
Query: 61 GTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDI 120
GT M RIWGRT CNFDG+GRG+CQTGDC G L CQG+G PNTLAEFALNQ N DF DI
Sbjct: 59 GTTMGRIWGRTNCNFDGSGRGKCQTGDCNGRLECQGFGTVPNTLAEFALNQPNNLDFVDI 118
Query: 121 SLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQ 180
SLVDGFNIPM+F P+NGGC KI C+ADINGQCP+ELRAPGGCNNPCTV+K EFCCTNG
Sbjct: 119 SLVDGFNIPMEFSPINGGCRKIRCSADINGQCPSELRAPGGCNNPCTVFKMKEFCCTNGP 178
Query: 181 GTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
G+CGPT++S+FFK RC D+YSYP DDPTS FTC AG+NYKVVFCP
Sbjct: 179 GSCGPTDYSKFFKQRCPDAYSYPLDDPTSMFTCHAGTNYKVVFCP 223
>gb|AAO13658.1| osmotin-like protein linusitin [Linum usitatissimum]
Length = 263
Score = 362 bits (930), Expect = 4e-99
Identities = 166/230 (72%), Positives = 188/230 (81%), Gaps = 8/230 (3%)
Query: 9 LLLMLTASL---VTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTAM 64
LL+ L +SL V++T AA +I NNCPYTVWAA++P GGGR+LD GQTW + AGT+M
Sbjct: 7 LLIPLFSSLLLWVSITNAAVIDIFNNCPYTVWAASTPIGGGRQLDHGQTWTIYPPAGTSM 66
Query: 65 ARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVD 124
ARIWGR CNFDG+GRG C+TGDC G LNCQGWGVPPNTLAE+ALNQ+ N DFYDISLVD
Sbjct: 67 ARIWGRRNCNFDGSGRGWCETGDCGGVLNCQGWGVPPNTLAEYALNQFSNLDFYDISLVD 126
Query: 125 GFNIPMDFFPL----NGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQ 180
GFNIPM F P +G C ++CTADIN QCP ELRAPGGCNNPCTV+KTNE+CCT G
Sbjct: 127 GFNIPMIFTPTANVGSGNCQSLTCTADINTQCPGELRAPGGCNNPCTVFKTNEYCCTQGY 186
Query: 181 GTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGSPH 230
GTCGPT FSRFFKDRC SYSYPQDDP+STFTCP G+NY+VVFCP GS H
Sbjct: 187 GTCGPTGFSRFFKDRCPTSYSYPQDDPSSTFTCPGGTNYRVVFCPYGSTH 236
>emb|CAB78208.1| osmotin precursor [Arabidopsis thaliana] gi|4539456|emb|CAB39936.1|
osmotin precursor [Arabidopsis thaliana]
gi|15234263|ref|NP_192902.1| osmotin-like protein
(OSM34) [Arabidopsis thaliana]
gi|21542444|sp|P50700|OSL3_ARATH Osmotin-like protein
OSM34 precursor gi|7442160|pir||T04212 osmotin precursor
- Arabidopsis thaliana
Length = 244
Score = 361 bits (927), Expect = 9e-99
Identities = 162/219 (73%), Positives = 183/219 (82%), Gaps = 7/219 (3%)
Query: 7 SSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMAR 66
S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W L V AGT MAR
Sbjct: 12 SALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWRLDVAAGTKMAR 65
Query: 67 IWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGF 126
IWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N DFYDISLVDGF
Sbjct: 66 IWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGQPPNTLAEYALNQFNNLDFYDISLVDGF 125
Query: 127 NIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPT 186
NIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++CCTNGQG+C T
Sbjct: 126 NIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYCCTNGQGSCSDT 185
Query: 187 NFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+SRFFK RC D+YSYPQDDPTSTFTC +NY+VVFCP
Sbjct: 186 EYSRFFKQRCPDAYSYPQDDPTSTFTC-TNTNYRVVFCP 223
>gb|AAM61750.1| osmotin precursor [Arabidopsis thaliana]
Length = 244
Score = 361 bits (927), Expect = 9e-99
Identities = 162/219 (73%), Positives = 184/219 (83%), Gaps = 7/219 (3%)
Query: 7 SSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMAR 66
S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W L V AGT MAR
Sbjct: 12 SALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWRLDVAAGTKMAR 65
Query: 67 IWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGF 126
IWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N DFYDISLVDGF
Sbjct: 66 IWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGHPPNTLAEYALNQFNNLDFYDISLVDGF 125
Query: 127 NIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPT 186
NIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++CCTNGQG+C T
Sbjct: 126 NIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYCCTNGQGSCSDT 185
Query: 187 NFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
++SRFFK RC D+YSYPQDDPTSTFTC +NY+VVFCP
Sbjct: 186 DYSRFFKQRCPDAYSYPQDDPTSTFTC-TNTNYRVVFCP 223
>emb|CAA61411.1| osmotin [Arabidopsis thaliana]
Length = 244
Score = 360 bits (924), Expect = 2e-98
Identities = 162/219 (73%), Positives = 183/219 (82%), Gaps = 7/219 (3%)
Query: 7 SSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMAR 66
S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W L V AGT MAR
Sbjct: 12 SALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWRLDVAAGTKMAR 65
Query: 67 IWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGF 126
IWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N DFYDISLVDGF
Sbjct: 66 IWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGQPPNTLAEYALNQFNNLDFYDISLVDGF 125
Query: 127 NIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPT 186
NIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++CCTNGQG+C T
Sbjct: 126 NIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYCCTNGQGSCSDT 185
Query: 187 NFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+SRFFK RC D+YSYPQDDPTSTFTC +NY+VVFCP
Sbjct: 186 VYSRFFKQRCPDAYSYPQDDPTSTFTC-TNTNYRVVFCP 223
>gb|AAQ22606.1| At4g11650 [Arabidopsis thaliana]
Length = 244
Score = 359 bits (921), Expect = 4e-98
Identities = 161/219 (73%), Positives = 182/219 (82%), Gaps = 7/219 (3%)
Query: 7 SSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMAR 66
S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W L V AGT MAR
Sbjct: 12 SALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWRLDVAAGTKMAR 65
Query: 67 IWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGF 126
IWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N DFYDISLVDGF
Sbjct: 66 IWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGQPPNTLAEYALNQFNNLDFYDISLVDGF 125
Query: 127 NIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPT 186
NIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++CCTNGQG+C T
Sbjct: 126 NIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYCCTNGQGSCSDT 185
Query: 187 NFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+SRFFK RC D+YSYPQDDPTS FTC +NY+VVFCP
Sbjct: 186 EYSRFFKQRCPDAYSYPQDDPTSIFTC-TNTNYRVVFCP 223
>gb|AAD55090.1| thaumatin [Vitis riparia]
Length = 247
Score = 356 bits (913), Expect = 4e-97
Identities = 160/234 (68%), Positives = 187/234 (79%), Gaps = 4/234 (1%)
Query: 9 LLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMARIW 68
+L + ++ AA F+I N C YTVWAAASPGGGRRLDRGQ+W L V AGT MARIW
Sbjct: 14 ILFIFFLCFISSIHAATFQITNQCSYTVWAAASPGGGRRLDRGQSWTLNVPAGTKMARIW 73
Query: 69 GRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNI 128
GRT C+FD +GRGRC TGDC G LNCQGWG PPNTLAE+ALNQ+GN+DF+DISLVDGFNI
Sbjct: 74 GRTNCHFDASGRGRCDTGDCGGVLNCQGWGSPPNTLAEYALNQFGNKDFFDISLVDGFNI 133
Query: 129 PMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNF 188
PMDF P +GGC I CTA+INGQCP L+A GGCNNPCTV+KT ++CC N + CGPT++
Sbjct: 134 PMDFSPTSGGCRGIKCTANINGQCPQALKASGGCNNPCTVFKTPQYCCNNIK--CGPTDY 191
Query: 189 SRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGSPH--VTLMMPGNDT 240
SRFFK RC D+YSYPQDDPTSTFTCP G+NY+VVFCP GS + L M G+D+
Sbjct: 192 SRFFKTRCPDAYSYPQDDPTSTFTCPGGANYRVVFCPTGSSNNIFPLEMYGSDS 245
>gb|AAK59277.1| thaumatin-like protein [Sambucus nigra]
Length = 226
Score = 351 bits (901), Expect = 9e-96
Identities = 158/226 (69%), Positives = 180/226 (78%), Gaps = 1/226 (0%)
Query: 1 MGYLTLSSLLLMLTASLV-TLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVN 59
MG+L + + SL+ + AANF I NNCP+TVWAAA PGGGR+L+RGQ W+L V
Sbjct: 1 MGFLKSLPISIFFVISLIFSSAHAANFNIRNNCPFTVWAAAVPGGGRQLNRGQVWSLDVP 60
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
A TA ARIW RT CNFDGAGRGRCQTGDC G L CQ +G PPNTLAE+ALNQ+ N DF+D
Sbjct: 61 ARTAGARIWARTNCNFDGAGRGRCQTGDCNGLLACQAYGTPPNTLAEYALNQFNNLDFFD 120
Query: 120 ISLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNG 179
ISLVDGFN+ MDF P G I CTADINGQCPN LRAPGGCNNPCTV++TNE+CCTNG
Sbjct: 121 ISLVDGFNVAMDFSPTGGCARGIQCTADINGQCPNVLRAPGGCNNPCTVFRTNEYCCTNG 180
Query: 180 QGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
QG+C TN SRFFK+RC D+YSYPQDD TSTFTCP G+NY+VVFCP
Sbjct: 181 QGSCSATNLSRFFKERCRDAYSYPQDDATSTFTCPGGTNYRVVFCP 226
>emb|CAC34055.2| osmotin-like protein [Capsicum annuum] gi|15419836|gb|AAK97184.1|
thaumatin-like protein [Capsicum annuum]
Length = 246
Score = 349 bits (895), Expect = 4e-95
Identities = 166/246 (67%), Positives = 188/246 (75%), Gaps = 11/246 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
MGYL S +L +L + VT T AA FE+ NNCPYTVWAA++P GGGRRLDRGQTW +
Sbjct: 1 MGYLRSSFVLFLL--AFVTYTYAATFEVRNNCPYTVWAASTPVGGGRRLDRGQTWTINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GTAMARIWGRT CNFDG+GRG CQTGDC G L C GWG PPNTLAE+ALNQ+ N DF+D
Sbjct: 59 PGTAMARIWGRTNCNFDGSGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALNQFNNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P N G CH I CTA+ING+CP LR PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTNPSGGKCHAIQCTANINGECPGSLRVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT S+FFK RC D+YSYPQDD TSTFTCP+GS NY+VVFCP G P+ L
Sbjct: 179 T--QGPCGPTELSKFFKKRCPDAYSYPQDDATSTFTCPSGSTNYRVVFCPNGVTGPNFPL 236
Query: 234 MMPGND 239
MPG+D
Sbjct: 237 EMPGSD 242
>emb|CAA33293.1| thaumatin-like protein [Nicotiana tabacum] gi|19980|emb|CAA31235.1|
unnamed protein product [Nicotiana tabacum]
gi|58200453|gb|AAW66482.1| thaumatin-like protein
[Nicotiana tabacum] gi|131015|sp|P13046|PRR1_TOBAC
Pathogenesis-related protein R major form precursor
(Thaumatin-like protein E22)
Length = 226
Score = 348 bits (892), Expect = 1e-94
Identities = 153/208 (73%), Positives = 173/208 (82%), Gaps = 1/208 (0%)
Query: 18 VTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDG 77
V +T AA F+IVN C YTVWAAASPGGGRRLD GQ+W++ VN GT ARIWGRT CNFDG
Sbjct: 20 VAVTHAATFDIVNKCTYTVWAAASPGGGRRLDSGQSWSINVNPGTVQARIWGRTNCNFDG 79
Query: 78 AGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNG 137
+GRG C+TGDC G L CQG+G PNTLAEFALNQ NQDF DISLVDGFNIPM+F P NG
Sbjct: 80 SGRGNCETGDCNGMLECQGYGKAPNTLAEFALNQ-PNQDFVDISLVDGFNIPMEFSPTNG 138
Query: 138 GCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRFFKDRCH 197
GC + CTA IN QCP +L+ GGCNNPCTV KTNE+CCTNG G+CGPT+ SRFFK+RC
Sbjct: 139 GCRNLRCTAPINEQCPAQLKTQGGCNNPCTVIKTNEYCCTNGPGSCGPTDLSRFFKERCP 198
Query: 198 DSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
D+YSYPQDDPTS FTCP+G+NY+VVFCP
Sbjct: 199 DAYSYPQDDPTSLFTCPSGTNYRVVFCP 226
>gb|AAB61590.1| VVTL1 [Vitis vinifera]
Length = 222
Score = 345 bits (885), Expect = 6e-94
Identities = 154/221 (69%), Positives = 177/221 (79%), Gaps = 4/221 (1%)
Query: 5 TLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAM 64
TL L+ +L + L T T AA F+I+N C YTVWAAASPGGGRRLD GQ+W + VN GT
Sbjct: 6 TLPILIPLLLSLLFTSTHAATFDILNKCTYTVWAAASPGGGRRLDSGQSWTITVNPGTTN 65
Query: 65 ARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVD 124
ARIWGRT C FD GRG+C+TGDC G L CQG+G PPNTLAEFALNQ N D+ DISLVD
Sbjct: 66 ARIWGRTSCTFDANGRGKCETGDCNGLLECQGYGSPPNTLAEFALNQPNNLDYIDISLVD 125
Query: 125 GFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCG 184
GFNIPMDF GC I C+ DINGQCP+EL+APGGCNNPCTV+KTNE+CCT+G G+CG
Sbjct: 126 GFNIPMDF----SGCRGIQCSVDINGQCPSELKAPGGCNNPCTVFKTNEYCCTDGPGSCG 181
Query: 185 PTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
PT +S+FFKDRC D+YSYPQDD TS FTCP+G+NYKV FCP
Sbjct: 182 PTTYSKFFKDRCPDAYSYPQDDKTSLFTCPSGTNYKVTFCP 222
>emb|CAA33292.1| thaumatin-like protein [Nicotiana tabacum] gi|20043|emb|CAA27548.1|
unnamed protein product [Nicotiana tabacum]
gi|131017|sp|P07052|PRR2_TOBAC Pathogenesis-related
protein R minor form precursor (PR-R) (PROB12)
(Thaumatin-like protein E2) gi|225022|prf||1206322A
protein,TMV induced
Length = 226
Score = 344 bits (882), Expect = 1e-93
Identities = 152/208 (73%), Positives = 171/208 (82%), Gaps = 1/208 (0%)
Query: 18 VTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDG 77
V +T AA F+IVN C YTVWAAASPGGGR+L+ GQ+W++ VN GT ARIWGRT CNFDG
Sbjct: 20 VAVTHAATFDIVNQCTYTVWAAASPGGGRQLNSGQSWSINVNPGTVQARIWGRTNCNFDG 79
Query: 78 AGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNG 137
+GRG C+TGDC G L CQG+G PPNTLAEFALNQ NQDF DISLVDGFNIPM+F P NG
Sbjct: 80 SGRGNCETGDCNGMLECQGYGKPPNTLAEFALNQ-PNQDFVDISLVDGFNIPMEFSPTNG 138
Query: 138 GCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRFFKDRCH 197
GC + CTA IN QCP +L+ GGCNNPCTV KTNEFCCTNG G+CGPT+ SRFFK RC
Sbjct: 139 GCRNLRCTAPINEQCPAQLKTQGGCNNPCTVIKTNEFCCTNGPGSCGPTDLSRFFKARCP 198
Query: 198 DSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
D+YSYPQDDP S FTCP G+NY+VVFCP
Sbjct: 199 DAYSYPQDDPPSLFTCPPGTNYRVVFCP 226
>gb|AAL79832.2| osmotin-like protein [Solanum nigrum]
Length = 247
Score = 343 bits (881), Expect = 2e-93
Identities = 164/245 (66%), Positives = 186/245 (74%), Gaps = 11/245 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
MGYL S + +LT VT T A +FE+ NNCPYTVWAA++P GGGRRLDRGQTW +
Sbjct: 1 MGYLRSSFVFFLLT--FVTYTYATSFEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT+MARIWGRT CNFDGAGRG CQTGDC G L C GWG PPNTLAE+ALNQ+ N DF+D
Sbjct: 59 RGTSMARIWGRTNCNFDGAGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALNQFSNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P N G CH I CTA+ING+CP LR PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTNPSGGKCHSIQCTANINGECPAALRVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT S+FFK RC D+YSYPQDDPTSTFTCP+ S NY+VVFCP G SP+ L
Sbjct: 179 T--QGPCGPTELSKFFKQRCPDAYSYPQDDPTSTFTCPSDSTNYRVVFCPNGVTSPNFPL 236
Query: 234 MMPGN 238
MP +
Sbjct: 237 EMPSS 241
>emb|CAA51432.1| osmotin-like protein [Solanum commersonii] gi|21195|emb|CAA47601.1|
osmotin-like protein [Solanum commersonii]
gi|419783|pir||S30144 osmotin-like protein precursor
(clone pA13) - Commerson's wild potato
gi|1709493|sp|P50701|OS13_SOLCO OSMOTIN-LIKE PROTEIN
OSML13 PRECURSOR (PA13)
Length = 246
Score = 343 bits (879), Expect = 3e-93
Identities = 164/246 (66%), Positives = 186/246 (74%), Gaps = 11/246 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
M YL S + +L + VT T AA E+ NNCPYTVWAA++P GGGRRLDRGQTW +
Sbjct: 1 MAYLRSSFVFFLL--AFVTYTYAATIEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT MARIWGRT CNFDGAGRG CQTGDC G L C GWG PPNTLAE+AL+Q+ N DF+D
Sbjct: 59 RGTKMARIWGRTNCNFDGAGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P N G CH I CTA+ING+CP LR PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTNPSGGKCHAIHCTANINGECPGSLRVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT+ SRFFK RC D+YSYPQDDPTSTFTCP+GS NY+VVFCP G SP+ L
Sbjct: 179 T--QGPCGPTDLSRFFKQRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGVTSPNFPL 236
Query: 234 MMPGND 239
MP +D
Sbjct: 237 EMPASD 242
>gb|AAU95237.1| osmotin-like protein [Solanum phureja]
Length = 246
Score = 343 bits (879), Expect = 3e-93
Identities = 164/246 (66%), Positives = 186/246 (74%), Gaps = 11/246 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
M YL S + +L + VT T AA E+ NNCPYTVWAA++P GGGRRLDRGQTW +
Sbjct: 1 MAYLRSSFVFFLL--AFVTYTYAATIEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT MARIWGRT CNFDGAGRG CQTGDC G L C GWG PPNTLAE+AL+Q+ N DF+D
Sbjct: 59 RGTKMARIWGRTNCNFDGAGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P N G CH I CTA+ING+CP LR PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTNPSGGKCHAIHCTANINGECPGSLRVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT+ SRFFK RC D+YSYPQDDPTSTFTCP+GS NY+VVFCP G SP+ L
Sbjct: 179 T--QGPCGPTDLSRFFKQRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGVTSPNFPL 236
Query: 234 MMPGND 239
MP +D
Sbjct: 237 EMPSSD 242
>emb|CAA50059.1| pathogenesis-related protein PR P23 [Lycopersicon esculentum]
Length = 233
Score = 342 bits (876), Expect = 7e-93
Identities = 160/231 (69%), Positives = 180/231 (77%), Gaps = 9/231 (3%)
Query: 16 SLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTAMARIWGRTGCN 74
+ VT T AA FE+ NNCPYTVWAA++P GGGRRLDRGQTW + GT MARIWGRT CN
Sbjct: 1 AFVTYTYAATFEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAPRGTKMARIWGRTNCN 60
Query: 75 FDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFP 134
FDGAGRG CQTGDC G L C GWG PPNTLAE+AL+Q+ N DF+DISLVDGFNIPM F P
Sbjct: 61 FDGAGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSNLDFWDISLVDGFNIPMTFAP 120
Query: 135 LN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRF 191
N G CH I CTA+ING+CP LR PGGCNNPCT + ++CCT QG CGPT+ SRF
Sbjct: 121 TNPSGGKCHAIHCTANINGECPGSLRVPGGCNNPCTTFGGQQYCCT--QGPCGPTDLSRF 178
Query: 192 FKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTLMMPGND 239
FK RC D+YSYPQDDPTSTFTCP+GS NY+VVFCP G SP+ L MP +D
Sbjct: 179 FKQRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGVTSPNFPLEMPSSD 229
>gb|AAL87640.1| osmotin-like protein precursor [Solanum nigrum]
Length = 247
Score = 341 bits (875), Expect = 9e-93
Identities = 163/245 (66%), Positives = 185/245 (74%), Gaps = 11/245 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
MGY S + +LT VT T A +FE+ NNCPYTVWAA++P GGGRRLDRGQTW +
Sbjct: 1 MGYSRSSFVFFLLT--FVTYTYATSFEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT+MARIWGRT CNFDGAGRG CQTGDC G L C GWG PPNTLAE+ALNQ+ N DF+D
Sbjct: 59 RGTSMARIWGRTNCNFDGAGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALNQFSNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P N G CH I CTA+ING+CP LR PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTNPSGGKCHSIQCTANINGECPAALRVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT S+FFK RC D+YSYPQDDPTSTFTCP+ S NY+VVFCP G SP+ L
Sbjct: 179 T--QGPCGPTELSKFFKQRCPDAYSYPQDDPTSTFTCPSDSTNYRVVFCPNGVTSPNFPL 236
Query: 234 MMPGN 238
MP +
Sbjct: 237 EMPSS 241
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.140 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 498,387,171
Number of Sequences: 2540612
Number of extensions: 24405026
Number of successful extensions: 39668
Number of sequences better than 10.0: 369
Number of HSP's better than 10.0 without gapping: 328
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 38231
Number of HSP's gapped (non-prelim): 432
length of query: 243
length of database: 863,360,394
effective HSP length: 124
effective length of query: 119
effective length of database: 548,324,506
effective search space: 65250616214
effective search space used: 65250616214
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0304.6


