

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0302a.1
(299 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
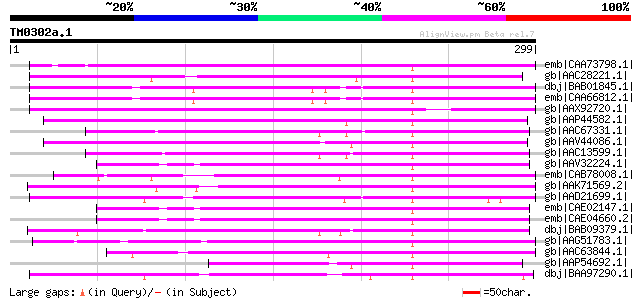
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulg... 171 3e-41
gb|AAC28221.1| similar to reverse transcriptases (PFam: rvt.hmm,... 110 6e-23
dbj|BAB01845.1| non-LTR retroelement reverse transcriptase-like ... 109 1e-22
emb|CAA66812.1| non-ltr retrotransposon reverse transcriptase-li... 109 1e-22
gb|AAX92720.1| retrotransposon protein, putative, unclassified [... 106 9e-22
gb|AAP44582.1| putative reverse transcriptase [Oryza sativa (jap... 104 3e-21
gb|AAC67331.1| putative non-LTR retroelement reverse transcripta... 103 5e-21
gb|AAV44086.1| hypothetical protein [Oryza sativa (japonica cult... 103 6e-21
gb|AAC13599.1| similar to reverse transcriptase (Pfam: transcrip... 103 8e-21
gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cult... 101 2e-20
emb|CAB78008.1| putative protein [Arabidopsis thaliana] gi|73210... 101 3e-20
gb|AAK71569.2| putative reverse transcriptase [Oryza sativa (jap... 101 3e-20
gb|AAD21699.1| Contains reverse transcriptase domain (rvt) PF|00... 101 3e-20
emb|CAE02147.1| OSJNBa0081G05.2 [Oryza sativa (japonica cultivar... 100 4e-20
emb|CAE04660.2| OSJNBa0061G20.16 [Oryza sativa (japonica cultiva... 100 4e-20
dbj|BAB09379.1| non-LTR retroelement reverse transcriptase-like ... 100 5e-20
gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Ara... 99 1e-19
gb|AAC63844.1| putative non-LTR retroelement reverse transcripta... 98 3e-19
gb|AAP54692.1| putative reverse transcriptase [Oryza sativa (jap... 98 3e-19
dbj|BAA97290.1| non-LTR retroelement reverse transcriptase-like ... 97 4e-19
>emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulgaris]
gi|7484649|pir||T14619 reverse transcriptase - beet
retrotransposon (fragment)
Length = 507
Score = 171 bits (432), Expect = 3e-41
Identities = 97/300 (32%), Positives = 155/300 (51%), Gaps = 16/300 (5%)
Query: 12 KPFRNRNCWLQNHAFGECVSTWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKI 71
KPFR NCWL + +C+ S + + + +KL+ LK L QW ++ G +D
Sbjct: 107 KPFRFLNCWLTHP---QCMKIISEIWTKNHNS-SVPDKLRTLKEALKQWNQKEFGIIDDN 162
Query: 72 CKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDS 131
K +I+ LD L + E ++RDD + W K ++S SR RW+K+GDS
Sbjct: 163 IKHCEEKIHFLDNIANSRLLCTNELQERDDAQLNLWTWLKRSESYWALNSRARWIKEGDS 222
Query: 132 NTKYFHNSVNWRRRTNAIRGLAVDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVFLEGTP 191
NT+YFH RR N ++ + +G+ + P +K + + +F+ F G
Sbjct: 223 NTRYFHTLATIRRHKNHLQYIKTEGKNISKPGEIKEEAVAYFQHIFAEEFSTRPVFNGLN 282
Query: 192 FRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVLK 239
F +S+ + +L F EEI AV C+ ++ GPD V+K DI ++++
Sbjct: 283 FNKLSESECSSLIAPFTHEEIDEAVDSCDAQKAPGPDGFNFKFIKSSWEVIKKDIYKIVE 342
Query: 240 DFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVMGHI 299
DF +G P G NS+FI LIP + N++ IS++GC+YKI++K+L RL +VM H+
Sbjct: 343 DFWASGSLPHGCNSAFIALIPKTQHPRGFNEFRPISMVGCLYKIIAKILARRLQRVMDHL 402
>gb|AAC28221.1| similar to reverse transcriptases (PFam: rvt.hmm, score: 60.13)
[Arabidopsis thaliana] gi|7488321|pir||T01871
RNA-directed DNA polymerase homolog T24M8.8 -
Arabidopsis thaliana
Length = 1164
Score = 110 bits (274), Expect = 6e-23
Identities = 77/304 (25%), Positives = 147/304 (48%), Gaps = 29/304 (9%)
Query: 12 KPFRNRNCWLQNHAFGECVST-WSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
KPFR N L++ F + W S + G Y KLK LK + + ++N D++K
Sbjct: 144 KPFRFNNFLLKDENFLSLICLKWFSTSVTGSAMYRVSVKLKALKKVIRDFSRDNYSDIEK 203
Query: 71 ICKDPVTRI----NALDIKEGDGNLSSQEQKQRDDFLVEYWKV-AKFNDSLLFQKSRDRW 125
K+ + + L N + + + QR W++ A+ S +Q+SR W
Sbjct: 204 RTKEAHDALLLAQSVLLASPCPSNAAIEAETQRK------WRILAEAEASFFYQRSRVNW 257
Query: 126 VKDGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIEFFEAQFTSNPEVG 184
+++GD N+ YFH + R+ N I L+ G+ +E + +++ +E+F++ S +
Sbjct: 258 LREGDMNSSYFHKMASARQSLNHIHFLSDPVGDRIEGQQNLENHCVEYFQSNLGSEQGLP 317
Query: 185 VFLEGTPFRSIS----DEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------ 228
+F + +S ++L F+ E+I+ A + +++ GPD
Sbjct: 318 LFEQADISNLLSYRCSPAQQVSLDTPFSSEQIKNAFFSLPRNKASGPDGFSPEFFCACWP 377
Query: 229 VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVL 288
++ ++ + +F +G K N++ + LIP I+N S++D+ IS + +YK++SK+L
Sbjct: 378 IIGGEVTEAIHEFFTSGKLLKQWNATNLVLIPKITNASSMSDFRPISCLNTVYKVISKLL 437
Query: 289 TTRL 292
T RL
Sbjct: 438 TDRL 441
>dbj|BAB01845.1| non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 893
Score = 109 bits (272), Expect = 1e-22
Identities = 87/317 (27%), Positives = 155/317 (48%), Gaps = 37/317 (11%)
Query: 12 KPFRNRNCWLQNHAFGECV-STWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PFR N +L N F + + W S ++ G Y +KLK LKL + + +EN D++K
Sbjct: 244 RPFRFFNYFLHNPDFLQLIRENWYSCNVSGSAMYRVSKKLKHLKLPICCFSRENYSDIEK 303
Query: 71 ICKDPVTRINALDIKEGDGNLSSQEQKQRDDFL--VEYWKV-AKFNDSLLFQKSRDRWVK 127
V+ +A+ + L++ L W++ AK +S QKS W+
Sbjct: 304 ----RVSEAHAIVLHRQRITLTNPSVVHATLELEATRKWQILAKAEESFFCQKSSISWLY 359
Query: 128 DGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIE----FFEAQFT---- 178
+GD+NT YFH + R+ N I L D GE +E + +K + E FFE+
Sbjct: 360 EGDNNTAYFHKMADMRKSINTINFLIDDFGERIETQQGIKEGIKEHSCNFFESLLCGVEG 419
Query: 179 ----SNPEVGVFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------ 228
+ ++ + L FR D+ N L ++F+ +I+ A + +++ GPD
Sbjct: 420 ENSLAQSDMNLLLS---FRCSVDQIN-DLERSFSDLDIQEAFFSLPRNKASGPDGYSSEF 475
Query: 229 ------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYK 282
V+ ++ +++F R+G K N++ + LIP I+N + D+ IS + +YK
Sbjct: 476 FKGVWFVVGPEVTEAVQEFFRSGQLLKQWNATTLVLIPKITNSSKMTDFRPISCLNTLYK 535
Query: 283 IVSKVLTTRLSKVMGHI 299
+++K+LT+RL K++ +
Sbjct: 536 VIAKLLTSRLKKLLNEV 552
>emb|CAA66812.1| non-ltr retrotransposon reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 893
Score = 109 bits (272), Expect = 1e-22
Identities = 87/317 (27%), Positives = 155/317 (48%), Gaps = 37/317 (11%)
Query: 12 KPFRNRNCWLQNHAFGECV-STWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PFR N +L N F + + W S ++ G Y +KLK LKL + + +EN D++K
Sbjct: 244 RPFRFFNYFLHNPDFLQLIRENWYSCNVSGSAMYRVSKKLKHLKLPICCFSRENYSDIEK 303
Query: 71 ICKDPVTRINALDIKEGDGNLSSQEQKQRDDFL--VEYWKV-AKFNDSLLFQKSRDRWVK 127
V+ +A+ + L++ L W++ AK +S QKS W+
Sbjct: 304 ----RVSEAHAIVLHRQRITLTNPSVVHATLELEATRKWQILAKAEESFFCQKSSISWLY 359
Query: 128 DGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIE----FFEAQFT---- 178
+GD+NT YFH + R+ N I L D GE +E + +K + E FFE+
Sbjct: 360 EGDNNTAYFHKMADMRKSINTINFLIDDFGERIETQQGIKEGIKEHSCNFFESLLCGVEG 419
Query: 179 ----SNPEVGVFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------ 228
+ ++ + L FR D+ N L ++F+ +I+ A + +++ GPD
Sbjct: 420 ENSLAQSDMNLLLS---FRCSVDQIN-DLERSFSDLDIQEAFFSLPRNKASGPDGYSSEF 475
Query: 229 ------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYK 282
V+ ++ +++F R+G K N++ + LIP I+N + D+ IS + +YK
Sbjct: 476 FKGVWFVVGPEVTEAVQEFFRSGQLLKQWNATTLVLIPKITNSSKMTDFRPISCLNTLYK 535
Query: 283 IVSKVLTTRLSKVMGHI 299
+++K+LT+RL K++ +
Sbjct: 536 VIAKLLTSRLKKLLNEV 552
>gb|AAX92720.1| retrotransposon protein, putative, unclassified [Oryza sativa
(japonica cultivar-group)] gi|62701880|gb|AAX92953.1|
retrotransposon protein, putative, unclassified [Oryza
sativa (japonica cultivar-group)]
Length = 838
Score = 106 bits (264), Expect = 9e-22
Identities = 73/290 (25%), Positives = 140/290 (48%), Gaps = 16/290 (5%)
Query: 12 KPFRNRNCWLQNHAFGECVSTWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKI 71
K F+ W E V + + G ++++ + L+ KLN W K G +
Sbjct: 230 KSFKFELSWFLREDLKEIVESVWKINYYGSSIERWQKRFRALRKKLNGWNKNWEGRYRRE 289
Query: 72 CKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDS 131
+ + +I LD K +S +++++R + + V + FQ+S+++ + +GD
Sbjct: 290 KESILEKIENLDAKVERCGMSREDREERKELEDKLNFVIREEKIKWFQRSKEKELLEGDC 349
Query: 132 NTKYFHNSVNWRRRTNAIRGLAVDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVFLEGTP 191
NTKY+H N R+R + I L + +E + + + + F++ F + +FL+
Sbjct: 350 NTKYYHAKANGRKRKSYIHSLTQEEGEIEGQEKLLTYITSFYKDLFGHPKQNTLFLDMRG 409
Query: 192 FRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD--VLKVDILRVLKDFHRNGVWPK 249
+I +E + LT+ F+ EE+R V+E + +++ GPD + VDI R+
Sbjct: 410 VTTIMEEHKVMLTKPFSEEEVRIVVFELKKNKAPGPDGFIGLVDIDRL------------ 457
Query: 250 GGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVMGHI 299
N I LIP I + + Y I ++ +KI++KVL R++++M +I
Sbjct: 458 --NFGVINLIPKIKDAAQIQKYRPICLLNVSFKIITKVLMNRITEIMTYI 505
>gb|AAP44582.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|34896522|ref|NP_909605.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1556
Score = 104 bits (260), Expect = 3e-21
Identities = 72/292 (24%), Positives = 136/292 (45%), Gaps = 16/292 (5%)
Query: 20 WLQNHAFGECVS-TWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKICKDPVTR 78
WL+ F + V W I+G ++ K++RL+ L W K G K K+ + +
Sbjct: 1029 WLRRDGFSDIVKEVWQGVSIEGTPLERWQRKIRRLRQFLRGWAKNITGAYKKEKKELLDK 1088
Query: 79 INALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHN 138
++ L K L+ E + ++ + + +Q+++ + + +GD+NTKY+H
Sbjct: 1089 LDTLVKKAEHTILNEFELNVKHVLNDRLAELLREEEIKWYQRAKVKHLLEGDANTKYYHL 1148
Query: 139 SVNWRRRTNAIRGLAVDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVFLEGT---PFRSI 195
N R R I L E + +K + ++F+ F + +FL+ + R +
Sbjct: 1149 LANGRHRKTRIFQLQDGNEVITGDAQLKEHITKYFKTLFGPSQTSSIFLDESQVDDIRQV 1208
Query: 196 SDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVLKDFHR 243
S E+N +LT F EEI+ A+++ + + + GPD ++K D+L + DF +
Sbjct: 1209 SAEENESLTADFTEEEIKCAIFQMKHNTAPGPDGFPPEFYQAFWTIIKDDLLALFSDFQQ 1268
Query: 244 NGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKV 295
+ N I L+P ++ ++ Y I ++ +KI +KV T RL+ V
Sbjct: 1269 GSLPLNSLNFGTIILLPKQKDVRTIQQYRPICLLNVSFKIFTKVATNRLTSV 1320
>gb|AAC67331.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25410913|pir||B84426 hypothetical protein
At2g01550 [imported] - Arabidopsis thaliana
Length = 1449
Score = 103 bits (258), Expect = 5e-21
Identities = 75/272 (27%), Positives = 125/272 (45%), Gaps = 21/272 (7%)
Query: 44 YTFKEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFL 103
+ F +KLK LK KL KE +G+L K ++ + + N S + + +
Sbjct: 700 FRFTKKLKALKPKLRGLAKEKMGNLVKRTREAYLSLCQAQ-QSNSQNPSQRAMEIESEAY 758
Query: 104 VEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAV-DGEWVEDP 162
V + ++A + L Q S+ W+K GD N K FH + R N+IR + DG
Sbjct: 759 VRWDRIASIEEKYLKQVSKLHWLKVGDKNNKTFHRAATARAAQNSIREIQKEDGSTATTK 818
Query: 163 KVVKSKMIEFFEA--QFTSNPEVGVFLEGT----PFRSISDEDNLALTQTFNLEEIRTAV 216
+K++ FF+ Q N G+ +E P+ E ++ LT + + +EIR A+
Sbjct: 819 DDIKNETERFFQEFLQLIPNDYEGITVEKLTSLLPYHCSPAEKDM-LTASVSAKEIRGAL 877
Query: 217 WECEGDRSLGPD------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISN 264
+ D+S GPD ++ + + +K F G PKG N++ + LIP
Sbjct: 878 FSMPNDKSPGPDGYTSEFYKRAWDIIGAEFVLAVKSFFEKGFLPKGVNTTILALIPKKLE 937
Query: 265 LMSLNDYISISVIGCMYKIVSKVLTTRLSKVM 296
+ DY IS +YK++SK++ RL V+
Sbjct: 938 AKEMKDYRPISCCNVIYKVISKIIANRLKHVL 969
>gb|AAV44086.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 1237
Score = 103 bits (257), Expect = 6e-21
Identities = 81/294 (27%), Positives = 133/294 (44%), Gaps = 20/294 (6%)
Query: 20 WLQNHAFGECVST-WSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKICKDPVTR 78
WL F E V WSS D ++ K++RL+ L W K + G K K + +
Sbjct: 890 WLLRDGFVEMVKEIWSSVDEGNTAMERWQAKIRRLRQHLRGWSKHSSGLYKKEKKKILDK 949
Query: 79 INALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHN 138
++ LD K + LS +E R + + + +Q+++ + + +GDSNTKYFH
Sbjct: 950 LDLLDKKAENSLLSPEELDIRWFCRNRLSSILREEEIKWYQRAKSKDILEGDSNTKYFHL 1009
Query: 139 SVNWRRRTNAIRGLAVDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVFLEGTPFR----- 193
N + R I L ++ +KS + +++ F P L+ R
Sbjct: 1010 VANGKHRKTRIFQLQDSDHLIDGDANLKSHITTYYKGLF--GPPDDSLLQFDESRVADIP 1067
Query: 194 SISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVLKDF 241
IS +N ALTQ F +EI+ A+++ E +++ GPD V+K D+L + K+F
Sbjct: 1068 QISLLENEALTQDFTEKEIKKAIFQMEHNKASGPDGFPAEFYQVFWNVIKNDLLDLFKEF 1127
Query: 242 HRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKV 295
H + N I L+P M + Y I ++ +KI +KV T R+ V
Sbjct: 1128 HNGSLPLFSLNFGTIILLPKCVEAMKIQQYRPICLLNVSFKIFTKVATNRVMSV 1181
>gb|AAC13599.1| similar to reverse transcriptase (Pfam: transcript_fact.hmm, score:
72.31) [Arabidopsis thaliana] gi|7488318|pir||T01191
RNA-directed DNA polymerase homolog F21E10.5 -
Arabidopsis thaliana
Length = 928
Score = 103 bits (256), Expect = 8e-21
Identities = 76/272 (27%), Positives = 122/272 (43%), Gaps = 21/272 (7%)
Query: 44 YTFKEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFL 103
+ F +KLK LK L KE +G+L K K+ + + N S ++ ++
Sbjct: 173 FRFSKKLKGLKPLLRNLGKERLGNLVKQTKEAFETLCQKQAMKM-ANPSPSSMQEENEAY 231
Query: 104 VEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAV-DGEWVEDP 162
++ +A + L Q+S+ W+ GD N K FH +V R N+IR + DG
Sbjct: 232 AKWDHIAVLEEKFLKQRSKLHWLDIGDRNNKAFHRAVVAREAQNSIREIICHDGSVASQE 291
Query: 163 KVVKSKMIEFFEA--QFTSNPEVGVFLEGT----PFRSISDEDNLALTQTFNLEEIRTAV 216
+ +K++ F Q N G+ +E P+R SD D LT + EEI V
Sbjct: 292 EKIKTEAEHHFREFLQLIPNDFEGIAVEELQDLLPYRC-SDSDKEMLTNHVSAEEIHKVV 350
Query: 217 WECEGDRSLGPD------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISN 264
+ D+S GPD ++ + + ++ F G PKG NS+ + LIP
Sbjct: 351 FSMPNDKSPGPDGYTAEFYKGAWNIIGAEFILAIQSFFAKGFLPKGINSTILALIPKKKE 410
Query: 265 LMSLNDYISISVIGCMYKIVSKVLTTRLSKVM 296
+ DY IS +YK++SK++ RL V+
Sbjct: 411 AKEMKDYRPISCCNVLYKVISKIIANRLKLVL 442
>gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|45267888|gb|AAS55787.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 1936
Score = 101 bits (252), Expect = 2e-20
Identities = 73/261 (27%), Positives = 123/261 (46%), Gaps = 21/261 (8%)
Query: 50 LKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKV 109
L R+ L W K V ++ K + ++ L N + +Q D + E +
Sbjct: 916 LGRVMSALRSWSKTKVKNVGKELEKARKKLEDLIAS----NAARSSIRQATDHMNE---M 968
Query: 110 AKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDGEWVEDP-KVVKSK 168
+ L Q+SR W+K+GD NT++FH+ WR + N I L + + V+++
Sbjct: 969 LYREEMLWLQRSRVNWLKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIHSTTSVLETM 1028
Query: 169 MIEFFEAQFTSNPEVGVFLEGTPFRS-ISDEDNLALTQTFNLEEIRTAVWECEGDRSLGP 227
E+F+ + ++P + F+ ++D N L Q F EEI A+++ +S P
Sbjct: 1029 ATEYFQGVYKADPSLNPESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPRP 1088
Query: 228 D------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISIS 275
D LK DI+ +++F ++G+ PKG N + I LIP + L DY IS
Sbjct: 1089 DGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPKGVNDTAIVLIPKKDQPIDLKDYRPIS 1148
Query: 276 VIGCMYKIVSKVLTTRLSKVM 296
+ +YK+VSK L RL ++
Sbjct: 1149 LCNVVYKVVSKCLVNRLRPIL 1169
>emb|CAB78008.1| putative protein [Arabidopsis thaliana] gi|7321072|emb|CAB82119.1|
putative protein [Arabidopsis thaliana]
gi|25407453|pir||H85088 hypothetical protein AT4g08830
[imported] - Arabidopsis thaliana
Length = 947
Score = 101 bits (251), Expect = 3e-20
Identities = 76/301 (25%), Positives = 141/301 (46%), Gaps = 45/301 (14%)
Query: 26 FGECVSTWSSFDIQGWG-AYTFKEK-------LKRLKLKLNQWEKENVGDLDKICKDPVT 77
FGE ++ S D+ G YT+K KRL +L +W ++ GD+ K ++ +
Sbjct: 8 FGEWINRVSLIDMGFSGNKYTWKRGRVENTFVAKRLD-RLKKWNRDVFGDVKKKKEELMR 66
Query: 78 RI----NALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNT 133
I +ALD+ + D L +E + F KSR++W+ GD NT
Sbjct: 67 EIQVVQDALDVHQSDDMLRKEEDLIK-----------------AFDKSREKWIALGDRNT 109
Query: 134 KYFHNSVNWRRRTNAIRGLA-VDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVF--LEGT 190
YFH + RRR N + L +G W+ D K +++ +++++ ++ + + V L
Sbjct: 110 NYFHTTTIIRRRQNRVERLHNEEGTWIFDAKELEALAVQYYQRLYSMDDVLQVVNPLPQD 169
Query: 191 PFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVL 238
++ E+ L+L + F E+ A+ ++ GPD V+ ++R
Sbjct: 170 GLERLTREEVLSLNKPFLATEVEVAIRSMGKYKAPGPDGYQPIFYQSCWEVVGNSVIRFA 229
Query: 239 KDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVMGH 298
DF +G+ P N + + LIP + +N + IS+ ++K+++K++ RL KV+G
Sbjct: 230 LDFFSSGILPPQTNDALVVLIPKVPKPERMNQFRPISLCNAIFKMITKMMVLRLKKVIGK 289
Query: 299 I 299
+
Sbjct: 290 L 290
>gb|AAK71569.2| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|50918647|ref|XP_469720.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1833
Score = 101 bits (251), Expect = 3e-20
Identities = 77/308 (25%), Positives = 140/308 (45%), Gaps = 29/308 (9%)
Query: 11 LKPFRNRNCWLQNHAFGECVSTWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+K R W + + E + + +K+K KL W K+ +G++ K
Sbjct: 220 MKIMRYEEVWERESSLPEVIQEAWTMGADASTLGDINDKMKVTMTKLVSWSKDKIGNVRK 279
Query: 71 ICKDPVTRINAL---DIKEGDGNLSSQEQKQRDDFLVE--YWKVAKFNDSLLFQKSRDRW 125
KD ++ L + + D + S +++ + E +WK Q+SR W
Sbjct: 280 KIKDLREKLGELRNIGLLDTDNEVHSVKKELEEMLHREEIWWK----------QRSRITW 329
Query: 126 VKDGDSNTKYFHNSVNWRRRTNAIRGLAV-DGEWVEDPKVVKSKMIEFFEAQFTSNPEVG 184
+K+GD NT+YFH +WR + N I+ L DG + K +K FF+ +T + +
Sbjct: 330 LKEGDLNTRYFHLKASWRAKKNKIKKLKKNDGSTTMNKKEMKEINRSFFQQLYTKDDNLN 389
Query: 185 -VFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLK 231
V L IS++ N L + F EEI A+++ ++ GPD +LK
Sbjct: 390 PVNLLNMFHEKISEQMNADLIKPFTNEEISDALFQIGPLKAPGPDGFPARFLQRNWGLLK 449
Query: 232 VDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTR 291
+++ +++F + V +G N + I +IP + + D+ IS+ +YK+V+K L R
Sbjct: 450 GEVIAAVRNFFEDEVMQEGVNDTVIVMIPKKNLAEDMKDFRPISLCNVVYKVVAKCLVNR 509
Query: 292 LSKVMGHI 299
+ ++ I
Sbjct: 510 MRPMLQEI 517
>gb|AAD21699.1| Contains reverse transcriptase domain (rvt) PF|00078. [Arabidopsis
thaliana] gi|25518658|pir||F86436 hypothetical protein
F28K20.4 - Arabidopsis thaliana
Length = 1253
Score = 101 bits (251), Expect = 3e-20
Identities = 82/315 (26%), Positives = 146/315 (46%), Gaps = 33/315 (10%)
Query: 12 KPFRNRNCWLQNHAFGECVST-WSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PFR N LQN F V W S ++ G + +KLK LK + + EN +L+K
Sbjct: 196 RPFRFYNFLLQNPDFISLVGELWYSINVVGSSMFKMSKKLKALKNPIRTFSMENFSNLEK 255
Query: 71 ICKDP----VTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWV 126
K+ + R N N + + + QR ++ + K +S Q+SR W+
Sbjct: 256 RVKEAHNLVLYRQNKTLSDPTIPNAALEMEAQR-----KWLILVKAEESFFCQRSRVTWM 310
Query: 127 KDGDSNTKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIEFFEAQFTSNPEVGV 185
+GDSNT YFH + R+ N I + D G ++ +K IE+F +
Sbjct: 311 GEGDSNTSYFHRMADSRKAVNTIHIIIDDNGVKIDTQLGIKEHCIEYFSNLLGGEVGPPM 370
Query: 186 FLEG-----TPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------ 228
++ PFR D+ L +F+ ++I++A + +++ GPD
Sbjct: 371 LIQEDFDLLLPFRCSHDQKK-ELAMSFSRQDIKSAFFSFPSNKTSGPDGFPVEFFKETWS 429
Query: 229 VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDY--ISISVIG--CMYKIV 284
V+ ++ + +F + V K N++ + LIP I+N +ND+ IS + G +YK++
Sbjct: 430 VIGTEVTDAVSEFFTSSVLLKQWNATTLVLIPKITNASKMNDFRPISCNDFGPITLYKVI 489
Query: 285 SKVLTTRLSKVMGHI 299
+++LT RL ++ +
Sbjct: 490 ARLLTNRLQCLLSQV 504
>emb|CAE02147.1| OSJNBa0081G05.2 [Oryza sativa (japonica cultivar-group)]
gi|50923489|ref|XP_472105.1| OSJNBa0081G05.2 [Oryza
sativa (japonica cultivar-group)]
Length = 1711
Score = 100 bits (250), Expect = 4e-20
Identities = 72/261 (27%), Positives = 124/261 (46%), Gaps = 21/261 (8%)
Query: 50 LKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKV 109
L R+ L W K V ++ + + ++ L N + +Q D + E +
Sbjct: 828 LGRVMSALRSWGKTKVKNVGRELEKARKKLEDLIAS----NAARSSIRQATDHMNE---M 880
Query: 110 AKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDGEWVED-PKVVKSK 168
+ L Q+SR W+K+GD NT++FH+ WR + N I L + + V+++
Sbjct: 881 LYREEMLWLQRSRVNWLKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIHSMTSVLETM 940
Query: 169 MIEFFEAQFTSNPEVGVFLEGTPFRS-ISDEDNLALTQTFNLEEIRTAVWECEGDRSLGP 227
E+F+ + ++P + F+ ++D N L Q F EEI A+++ +S GP
Sbjct: 941 ATEYFQEVYKADPSLNPESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPGP 1000
Query: 228 D------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISIS 275
D LK DI+ +++F ++G+ P+G N + I LIP + L DY IS
Sbjct: 1001 DGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPEGVNDTAIVLIPKKDQPIDLKDYRPIS 1060
Query: 276 VIGCMYKIVSKVLTTRLSKVM 296
+ +YK+VSK L RL ++
Sbjct: 1061 LCNVVYKVVSKCLVNRLRPIL 1081
>emb|CAE04660.2| OSJNBa0061G20.16 [Oryza sativa (japonica cultivar-group)]
Length = 1285
Score = 100 bits (250), Expect = 4e-20
Identities = 72/261 (27%), Positives = 124/261 (46%), Gaps = 21/261 (8%)
Query: 50 LKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKV 109
L R+ L W K V ++ + + ++ L N + +Q D + E +
Sbjct: 828 LGRVMSALRSWGKTKVKNVGRELEKARKKLEDLIAS----NAARSSIRQATDHMNE---M 880
Query: 110 AKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDGEWVED-PKVVKSK 168
+ L Q+SR W+K+GD NT++FH+ WR + N I L + + V+++
Sbjct: 881 LYREEMLWLQRSRVNWLKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIHSMTSVLETM 940
Query: 169 MIEFFEAQFTSNPEVGVFLEGTPFRS-ISDEDNLALTQTFNLEEIRTAVWECEGDRSLGP 227
E+F+ + ++P + F+ ++D N L Q F EEI A+++ +S GP
Sbjct: 941 ATEYFQEVYKADPSLNPESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPGP 1000
Query: 228 D------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISIS 275
D LK DI+ +++F ++G+ P+G N + I LIP + L DY IS
Sbjct: 1001 DGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPEGVNDTAIVLIPKKDQPIDLKDYRPIS 1060
Query: 276 VIGCMYKIVSKVLTTRLSKVM 296
+ +YK+VSK L RL ++
Sbjct: 1061 LCNVVYKVVSKCLVNRLRPIL 1081
>dbj|BAB09379.1| non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 1223
Score = 100 bits (249), Expect = 5e-20
Identities = 87/309 (28%), Positives = 134/309 (43%), Gaps = 25/309 (8%)
Query: 11 LKPFRNRNCWLQNHAFGECVST-WSSFD---IQGWGAYTFKEKLKRLKLKLNQWEKENVG 66
LKPF+ N F VST W + + + F + LK LK K+ ++ +G
Sbjct: 248 LKPFKFVNALTDMEDFKPMVSTYWKDTEPLILSTSTLFRFSKNLKGLKPKIRSMARDRLG 307
Query: 67 DLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWV 126
+L K + +I N SS ++ + + +VA + L QKS+ W
Sbjct: 308 NLSKKANEAY-KILCAKQHVNLTNPSSMAMEEENAAYSRWDRVAILEEKYLKQKSKLHWC 366
Query: 127 KDGDSNTKYFHNSVNWRRRTNAIRG-LAVDGEWVEDPKVVKSKMIEFFEA--QFTSNPEV 183
+ GD NTK FH + R N IR L+ DG +K++ FF Q N
Sbjct: 367 QVGDQNTKAFHRAAAAREAHNTIREILSNDGIVKTKGDEIKAEAERFFREFLQLIPNDFE 426
Query: 184 GVFL----EGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD----------- 228
GV + + P R SD D +L + EEIR ++ D+S GPD
Sbjct: 427 GVTITELQQLLPVRC-SDADQQSLIRPVTAEEIRKVLFRMPSDKSPGPDGYTSEFFKATW 485
Query: 229 -VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKV 287
++ + ++ F G PKG NS+ + LIP + + DY IS +YK++SK+
Sbjct: 486 EIIGDEFTLAVQSFFTKGFLPKGINSTILALIPKKTEAREMKDYRPISCCNVLYKVISKI 545
Query: 288 LTTRLSKVM 296
+ RL V+
Sbjct: 546 IANRLKLVL 554
>gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
gi|25405244|pir||E96519 probable reverse transcriptase,
16838-20266 [imported] - Arabidopsis thaliana
Length = 1142
Score = 99.0 bits (245), Expect = 1e-19
Identities = 82/301 (27%), Positives = 141/301 (46%), Gaps = 23/301 (7%)
Query: 14 FRNRNCWLQNHAFGECVSTWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKICK 73
FR W+ E +S + D G+ F EKL + +++W K L +
Sbjct: 24 FRFDKRWIGKDGLLEAISQGWNLD-SGFREGQFVEKLTNCRRAISKWRKS----LIPFGR 78
Query: 74 DPVTRINA-LDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSN 132
+ + A LD+ + D + S +E + L E ++ + +QKSR W+K GD+N
Sbjct: 79 QTIEDLKAELDVAQRDDDRSREEITELTLRLKEAYRD---EEQYWYQKSRSLWMKLGDNN 135
Query: 133 TKYFHNSVNWRRRTNAIRGLAVD-GEWVEDPKVVKSKMIEFFEAQFTS-NPEVGVFLEGT 190
+K+FH RR N I GL + G W + +++ + +F+ FT+ NP+V G
Sbjct: 136 SKFFHALTKQRRARNRITGLHDENGIWSIEDDDIQNIAVSYFQNLFTTANPQVFDEALGE 195
Query: 191 PFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD------------VLKVDILRVL 238
I+D N LT E+R A++ +++ GPD ++K D+L ++
Sbjct: 196 VQVLITDRINDLLTADATECEVRAALFMIHPEKAPGPDGMTALFFQKSWAIIKSDLLSLV 255
Query: 239 KDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVSKVLTTRLSKVMGH 298
F + GV+ K N++ I LIP + + IS+ YK++SK+L RL V+ +
Sbjct: 256 NSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLCNVGYKVISKILCQRLKTVLPN 315
Query: 299 I 299
+
Sbjct: 316 L 316
>gb|AAC63844.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25408124|pir||C84716 hypothetical protein
At2g31080 [imported] - Arabidopsis thaliana
Length = 1231
Score = 98.2 bits (243), Expect = 3e-19
Identities = 71/263 (26%), Positives = 125/263 (46%), Gaps = 24/263 (9%)
Query: 56 KLNQWEKENVGDL----DKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAK 111
KL +W +E GD+ +K+ D + L + D L+ +E L E V +
Sbjct: 139 KLRKWNREVFGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEV-----LLKEMDLVLE 193
Query: 112 FNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDGE-WVEDPKVVKSKMI 170
++L FQKSR+++++ GD NT +FH S RRR N I L D + WV D +++ +
Sbjct: 194 QEETLWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMAL 253
Query: 171 EFFEAQFTSN--PEVGVFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPD 228
+++ ++ EV L F SIS+ + AL Q F E+ +AV ++ GPD
Sbjct: 254 TYYKRLYSLEDVSEVRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPD 313
Query: 229 ------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISV 276
+ + R + +F GV P N + + LI ++ + + +S+
Sbjct: 314 GYQPVFYQQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQFRPVSL 373
Query: 277 IGCMYKIVSKVLTTRLSKVMGHI 299
++KI++K++ TRL V+ +
Sbjct: 374 CNVLFKIITKMMVTRLKNVISKL 396
>gb|AAP54692.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37536206|ref|NP_922405.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|27311287|gb|AAO00713.1|
retrotransposon protein, putative, unclassified [Oryza
sativa (japonica cultivar-group)]
Length = 1557
Score = 98.2 bits (243), Expect = 3e-19
Identities = 62/197 (31%), Positives = 101/197 (50%), Gaps = 22/197 (11%)
Query: 114 DSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLA-VDGEWVEDPKVVKSKMIEF 172
+ L Q+SR W+K+GD NTK+FHN V WR + N I L G + V++ ++
Sbjct: 721 EMLWLQRSRVNWLKEGDRNTKFFHNRVAWRAKKNKITKLRDAAGTFHRSKMVMEQMATQY 780
Query: 173 FEAQFTSNPEVGVFLEGTPFR-----SISDEDNLALTQTFNLEEIRTAVWECEGDRSLGP 227
F+ FT++P L+ +P +++E N L F+ +EI A+++ ++ G
Sbjct: 781 FKDMFTADPN----LDHSPVTRLIQPKVTEEMNNRLCSEFSEDEIANALFQIGPLKAPGT 836
Query: 228 D------------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISIS 275
D +K DI+R +K+F +G+ +G N + I LIP I L D+ IS
Sbjct: 837 DGFLARFYQRNWGCIKNDIVRAVKEFFHSGIMLEGVNDTAIVLIPKIEQPRDLRDFRPIS 896
Query: 276 VIGCMYKIVSKVLTTRL 292
+ +YK++SK L RL
Sbjct: 897 LCNVIYKVLSKCLVNRL 913
>dbj|BAA97290.1| non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 1072
Score = 97.4 bits (241), Expect = 4e-19
Identities = 81/321 (25%), Positives = 145/321 (44%), Gaps = 47/321 (14%)
Query: 12 KPFRNRNCWLQNHAFGECV-STWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDK 70
+PF+ N L+N F V W S ++ G Y +KLK +K + + + N ++
Sbjct: 107 RPFKFFNFLLKNEDFLNVVMDNWFSTNVVGSSMYRVSKKLKAMKKPIKDFSRLNYSGIEL 166
Query: 71 ICKDP----VTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWV 126
K+ +T N N + + + QR L+ + +S Q+SR W
Sbjct: 167 RTKEAHELLITCQNLTLANPSVSNAALELEAQRKWVLLSCAE-----ESFFHQRSRVSWF 221
Query: 127 KDGDSNTKYFHNSVNWRRRTNAIRGLA-VDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGV 185
+GDSNT YFH V+ R+ N I L +G ++ + + + ++E S
Sbjct: 222 AEGDSNTHYFHRMVDSRKSFNTINSLVDSNGLLIDSQQGILDHCVTYYERLLGSIE---- 277
Query: 186 FLEGTPFRSISDEDNLALT------------QTFNLEEIRTAVWECEGDRSLGPD----- 228
+PF ++ NL LT ++F +EI+ A +++ GPD
Sbjct: 278 ----SPFSMEQEDMNLLLTYRCSQDQCSELEKSFTDDEIKAAFKSLPRNKTSGPDGYSVE 333
Query: 229 -------VLKVDILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMY 281
++ ++L + +F +G K N++ + LIP SN +++++ IS + +Y
Sbjct: 334 FFRDTWSIIGPEVLAAIHEFFDSGQLLKQWNATTLVLIPKTSNACTISEFRPISCLNTLY 393
Query: 282 KIVSKVLTTR----LSKVMGH 298
K++SK+LT+R LS V+GH
Sbjct: 394 KVISKLLTSRLQGLLSAVIGH 414
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.140 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 516,551,611
Number of Sequences: 2540612
Number of extensions: 21463698
Number of successful extensions: 64356
Number of sequences better than 10.0: 309
Number of HSP's better than 10.0 without gapping: 191
Number of HSP's successfully gapped in prelim test: 118
Number of HSP's that attempted gapping in prelim test: 63848
Number of HSP's gapped (non-prelim): 342
length of query: 299
length of database: 863,360,394
effective HSP length: 127
effective length of query: 172
effective length of database: 540,702,670
effective search space: 93000859240
effective search space used: 93000859240
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0302a.1


