

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0283.14
(461 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
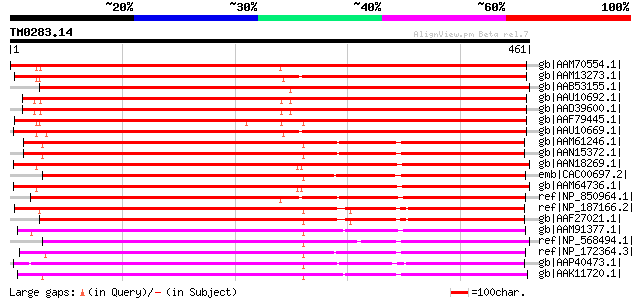
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM70554.1| At1g75220/F22H5_6 [Arabidopsis thaliana] gi|15222... 580 e-164
gb|AAM13273.1| similar to integral membrane protein [Arabidopsis... 578 e-163
gb|AAB53155.1| integral membrane protein [Beta vulgaris] gi|7446... 569 e-161
gb|AAU10692.1| putative sugar transporter [Oryza sativa (japonic... 568 e-160
gb|AAD39600.1| 10A19I.3 [Oryza sativa (japonica cultivar-group)] 568 e-160
gb|AAF79445.1| F18O14.22 [Arabidopsis thaliana] 563 e-159
gb|AAU10669.1| putative integral membrane protein [Oryza sativa ... 551 e-155
gb|AAM61246.1| putative sugar transporter [Arabidopsis thaliana] 382 e-104
gb|AAN15372.1| putative putative sister-chromatide cohesion prot... 379 e-103
gb|AAN18269.1| At5g18840/F17K4_90 [Arabidopsis thaliana] gi|1738... 378 e-103
emb|CAC00697.2| putative sugar transporter [Lycopersicon esculen... 377 e-103
gb|AAM64736.1| sugar transporter-like protein [Arabidopsis thali... 375 e-102
ref|NP_850964.1| sugar transporter, putative [Arabidopsis thaliana] 370 e-101
ref|NP_187166.2| sugar transporter family protein [Arabidopsis t... 358 1e-97
gb|AAF27021.1| putative sugar transporter [Arabidopsis thaliana] 357 3e-97
gb|AAM91377.1| At1g08920/F7G19_20 [Arabidopsis thaliana] gi|1839... 333 9e-90
ref|NP_568494.1| sugar-porter family protein 2 (SFP2) [Arabidops... 332 1e-89
ref|NP_172364.3| sugar transporter family protein [Arabidopsis t... 332 2e-89
gb|AAP40473.1| putative zinc finger protein ATZF1 [Arabidopsis t... 330 4e-89
gb|AAK11720.1| sugar-porter family protein 1 [Arabidopsis thaliana] 330 6e-89
>gb|AAM70554.1| At1g75220/F22H5_6 [Arabidopsis thaliana]
gi|15222158|ref|NP_177658.1| integral membrane protein,
putative [Arabidopsis thaliana]
gi|15724240|gb|AAL06513.1| At1g75220/F22H5_6
[Arabidopsis thaliana] gi|10092276|gb|AAG12689.1|
integral membrane protein, putative; 33518-36712
[Arabidopsis thaliana] gi|25308944|pir||E96782
hypothetical protein F22H5.6 [imported] - Arabidopsis
thaliana
Length = 487
Score = 580 bits (1496), Expect = e-164
Identities = 300/486 (61%), Positives = 371/486 (75%), Gaps = 27/486 (5%)
Query: 1 MPLGEDYEDSRN-IRKPFINNNN-----------AGSG------SFFVVLCVLVVALGPI 42
M +D E++RN +R+PFI+ + GS S V+ CVL+VALGPI
Sbjct: 1 MSFRDDNEEARNDLRRPFIHTGSWYRMGSRQSSMMGSSQVIRDSSISVLACVLIVALGPI 60
Query: 43 QFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLV 102
QFGFTCGYSSPTQ + +DL L++S +S+FGSLSNVGAMVGA SG +AEY GRKGSL++
Sbjct: 61 QFGFTCGYSSPTQAAITKDLGLTVSEYSVFGSLSNVGAMVGAIASGQIAEYIGRKGSLMI 120
Query: 103 AAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQ 162
AA+PNI GW+ IS AKD+S L++GRLLEGFGVGIISY VPVYIAEI P+ MRG LG+VNQ
Sbjct: 121 AAIPNIIGWLCISFAKDTSFLYMGRLLEGFGVGIISYTVPVYIAEIAPQNMRGGLGSVNQ 180
Query: 163 LSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSL 222
LSVTIGIMLAYLLGLF WR LA+LG+ PC +LIPGL+FIPESPRWLA+MG+ + FE+SL
Sbjct: 181 LSVTIGIMLAYLLGLFVPWRILAVLGILPCTLLIPGLFFIPESPRWLAKMGMTDEFETSL 240
Query: 223 QTLRGSNVDITMEAQEI---------QNRFYTVCRPQEEKILVSFDGIGLLVLQQLSGIN 273
Q LRG DIT+E EI +N V + GIGLLVLQQL GIN
Sbjct: 241 QVLRGFETDITVEVNEIKRSVASSTKRNTVRFVDLKRRRYYFPLMVGIGLLVLQQLGGIN 300
Query: 274 GVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISL 333
GV FY+S IF SAG++SS+AATFG+GAIQVV T ++TWLVDK+GRR+LL +SS MTISL
Sbjct: 301 GVLFYSSTIFESAGVTSSNAATFGVGAIQVVATAISTWLVDKAGRRLLLTISSVGMTISL 360
Query: 334 LLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKG 393
++VA+AFYL+ VS S++++ L ILSVVG+VA+++ F+LG+GPIPWLIMSEILP NIKG
Sbjct: 361 VIVAAAFYLKEFVSPDSDMYSWLSILSVVGVVAMVVFFSLGMGPIPWLIMSEILPVNIKG 420
Query: 394 LAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLE 453
LAGS+AT +NWF + +IT+TAN LL WSS GTFT+Y + AFTV F L+VPETK +TLE
Sbjct: 421 LAGSIATLANWFFSWLITMTANLLLAWSSGGTFTLYGLVCAFTVVFVTLWVPETKGKTLE 480
Query: 454 EIQASF 459
E+Q+ F
Sbjct: 481 ELQSLF 486
>gb|AAM13273.1| similar to integral membrane protein [Arabidopsis thaliana]
gi|15223557|ref|NP_173377.1| integral membrane protein,
putative / sugar transporter family protein [Arabidopsis
thaliana] gi|16648957|gb|AAL24330.1| similar to integral
membrane protein [Arabidopsis thaliana]
Length = 488
Score = 578 bits (1490), Expect = e-163
Identities = 301/484 (62%), Positives = 372/484 (76%), Gaps = 31/484 (6%)
Query: 5 EDYEDSRN-IRKPFINNNN---AGS--------------GSFFVVLCVLVVALGPIQFGF 46
++ E+ RN +R+PF++ + GS S V+ CVL+VALGPIQFGF
Sbjct: 6 DNTEEGRNDLRRPFLHTGSWYRMGSRQSSMLESSQVIRDSSISVLACVLIVALGPIQFGF 65
Query: 47 TCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVP 106
TCGYSSPTQ + +DL L++S +S+FGSLSNVGAMVGA SG +AEY GRKGSL++AA+P
Sbjct: 66 TCGYSSPTQAAITKDLGLTVSEYSVFGSLSNVGAMVGAIASGQIAEYVGRKGSLMIAAIP 125
Query: 107 NIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVT 166
NI GW++IS AKD+S L++GRLLEGFGVGIISY VPVYIAEI P+TMRG+LG+VNQLSVT
Sbjct: 126 NIIGWLSISFAKDTSFLYMGRLLEGFGVGIISYTVPVYIAEIAPQTMRGALGSVNQLSVT 185
Query: 167 IGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLR 226
IGIMLAYLLGLF WR LA+LGV PC +LIPGL+FIPESPRWLA+MGL + FE+SLQ LR
Sbjct: 186 IGIMLAYLLGLFVPWRILAVLGVLPCTLLIPGLFFIPESPRWLAKMGLTDDFETSLQVLR 245
Query: 227 GSNVDITMEAQEIQN-----------RFYTVCRPQEEKILVSFDGIGLLVLQQLSGINGV 275
G DIT+E EI+ RF + R + L+ GIGLL LQQL GINGV
Sbjct: 246 GFETDITVEVNEIKRSVASSSKRSAVRFVDLKRRRYYFPLMV--GIGLLALQQLGGINGV 303
Query: 276 FFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLL 335
FY+S IF SAG++SS+ ATFG+G +QVV TG+ATWLVDK+GRR+LL++SS MTISL++
Sbjct: 304 LFYSSTIFESAGVTSSNVATFGVGVVQVVATGIATWLVDKAGRRLLLMISSIGMTISLVI 363
Query: 336 VASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLA 395
VA AFYL+ VS S ++NIL ++SVVG+VA++I +LG+GPIPWLIMSEILP NIKGLA
Sbjct: 364 VAVAFYLKEFVSPDSNMYNILSMVSVVGVVAMVISCSLGMGPIPWLIMSEILPVNIKGLA 423
Query: 396 GSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEI 455
GS+AT NWF + ++T+TAN LL WSS GTFT+YA+ FTV F L+VPETK +TLEEI
Sbjct: 424 GSIATLLNWFVSWLVTMTANMLLAWSSGGTFTLYALVCGFTVVFVSLWVPETKGKTLEEI 483
Query: 456 QASF 459
QA F
Sbjct: 484 QALF 487
>gb|AAB53155.1| integral membrane protein [Beta vulgaris] gi|7446725|pir||T14545
probable sugar transporter protein - beet
Length = 490
Score = 569 bits (1466), Expect = e-161
Identities = 285/444 (64%), Positives = 355/444 (79%), Gaps = 9/444 (2%)
Query: 27 SFFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATV 86
S V+ CVL+VALGPIQFGFT GYSSPTQ + +L LS++ +S FGSLSNVGAMVGA
Sbjct: 47 SISVLACVLIVALGPIQFGFTAGYSSPTQSAITNELGLSVAEYSWFGSLSNVGAMVGAIA 106
Query: 87 SGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIA 146
SG ++EY GRKGSL++AA+PNI GW+AIS AKDSS L++GR+LEGFGVGIISY VPVYI+
Sbjct: 107 SGQISEYIGRKGSLMIAAIPNIIGWLAISFAKDSSFLYMGRMLEGFGVGIISYTVPVYIS 166
Query: 147 EIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESP 206
EI P+ +RG+LG+VNQLSVTIGIML+Y+LGLF WR LA+LG+ PC ILIPGL+FIPESP
Sbjct: 167 EIAPQNLRGALGSVNQLSVTIGIMLSYMLGLFVPWRILAVLGILPCTILIPGLFFIPESP 226
Query: 207 RWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFYTVCR---------PQEEKILVS 257
RWLA+MG+ME FE SLQ LRG + DI++E EI+ + + Q L
Sbjct: 227 RWLAKMGMMEEFEVSLQVLRGFDTDISLEVNEIKRSVASSSKRTTIRFAELRQRRYWLPL 286
Query: 258 FDGIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSG 317
G GLL+LQQLSGINGV FY+S IF AG++SS+AATFGLGA+QV+ T V TWLVDKSG
Sbjct: 287 MIGNGLLILQQLSGINGVLFYSSTIFKEAGVTSSNAATFGLGAVQVIATVVTTWLVDKSG 346
Query: 318 RRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGP 377
RR+LLIVSSS MT+SLL+VA +F+L+ +VS S +++ ILSVVG+VA+++ F+LGIG
Sbjct: 347 RRLLLIVSSSGMTLSLLVVAMSFFLKEMVSDESTWYSVFSILSVVGVVAMVVTFSLGIGA 406
Query: 378 IPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTV 437
IPW+IMSEILP NIKGLAGS+AT +NWF A ++T+TAN +L+W+S GTF+IY + AFTV
Sbjct: 407 IPWIIMSEILPINIKGLAGSIATLANWFVAWIVTMTANIMLSWNSGGTFSIYMVVCAFTV 466
Query: 438 AFALLFVPETKDRTLEEIQASFIR 461
AF +++VPETK RTLEEIQ SF R
Sbjct: 467 AFVVIWVPETKGRTLEEIQWSFRR 490
>gb|AAU10692.1| putative sugar transporter [Oryza sativa (japonica cultivar-group)]
Length = 501
Score = 568 bits (1463), Expect = e-160
Identities = 291/480 (60%), Positives = 369/480 (76%), Gaps = 32/480 (6%)
Query: 12 NIRKPFINN------NNAGSG-----------------SFFVVLCVLVVALGPIQFGFTC 48
++RKPF++ ++AG G S VLC L+VALGPIQFGFTC
Sbjct: 21 DLRKPFLHTGSWYKMSSAGGGGGMGSRLGSSAYSLRDSSVSAVLCTLIVALGPIQFGFTC 80
Query: 49 GYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNI 108
G+SSPTQ+ +I DL L++S FSLFGSLSNVGAMVGA SG +AEY GRKGSL++AA+PNI
Sbjct: 81 GFSSPTQDAIISDLGLTLSEFSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIAAIPNI 140
Query: 109 FGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIG 168
GW+AIS AKDSS LF+GRLLEGFGVG+ISYVVPVYIAEI P+TMRG+LG+VNQLSVTIG
Sbjct: 141 IGWLAISFAKDSSFLFMGRLLEGFGVGVISYVVPVYIAEIAPQTMRGALGSVNQLSVTIG 200
Query: 169 IMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGS 228
I+LAYLLG+F WR L++LG+ PC+ILIPGL+FIPESPRWLA+MG ME FESSLQ LRG
Sbjct: 201 ILLAYLLGMFVPWRILSVLGILPCSILIPGLFFIPESPRWLAKMGKMEDFESSLQVLRGF 260
Query: 229 NVDITMEAQEIQ-----NRFYTVCR----PQEEKILVSFDGIGLLVLQQLSGINGVFFYA 279
DI +E EI+ +R T R Q+ + GIGLLVLQQLSG+NG+ FYA
Sbjct: 261 ETDIAVEVNEIKRTVQSSRRRTTIRFADIKQKRYSVPLMIGIGLLVLQQLSGVNGILFYA 320
Query: 280 SKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASA 339
+ IF +AG+++S+ ATFGLG +QVV TGV TWL DK+GRR+LLI+S++ MTI+L++V+ +
Sbjct: 321 ASIFKAAGLTNSNLATFGLGVVQVVATGVTTWLTDKAGRRLLLIISTTGMTITLVVVSVS 380
Query: 340 FYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVA 399
F+++ ++ GS L++++ +LS+VGLVA +I F+LG+G IPW+IMSEILP NIK LAGSVA
Sbjct: 381 FFVKDNITNGSHLYSVMSMLSLVGLVAFVISFSLGLGAIPWIIMSEILPVNIKSLAGSVA 440
Query: 400 TFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASF 459
T +NW TA +IT+TA+ +L+WS+ GTF IYA A T+ F L+VPETK RTLEEI SF
Sbjct: 441 TLANWLTAWLITMTASLMLSWSNGGTFAIYAAVCAGTLVFVCLWVPETKGRTLEEIAFSF 500
>gb|AAD39600.1| 10A19I.3 [Oryza sativa (japonica cultivar-group)]
Length = 501
Score = 568 bits (1463), Expect = e-160
Identities = 291/480 (60%), Positives = 369/480 (76%), Gaps = 32/480 (6%)
Query: 12 NIRKPFINN------NNAGSG-----------------SFFVVLCVLVVALGPIQFGFTC 48
++RKPF++ ++AG G S VLC L+VALGPIQFGFTC
Sbjct: 21 DLRKPFLHTGSWYKMSSAGGGGGMGSRLGSSAYSLRDSSVSAVLCTLIVALGPIQFGFTC 80
Query: 49 GYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNI 108
G+SSPTQ+ +I DL L++S FSLFGSLSNVGAMVGA SG +AEY GRKGSL++AA+PNI
Sbjct: 81 GFSSPTQDAIISDLGLTLSEFSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIAAIPNI 140
Query: 109 FGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIG 168
GW+AIS AKDSS LF+GRLLEGFGVG+ISYVVPVYIAEI P+TMRG+LG+VNQLSVTIG
Sbjct: 141 IGWLAISFAKDSSFLFMGRLLEGFGVGVISYVVPVYIAEIAPQTMRGALGSVNQLSVTIG 200
Query: 169 IMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGS 228
I+LAYLLG+F WR L++LG+ PC+ILIPGL+FIPESPRWLA+MG ME FESSLQ LRG
Sbjct: 201 ILLAYLLGMFVPWRILSVLGILPCSILIPGLFFIPESPRWLAKMGKMEDFESSLQVLRGF 260
Query: 229 NVDITMEAQEIQ-----NRFYTVCR----PQEEKILVSFDGIGLLVLQQLSGINGVFFYA 279
DI +E EI+ +R T R Q+ + GIGLLVLQQLSG+NG+ FYA
Sbjct: 261 ETDIAVEVNEIKRSVQSSRRRTTIRFADIKQKRYSVPLMVGIGLLVLQQLSGVNGILFYA 320
Query: 280 SKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASA 339
+ IF +AG+++S+ ATFGLG +QVV TGV TWL DK+GRR+LLI+S++ MTI+L++V+ +
Sbjct: 321 ASIFKAAGLTNSNLATFGLGVVQVVATGVTTWLTDKAGRRLLLIISTTGMTITLVVVSVS 380
Query: 340 FYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVA 399
F+++ ++ GS L++++ +LS+VGLVA +I F+LG+G IPW+IMSEILP NIK LAGSVA
Sbjct: 381 FFVKDNITNGSHLYSVMSMLSLVGLVAFVISFSLGLGAIPWIIMSEILPVNIKSLAGSVA 440
Query: 400 TFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASF 459
T +NW TA +IT+TA+ +L+WS+ GTF IYA A T+ F L+VPETK RTLEEI SF
Sbjct: 441 TLANWLTAWLITMTASLMLSWSNGGTFAIYAAVCAGTLVFVCLWVPETKGRTLEEIAFSF 500
>gb|AAF79445.1| F18O14.22 [Arabidopsis thaliana]
Length = 515
Score = 563 bits (1452), Expect = e-159
Identities = 300/509 (58%), Positives = 370/509 (71%), Gaps = 54/509 (10%)
Query: 5 EDYEDSRN-IRKPFINNNN---AGS--------------GSFFVVLCVLVVALGPIQFGF 46
++ E+ RN +R+PF++ + GS S V+ CVL+VALGPIQFGF
Sbjct: 6 DNTEEGRNDLRRPFLHTGSWYRMGSRQSSMLESSQVIRDSSISVLACVLIVALGPIQFGF 65
Query: 47 TCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVP 106
TCGYSSPTQ + +DL L++S +S+FGSLSNVGAMVGA SG +AEY GRKGSL++AA+P
Sbjct: 66 TCGYSSPTQAAITKDLGLTVSEYSVFGSLSNVGAMVGAIASGQIAEYVGRKGSLMIAAIP 125
Query: 107 NIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVT 166
NI GW++IS AKD+S L++GRLLEGFGVGIISY VPVYIAEI P+TMRG+LG+VNQLSVT
Sbjct: 126 NIIGWLSISFAKDTSFLYMGRLLEGFGVGIISYTVPVYIAEIAPQTMRGALGSVNQLSVT 185
Query: 167 IGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWL----------------- 209
IGIMLAYLLGLF WR LA+LGV PC +LIPGL+FIPESPRWL
Sbjct: 186 IGIMLAYLLGLFVPWRILAVLGVLPCTLLIPGLFFIPESPRWLVCLYNRIAYIVEFVALL 245
Query: 210 AEMGLMERFESSLQTLRGSNVDITMEAQEIQ------NRFYTVCRPQEEKILVSFD---- 259
A+MGL + FE+SLQ LRG DIT+E EI+ F ++ V F
Sbjct: 246 AKMGLTDDFETSLQVLRGFETDITVEVNEIKVVTKLKKCFDRSVASSSKRSAVRFVDLKR 305
Query: 260 ---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVAT 310
GIGLL LQQL GINGV FY+S IF SAG++SS+ ATFG+G +QVV TG+AT
Sbjct: 306 RRYYFPLMVGIGLLALQQLGGINGVLFYSSTIFESAGVTSSNVATFGVGVVQVVATGIAT 365
Query: 311 WLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIG 370
WLVDK+GRR+LL++SS MTISL++VA AFYL+ VS S ++NIL ++SVVG+VA++I
Sbjct: 366 WLVDKAGRRLLLMISSIGMTISLVIVAVAFYLKEFVSPDSNMYNILSMVSVVGVVAMVIS 425
Query: 371 FALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYA 430
+LG+GPIPWLIMSEILP NIKGLAGS+AT NWF + ++T+TAN LL WSS GTFT+YA
Sbjct: 426 CSLGMGPIPWLIMSEILPVNIKGLAGSIATLLNWFVSWLVTMTANMLLAWSSGGTFTLYA 485
Query: 431 IFSAFTVAFALLFVPETKDRTLEEIQASF 459
+ FTV F L+VPETK +TLEEIQA F
Sbjct: 486 LVCGFTVVFVSLWVPETKGKTLEEIQALF 514
>gb|AAU10669.1| putative integral membrane protein [Oryza sativa (japonica
cultivar-group)]
Length = 501
Score = 551 bits (1419), Expect = e-155
Identities = 282/489 (57%), Positives = 362/489 (73%), Gaps = 35/489 (7%)
Query: 4 GEDYEDSRNIRKPFINNNN--------------AGSGSFFVV--------LCVLVVALGP 41
G DYE +RKP + + AG+ S ++ LC L+VALGP
Sbjct: 14 GSDYESGGGMRKPLLMHTGSWYRMGSRQGSLTGAGTSSMAILRESHVSAFLCTLIVALGP 73
Query: 42 IQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLL 101
IQFGFT G+SSPTQ+ +IRDL+L++S FS+FGSLSNVGAMVGA SG +AEY GRKGSL+
Sbjct: 74 IQFGFTGGFSSPTQDAIIRDLDLTLSEFSVFGSLSNVGAMVGAIASGQMAEYIGRKGSLM 133
Query: 102 VAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVN 161
+AA+PNI GW+AIS AKDSS L++GRLLEGFGVG+ISY VPVYIAEI P+ MRG+LG+VN
Sbjct: 134 IAAIPNIIGWLAISFAKDSSFLYMGRLLEGFGVGVISYTVPVYIAEISPQNMRGALGSVN 193
Query: 162 QLSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESS 221
QLSVT+GI+LAYLLG+F WR LA++G+ PC +LIPGL+FIPESPRWLA+M +M+ FE+S
Sbjct: 194 QLSVTVGILLAYLLGMFVPWRLLAVIGILPCTVLIPGLFFIPESPRWLAKMNMMDDFETS 253
Query: 222 LQTLRGSNVDITMEAQEIQN-----------RFYTVCRPQEEKILVSFDGIGLLVLQQLS 270
LQ LRG DI+ E +I+ RF + + + L+ GIGLLVLQQLS
Sbjct: 254 LQVLRGFETDISAEVNDIKRAVASANKRTTIRFQELNQKKYRTPLIL--GIGLLVLQQLS 311
Query: 271 GINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMT 330
GING+ FYA IF +AG+++SD AT LGAIQV+ TGV TWL+D++GRRILLI+SS+ MT
Sbjct: 312 GINGILFYAGSIFKAAGLTNSDLATCALGAIQVLATGVTTWLLDRAGRRILLIISSAGMT 371
Query: 331 ISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPN 390
+SLL VA F+L+ +S+ S ++ L ++S+V LVA +I F+ G+G IPW+IMSEILP +
Sbjct: 372 LSLLAVAVVFFLKDSISQDSHMYYTLSMISLVALVAFVIAFSFGMGAIPWIIMSEILPVS 431
Query: 391 IKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDR 450
IK LAGS AT +NW T+ IT+TAN +L+WS+ GTF Y + SAFT+ F +L+VPETK R
Sbjct: 432 IKSLAGSFATLANWLTSFGITMTANLMLSWSAGGTFVSYMVVSAFTLVFVILWVPETKGR 491
Query: 451 TLEEIQASF 459
TLEEIQ SF
Sbjct: 492 TLEEIQWSF 500
>gb|AAM61246.1| putative sugar transporter [Arabidopsis thaliana]
Length = 463
Score = 382 bits (981), Expect = e-104
Identities = 202/457 (44%), Positives = 287/457 (62%), Gaps = 17/457 (3%)
Query: 13 IRKPFINNNNAGSGS---FFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRF 69
+R+P ++ N AGS + V L V GP FG GYSSP Q + DL+L+I+ F
Sbjct: 8 VREPLVDKNMAGSKPDQPWMVYLSTFVAVCGPFAFGSCAGYSSPAQAAIRNDLSLTIAEF 67
Query: 70 SLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLL 129
SLFGSL GAM+GA SG +A+ GRKG++ V++ + GW+AI AK L +GRL
Sbjct: 68 SLFGSLLTFGAMIGAITSGPIADLVGRKGAMRVSSAFCVVGWLAIIFAKGVVALDLGRLA 127
Query: 130 EGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGV 189
G+G+G SYVVP++IAEI P+T RG+L +NQ+ + G+ +++++G WR LAL+G+
Sbjct: 128 TGYGMGAFSYVVPIFIAEIAPKTFRGALTTLNQILICTGVSVSFIIGTLVTWRVLALIGI 187
Query: 190 FPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFYTVCRP 249
PCA GL+FIPESPRWLA++G FE++L+ LRG DI+ EA EIQ+ T+ R
Sbjct: 188 IPCAASFLGLFFIPESPRWLAKVGRDTEFEAALRKLRGKKADISEEAAEIQDYIETLERL 247
Query: 250 QEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGA 300
+ K+L F GL+V QQ GING+ FY S IF AG +
Sbjct: 248 PKAKMLDLFQRRYIRSVLIAFGLMVFQQFGGINGICFYTSSIFEQAGFPTR-LGMIIYAV 306
Query: 301 IQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILS 360
+QVV+T + +VD++GR+ LL+VS++ + I L+ A +FYL+ H + +L+
Sbjct: 307 LQVVITALNAPIVDRAGRKPLLLVSATGLVIGCLIAAVSFYLK----VHDMAHEAVPVLA 362
Query: 361 VVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNW 420
VVG++ I F+ G+G +PW++MSEI P NIKG+AG +AT NWF A ++ T NFL++W
Sbjct: 363 VVGIMVYIGSFSAGMGAMPWVVMSEIFPINIKGVAGGMATLVNWFGAWAVSYTFNFLMSW 422
Query: 421 SSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQA 457
SS GTF IYA +A + F + VPETK +TLE+IQA
Sbjct: 423 SSYGTFLIYAAINALAIVFVIAIVPETKGKTLEQIQA 459
>gb|AAN15372.1| putative putative sister-chromatide cohesion protein [Arabidopsis
thaliana] gi|21539441|gb|AAM53273.1| putative putative
sister-chromatide cohesion protein [Arabidopsis
thaliana] gi|20197560|gb|AAD13706.2| putative sugar
transporter [Arabidopsis thaliana]
gi|30690868|ref|NP_850483.1| sugar transporter, putative
[Arabidopsis thaliana] gi|18407508|ref|NP_566120.1|
sugar transporter, putative [Arabidopsis thaliana]
Length = 463
Score = 379 bits (973), Expect = e-103
Identities = 201/457 (43%), Positives = 286/457 (61%), Gaps = 17/457 (3%)
Query: 13 IRKPFINNNNAGSGS---FFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRF 69
+R+P ++ N AGS + V L V G FG GYSSP Q + DL+L+I+ F
Sbjct: 8 VREPLVDKNMAGSKPDQPWMVYLSTFVAVCGSFAFGSCAGYSSPAQAAIRNDLSLTIAEF 67
Query: 70 SLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLL 129
SLFGSL GAM+GA SG +A+ GRKG++ V++ + GW+AI AK L +GRL
Sbjct: 68 SLFGSLLTFGAMIGAITSGPIADLVGRKGAMRVSSAFCVVGWLAIIFAKGVVALDLGRLA 127
Query: 130 EGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGV 189
G+G+G SYVVP++IAEI P+T RG+L +NQ+ + G+ +++++G WR LAL+G+
Sbjct: 128 TGYGMGAFSYVVPIFIAEIAPKTFRGALTTLNQILICTGVSVSFIIGTLVTWRVLALIGI 187
Query: 190 FPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFYTVCRP 249
PCA GL+FIPESPRWLA++G FE++L+ LRG DI+ EA EIQ+ T+ R
Sbjct: 188 IPCAASFLGLFFIPESPRWLAKVGRDTEFEAALRKLRGKKADISEEAAEIQDYIETLERL 247
Query: 250 QEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGA 300
+ K+L F GL+V QQ GING+ FY S IF AG +
Sbjct: 248 PKAKMLDLFQRRYIRSVLIAFGLMVFQQFGGINGICFYTSSIFEQAGFPTR-LGMIIYAV 306
Query: 301 IQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILS 360
+QVV+T + +VD++GR+ LL+VS++ + I L+ A +FYL+ H + +L+
Sbjct: 307 LQVVITALNAPIVDRAGRKPLLLVSATGLVIGCLIAAVSFYLK----VHDMAHEAVPVLA 362
Query: 361 VVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNW 420
VVG++ I F+ G+G +PW++MSEI P NIKG+AG +AT NWF A ++ T NFL++W
Sbjct: 363 VVGIMVYIGSFSAGMGAMPWVVMSEIFPINIKGVAGGMATLVNWFGAWAVSYTFNFLMSW 422
Query: 421 SSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQA 457
SS GTF IYA +A + F + VPETK +TLE+IQA
Sbjct: 423 SSYGTFLIYAAINALAIVFVIAIVPETKGKTLEQIQA 459
>gb|AAN18269.1| At5g18840/F17K4_90 [Arabidopsis thaliana]
gi|17381265|gb|AAL36051.1| AT5g18840/F17K4_90
[Arabidopsis thaliana] gi|18419741|ref|NP_568367.1|
sugar transporter, putative [Arabidopsis thaliana]
Length = 482
Score = 378 bits (971), Expect = e-103
Identities = 203/473 (42%), Positives = 299/473 (62%), Gaps = 19/473 (4%)
Query: 4 GEDYEDSRNIRKPFINNNN-----AGSGSFFVVLCVLVVAL-GPIQFGFTCGYSSPTQEE 57
GE ++ KPF+ + + + S+ +VL VA+ G +FG GYS+PTQ
Sbjct: 12 GEIVNKVEDLGKPFLTHEDDEKESENNESYLMVLFSTFVAVCGSFEFGSCVGYSAPTQSS 71
Query: 58 MIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLA 117
+ +DLNLS++ FS+FGS+ +GAM+GA +SG ++++ GRKG++ +A I GW+A+
Sbjct: 72 IRQDLNLSLAEFSMFGSILTIGAMLGAVMSGKISDFSGRKGAMRTSACFCITGWLAVFFT 131
Query: 118 KDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGL 177
K + LL +GR G+G+G+ SYVVPVYIAEI P+ +RG L +NQL + IG +++L+G
Sbjct: 132 KGALLLDVGRFFTGYGIGVFSYVVPVYIAEISPKNLRGGLTTLNQLMIVIGSSVSFLIGS 191
Query: 178 FFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQ 237
+W+TLAL G+ PC +L+ GL FIPESPRWLA+ G + F +LQ LRG + DIT EA
Sbjct: 192 LISWKTLALTGLAPCIVLLFGLCFIPESPRWLAKAGHEKEFRVALQKLRGKDADITNEAD 251
Query: 238 EIQNRFYTVCRPQEEKI--LVS-------FDGIGLLVLQQLSGINGVFFYASKIFASAGI 288
IQ + + +I LVS G+ L+V QQ GING+ FYAS+ F AG
Sbjct: 252 GIQVSIQALEILPKARIQDLVSKKYGRSVIIGVSLMVFQQFVGINGIGFYASETFVKAGF 311
Query: 289 SSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSK 348
+S T + +QV +T + T L+DKSGRR L+++S+ + + +L ++F L+G
Sbjct: 312 TSGKLGTIAIACVQVPITVLGTILIDKSGRRPLIMISAGGIFLGCILTGTSFLLKG---- 367
Query: 349 GSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTAS 408
S L + L+V G++ + F++G+GP+PW+IMSEI P N+KG+AGS+ NW A
Sbjct: 368 QSLLLEWVPSLAVGGVLIYVAAFSIGMGPVPWVIMSEIFPINVKGIAGSLVVLVNWSGAW 427
Query: 409 VITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASFIR 461
++ T NFL++WSS GTF +Y+ F+A T+ F VPETK +TLEEIQA R
Sbjct: 428 AVSYTFNFLMSWSSPGTFYLYSAFAAATIIFVAKMVPETKGKTLEEIQACIRR 480
>emb|CAC00697.2| putative sugar transporter [Lycopersicon esculentum]
Length = 480
Score = 377 bits (968), Expect = e-103
Identities = 199/439 (45%), Positives = 283/439 (64%), Gaps = 16/439 (3%)
Query: 30 VVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGH 89
V L LV G G GYSSPTQ + DLNLSI++ SLFGS+ GAM+GA SG
Sbjct: 44 VYLSTLVAVRGSYSLGSCAGYSSPTQSAIREDLNLSIAQISLFGSIWTFGAMIGAITSGP 103
Query: 90 LAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIP 149
+A+Y GRKG++ +++ + GW+AI A+ + L IGRL G+G+G+ SYVVPV+IAEI
Sbjct: 104 IADYIGRKGAMRMSSGFCVAGWLAIFFAQGALALDIGRLATGYGMGVFSYVVPVFIAEIA 163
Query: 150 PRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWL 209
P+ +RG+L +NQL + G+ +++++G WRTLAL G+ PCAIL+ GL+ IPESPRWL
Sbjct: 164 PKDLRGALTTINQLMICCGVSVSFIIGTMMTWRTLALTGLIPCAILLFGLFIIPESPRWL 223
Query: 210 AEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFYTVCRPQEEKILVSFD---------G 260
A++G + FE +L+ LRG + DI+ EA EI++ T+ + + + F G
Sbjct: 224 AKIGHQKEFELALRKLRGKDADISEEAAEIKDYIETLEKLPKVNLFDLFQRRYSSSLIVG 283
Query: 261 IGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRI 320
+GL+V QQ GING+ FY IF S+G SSD T IQV +T + L+D++GR+
Sbjct: 284 VGLMVFQQFGGINGICFYTGSIFESSGF-SSDIGTIIYAIIQVPITALGAALIDRTGRKP 342
Query: 321 LLIVSSSIMTISLLLVASAFYLEGVVSKGSELH-NILGILSVVGLVALIIGFALGIGPIP 379
LL+VS + + I +L +FY+ KG E+ IL+V G++ I F++G+G +P
Sbjct: 343 LLLVSGTGLVIGCILTGISFYM-----KGHEMAIKAAPILAVTGILVYIGSFSVGMGAVP 397
Query: 380 WLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAF 439
W++MSEI P NIKG AGS+AT NWF A + T NFL+ W+S GTF +YA +A ++ F
Sbjct: 398 WVVMSEIYPINIKGAAGSLATLVNWFGAWACSYTFNFLMTWNSFGTFVLYAAVNALSILF 457
Query: 440 ALLFVPETKDRTLEEIQAS 458
+ VPETK RTLE+IQA+
Sbjct: 458 VIKIVPETKGRTLEQIQAA 476
>gb|AAM64736.1| sugar transporter-like protein [Arabidopsis thaliana]
Length = 482
Score = 375 bits (962), Expect = e-102
Identities = 202/473 (42%), Positives = 298/473 (62%), Gaps = 19/473 (4%)
Query: 4 GEDYEDSRNIRKPFINNNN-----AGSGSFFVVLCVLVVAL-GPIQFGFTCGYSSPTQEE 57
GE ++ KPF+ + + + S+ +VL VA+ G +FG GYS+PTQ
Sbjct: 12 GEIVNKVEDLGKPFLTHEDDEKESENNESYLMVLFSTFVAVCGSFEFGSCVGYSAPTQSS 71
Query: 58 MIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLA 117
+ +DLNLS++ FS+FGS+ +GAM+GA +SG ++++ GRKG++ +A I GW+A+
Sbjct: 72 IRQDLNLSLAEFSMFGSILTIGAMLGAVMSGKISDFSGRKGAMRTSACFCITGWLAVFFT 131
Query: 118 KDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGL 177
K + LL +GR G+G+G+ SYVVPVYIAEI P+ +RG L +NQL + IG +++L+G
Sbjct: 132 KGALLLDVGRFFTGYGIGVFSYVVPVYIAEISPKNLRGGLTTLNQLMIVIGSSVSFLIGS 191
Query: 178 FFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQ 237
+W+TLAL + PC +L+ GL FIPESPRWLA+ G + F +LQ LRG + DIT EA
Sbjct: 192 LISWKTLALTVLAPCIVLLFGLCFIPESPRWLAKAGHEKEFRVALQKLRGKDADITNEAD 251
Query: 238 EIQNRFYTVCRPQEEKI--LVS-------FDGIGLLVLQQLSGINGVFFYASKIFASAGI 288
IQ + + +I LVS G+ L+V QQ GING+ FYAS+ F AG
Sbjct: 252 GIQVSIQALEILPKARIQDLVSKKYGRSVIIGVSLMVFQQFVGINGIGFYASETFVKAGF 311
Query: 289 SSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSK 348
+S T + +QV +T + T L+DKSGRR L+++S+ + + +L ++F L+G
Sbjct: 312 TSGKLGTIAIACVQVPITVLGTILIDKSGRRPLIMISAGGIFLGCILTGTSFLLKG---- 367
Query: 349 GSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTAS 408
S L + L+V G++ + F++G+GP+PW+IMSEI P N+KG+AGS+ NW A
Sbjct: 368 QSLLLEWVPSLAVGGVLIYVAAFSIGMGPVPWVIMSEIFPINVKGIAGSLVVLVNWSGAW 427
Query: 409 VITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASFIR 461
++ T NFL++WSS GTF +Y+ F+A T+ F VPETK +TLEEIQA R
Sbjct: 428 AVSYTFNFLMSWSSPGTFYLYSAFAAATIIFVAKMVPETKGKTLEEIQACIRR 480
>ref|NP_850964.1| sugar transporter, putative [Arabidopsis thaliana]
Length = 470
Score = 370 bits (949), Expect = e-101
Identities = 197/451 (43%), Positives = 285/451 (62%), Gaps = 18/451 (3%)
Query: 19 NNNNAGSGSFFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNV 78
N +++ + + ++L V G FG GYSSP Q ++ ++LNLS++ +SLFGS+ +
Sbjct: 21 NQDSSATITTTLLLTTFVAVSGSFVFGSAIGYSSPVQSDLTKELNLSVAEYSLFGSILTI 80
Query: 79 GAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIIS 138
GAM+GA +SG +A+ GR+ ++ + + I GW+AI L+K + L +GR L G+G+G+ S
Sbjct: 81 GAMIGAAMSGRIADMIGRRATMGFSEMFCILGWLAIYLSKVAIWLDVGRFLVGYGMGVFS 140
Query: 139 YVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPG 198
+VVPVYIAEI P+ +RG V+QL + +G+ + YLLG F WR LAL+G+ PC + + G
Sbjct: 141 FVVPVYIAEITPKGLRGGFTTVHQLLICLGVSVTYLLGSFIGWRILALIGMIPCVVQMMG 200
Query: 199 LYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEI-----------QNRFYTVC 247
L+ IPESPRWLA++G E FE +LQ LRG + DI+ E+ EI + +
Sbjct: 201 LFVIPESPRWLAKVGKWEEFEIALQRLRGESADISYESNEIKDYTRRLTDLSEGSIVDLF 260
Query: 248 RPQEEKILVSFDGIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTG 307
+PQ K LV G+GL+VLQQ G+NG+ FYAS IF SAG+SS + +Q+ MT
Sbjct: 261 QPQYAKSLVV--GVGLMVLQQFGGVNGIAFYASSIFESAGVSSK-IGMIAMVVVQIPMTT 317
Query: 308 VATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVAL 367
+ L+DKSGRR LL++S++ I LV +F L+ V +L L++ G++
Sbjct: 318 LGVLLMDKSGRRPLLLISATGTCIGCFLVGLSFSLQFV----KQLSGDASYLALTGVLVY 373
Query: 368 IIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFT 427
F+LG+G IPW+IMSEI P +IKG AGS+ T +W + +I+ T NFL+NW+ AGTF
Sbjct: 374 TGSFSLGMGGIPWVIMSEIFPIDIKGSAGSLVTVVSWVGSWIISFTFNFLMNWNPAGTFY 433
Query: 428 IYAIFSAFTVAFALLFVPETKDRTLEEIQAS 458
++A TV F VPETK RTLEEIQ S
Sbjct: 434 VFATVCGATVIFVAKLVPETKGRTLEEIQYS 464
>ref|NP_187166.2| sugar transporter family protein [Arabidopsis thaliana]
Length = 470
Score = 358 bits (920), Expect = 1e-97
Identities = 203/472 (43%), Positives = 288/472 (61%), Gaps = 29/472 (6%)
Query: 5 EDYEDSRNIRKPFINNNNAGS----GSFFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIR 60
+D E + +P + N S+ V L ++ G +FG GYS+PTQ ++
Sbjct: 6 DDMEKRNDKSEPLLLPENGSDVSEEASWMVYLSTIIAVCGSYEFGTCVGYSAPTQFGIME 65
Query: 61 DLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDS 120
+LNLS S+FS+FGS+ N+GA++GA SG ++++ GRKG++ +++V + GW+ I LAK
Sbjct: 66 ELNLSYSQFSVFGSILNMGAVLGAITSGKISDFIGRKGAMRLSSVISAIGWLIIYLAKGD 125
Query: 121 SLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFN 180
L GR L G+G G +S+VVPV+IAEI PR +RG+L +NQL + IG+ +L+G N
Sbjct: 126 VPLDFGRFLTGYGCGTLSFVVPVFIAEISPRKLRGALATLNQLFIVIGLASMFLIGAVVN 185
Query: 181 WRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQ 240
WRTLAL GV PC +L G +FIPESPRWL +G FE +LQ LRG +IT EA EIQ
Sbjct: 186 WRTLALTGVAPCVVLFFGTWFIPESPRWLEMVGRHSDFEIALQKLRGPQANITREAGEIQ 245
Query: 241 NRFYTVCRPQEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSS 291
++ + ++ D G+GL+ QQ GINGV FYA +IF SAG S +
Sbjct: 246 EYLASLAHLPKATLMDLIDKKNIRFVIVGVGLMFFQQFVGINGVIFYAQQIFVSAGASPT 305
Query: 292 DAATFGLGAI-----QVVMTGV-ATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGV 345
LG+I QVV+T + AT L+D+ GRR LL+ S+ M I LL+ ++F L+
Sbjct: 306 ------LGSILYSIEQVVLTALGATLLIDRLGRRPLLMASAVGMLIGCLLIGNSFLLK-- 357
Query: 346 VSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWF 405
+ G L +I+ L+V G++ I F++G+G IPW+IMSEI P N+KG AG + T NW
Sbjct: 358 -AHGLAL-DIIPALAVSGVLVYIGSFSIGMGAIPWVIMSEIFPINLKGTAGGLVTVVNWL 415
Query: 406 TASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQA 457
++ +++ T NFL+ WS GTF +Y + F VPETK RTLEEIQA
Sbjct: 416 SSWLVSFTFNFLMIWSPHGTFYVYGGVCVLAIIFIAKLVPETKGRTLEEIQA 467
>gb|AAF27021.1| putative sugar transporter [Arabidopsis thaliana]
Length = 463
Score = 357 bits (917), Expect = 3e-97
Identities = 199/446 (44%), Positives = 280/446 (62%), Gaps = 25/446 (5%)
Query: 27 SFFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATV 86
S+ V L ++ G +FG GYS+PTQ ++ +LNLS S+FS+FGS+ N+GA++GA
Sbjct: 25 SWMVYLSTIIAVCGSYEFGTCVGYSAPTQFGIMEELNLSYSQFSVFGSILNMGAVLGAIT 84
Query: 87 SGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIA 146
SG ++++ GRKG++ +++V + GW+ I LAK L GR L G+G G +S+VVPV+IA
Sbjct: 85 SGKISDFIGRKGAMRLSSVISAIGWLIIYLAKGDVPLDFGRFLTGYGCGTLSFVVPVFIA 144
Query: 147 EIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESP 206
EI PR +RG+L +NQL + IG+ +L+G NWRTLAL GV PC +L G +FIPESP
Sbjct: 145 EISPRKLRGALATLNQLFIVIGLASMFLIGAVVNWRTLALTGVAPCVVLFFGTWFIPESP 204
Query: 207 RWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFYTVCRPQEEKILVSFD------- 259
RWL +G FE +LQ LRG +IT EA EIQ ++ + ++ D
Sbjct: 205 RWLEMVGRHSDFEIALQKLRGPQANITREAGEIQEYLASLAHLPKATLMDLIDKKNIRFV 264
Query: 260 --GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAI-----QVVMTGV-ATW 311
G+GL+ QQ GINGV FYA +IF SAG S + LG+I QVV+T + AT
Sbjct: 265 IVGVGLMFFQQFVGINGVIFYAQQIFVSAGASPT------LGSILYSIEQVVLTALGATL 318
Query: 312 LVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGF 371
L+D+ GRR LL+ S+ M I LL+ ++F L+ + G L +I+ L+V G++ I F
Sbjct: 319 LIDRLGRRPLLMASAVGMLIGCLLIGNSFLLK---AHGLAL-DIIPALAVSGVLVYIGSF 374
Query: 372 ALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAI 431
++G+G IPW+IMSEI P N+KG AG + T NW ++ +++ T NFL+ WS GTF +Y
Sbjct: 375 SIGMGAIPWVIMSEIFPINLKGTAGGLVTVVNWLSSWLVSFTFNFLMIWSPHGTFYVYGG 434
Query: 432 FSAFTVAFALLFVPETKDRTLEEIQA 457
+ F VPETK RTLEEIQA
Sbjct: 435 VCVLAIIFIAKLVPETKGRTLEEIQA 460
>gb|AAM91377.1| At1g08920/F7G19_20 [Arabidopsis thaliana]
gi|18390957|ref|NP_563829.1| sugar transporter, putative
[Arabidopsis thaliana] gi|14194109|gb|AAK56249.1|
At1g08920/F7G19_20 [Arabidopsis thaliana]
Length = 470
Score = 333 bits (853), Expect = 9e-90
Identities = 185/465 (39%), Positives = 270/465 (57%), Gaps = 19/465 (4%)
Query: 8 EDSRNIRKPFI-----NNNNAGSGSFFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDL 62
E+SRN+ + N+ N + V+ V G FG GYSS Q +I DL
Sbjct: 5 ENSRNLEAGLLLRKNQNDINECRITAVVLFSTFVSVCGSFCFGCAAGYSSVAQTGIINDL 64
Query: 63 NLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSL 122
LS++++S+FGS+ G M+GA SG +A+ GRKG++ A + IFGWVA++LAKDS
Sbjct: 65 GLSVAQYSMFGSIMTFGGMIGAIFSGKVADLMGRKGTMWFAQIFCIFGWVAVALAKDSMW 124
Query: 123 LFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWR 182
L IGRL GF VG++SYV+PVYIAEI P+ +RG+ NQL + G+ L Y++G F +WR
Sbjct: 125 LDIGRLSTGFAVGLLSYVIPVYIAEITPKHVRGAFVFANQLMQSCGLSLFYVIGNFVHWR 184
Query: 183 TLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNR 242
LAL+G+ PCA+ + L+FIPESPR L + G + +SLQ+LRG + DI+ EA I+
Sbjct: 185 NLALIGLIPCALQVVTLFFIPESPRLLGKWGHEKECRASLQSLRGDDADISEEANTIKET 244
Query: 243 FYTVCRPQEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDA 293
+ +++ F G+GL++LQQLSG +G+ +Y +F G SS
Sbjct: 245 MILFDEGPKSRVMDLFQRRYAPSVVIGVGLMLLQQLSGSSGLMYYVGSVFDKGGFPSSIG 304
Query: 294 ATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELH 353
+ L I + + LV+K GRR LL+ S+ M LL++ +F L
Sbjct: 305 SMI-LAVIMIPKALLGLILVEKMGRRPLLLASTGGMCFFSLLLSFSFCFRSY----GMLD 359
Query: 354 NILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLT 413
+ I + +G+V I FA+G+G +PW+IMSEI P N+K AG++ T +NW ++
Sbjct: 360 ELTPIFTCIGVVGFISSFAVGMGGLPWIIMSEIFPMNVKVSAGTLVTLANWSFGWIVAFA 419
Query: 414 ANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQAS 458
NF+L W+++GTF I+ + F VPETK RTLE+IQAS
Sbjct: 420 YNFMLEWNASGTFLIFFTICGAGIVFIYAMVPETKGRTLEDIQAS 464
>ref|NP_568494.1| sugar-porter family protein 2 (SFP2) [Arabidopsis thaliana]
gi|14585701|gb|AAK11721.1| sugar-porter family protein 2
[Arabidopsis thaliana]
Length = 478
Score = 332 bits (852), Expect = 1e-89
Identities = 190/442 (42%), Positives = 262/442 (58%), Gaps = 16/442 (3%)
Query: 30 VVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGH 89
V+L + G FG + GY+S + +++DL+LSI++FS F SLS +GA +GA SG
Sbjct: 35 VILSTFIAVCGSFSFGVSLGYTSGAEIGIMKDLDLSIAQFSAFASLSTLGAAIGALFSGK 94
Query: 90 LAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIP 149
+A GR+ ++ V+ + I GW +I+ AKD L GR+ G G+G+ISYVVPVYIAEI
Sbjct: 95 MAIILGRRKTMWVSDLLCIIGWFSIAFAKDVMWLNFGRISSGIGLGLISYVVPVYIAEIS 154
Query: 150 PRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWL 209
P+ +RG+ NQL G+ + Y G F NWR LALLG PC I + GL+F+PESPRWL
Sbjct: 155 PKHVRGTFTFTNQLLQNSGLAMVYFSGNFLNWRILALLGALPCFIQVIGLFFVPESPRWL 214
Query: 210 AEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFYTVCRPQEEKILVSFD---------G 260
A++G + E+SL LRG N DI+ EA +I+ V + F G
Sbjct: 215 AKVGSDKELENSLLRLRGGNADISREASDIEVMTKMVENDSKSSFCDLFQRKYRYTLVVG 274
Query: 261 IGLLVLQQLSGINGVFFYASKIFASAGISSSDAAT-FGLGAIQVVMTGVATWLVDKSGRR 319
IGL+++QQ SG + V YAS I AG S + +T GL I M GV LVDK GRR
Sbjct: 275 IGLMLIQQFSGSSAVLSYASTILRKAGFSVTIGSTLLGLFMIPKAMIGVI--LVDKWGRR 332
Query: 320 ILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIP 379
LL+ S S M I+ +L+ AF L+ K L + + + + + I +A+G+G +P
Sbjct: 333 PLLLTSVSGMCITSMLIGVAFTLQ----KMQLLPELTPVFTFICVTLYIGTYAIGLGGLP 388
Query: 380 WLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAF 439
W+IMSEI P NIK AGS+ T +W ++S++T NFLL WS+ GTF ++ + F
Sbjct: 389 WVIMSEIFPMNIKVTAGSIVTLVSWSSSSIVTYAFNFLLEWSTQGTFYVFGAVGGLALLF 448
Query: 440 ALLFVPETKDRTLEEIQASFIR 461
L VPETK +LEEIQAS IR
Sbjct: 449 IWLLVPETKGLSLEEIQASLIR 470
>ref|NP_172364.3| sugar transporter family protein [Arabidopsis thaliana]
Length = 464
Score = 332 bits (850), Expect = 2e-89
Identities = 187/463 (40%), Positives = 265/463 (56%), Gaps = 18/463 (3%)
Query: 9 DSRNIRKPFINNNNAGSGSFFV----VLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNL 64
+S +++ P +NN S + +L V G +G YSSP Q +++ +L L
Sbjct: 2 ESGSMKTPLVNNQEEARSSSSITCGLLLSTSVAVTGSFVYGCAMSYSSPAQSKIMEELGL 61
Query: 65 SISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLF 124
S++ +S F S+ +G M+ A SG +A GR+ ++ +A V IFGW+A++ A D LL
Sbjct: 62 SVADYSFFTSVMTLGGMITAAFSGKIAAVIGRRQTMWIADVFCIFGWLAVAFAHDKMLLN 121
Query: 125 IGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTL 184
IGR GFGVG+ISYVVPVYIAEI P+ RG NQL + GI L + G FF+WRTL
Sbjct: 122 IGRGFLGFGVGLISYVVPVYIAEITPKAFRGGFSFSNQLLQSFGISLMFFTGNFFHWRTL 181
Query: 185 ALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFY 244
ALL PC I + L+FIPESPRWLA G E +L+ LRG N DI EA EI+
Sbjct: 182 ALLSAIPCGIQMICLFFIPESPRWLAMYGRERELEVTLKRLRGENGDILEEAAEIRETVE 241
Query: 245 TVCRPQEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAAT 295
T R + F+ G+GL++LQQ G + + YA++IF +AG SD T
Sbjct: 242 TSRRESRSGLKDLFNMKNAHPLIIGLGLMLLQQFCGSSAISAYAARIFDTAGF-PSDIGT 300
Query: 296 FGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNI 355
L I V + + + VD+ GRR LL+ SS + I L+ ++YL+ +
Sbjct: 301 SILAVILVPQSIIVMFAVDRCGRRPLLMSSSIGLCICSFLIGLSYYLQ----NHGDFQEF 356
Query: 356 LGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTAN 415
+ +VGLV ++ F +G+G +PW+IMSE+ P N+K AGS+ T SNWF + +I + N
Sbjct: 357 CSPILIVGLVGYVLSFGIGLGGLPWVIMSEVFPVNVKITAGSLVTVSNWFFSWIIIFSFN 416
Query: 416 FLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQAS 458
F++ WS+ GT+ I+A S + F VPETK RTLE+IQ S
Sbjct: 417 FMMQWSAFGTYFIFAGVSLMSFVFVWTLVPETKGRTLEDIQQS 459
>gb|AAP40473.1| putative zinc finger protein ATZF1 [Arabidopsis thaliana]
gi|3123712|dbj|BAA25989.1| ERD6 protein [Arabidopsis
thaliana] gi|6686825|emb|CAB64732.1| putative sugar
transporter [Arabidopsis thaliana]
gi|18390959|ref|NP_563830.1| early-responsive to
dehydration stress protein (ERD6) / sugar transporter
family protein [Arabidopsis thaliana]
gi|25308964|pir||T52132 probable sugar transporter
protein ERD6 [imported] - Arabidopsis thaliana
Length = 496
Score = 330 bits (847), Expect = 4e-89
Identities = 190/463 (41%), Positives = 277/463 (59%), Gaps = 15/463 (3%)
Query: 4 GEDYEDSRNIRKPFINNNNAGSGSFFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLN 63
G + R ++KP N++ + V L V G G G+SS Q + +DL+
Sbjct: 33 GLSRKSPREVKKPQ-NDDGECRVTASVFLSTFVAVSGSFCTGCGVGFSSGAQAGITKDLS 91
Query: 64 LSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLL 123
LS++ +S+FGS+ +G ++GA SG +A+ GRK ++L I GW+ ++LA+++ L
Sbjct: 92 LSVAEYSMFGSILTLGGLIGAVFSGKVADVLGRKRTMLFCEFFCITGWLCVALAQNAMWL 151
Query: 124 FIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRT 183
GRLL G GVGI SYV+PVYIAEI P+ +RGS NQL GI L +++G F WR
Sbjct: 152 DCGRLLLGIGVGIFSYVIPVYIAEIAPKHVRGSFVFANQLMQNCGISLFFIIGNFIPWRL 211
Query: 184 LALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRF 243
L ++G+ PC + L+FIPESPRWLA++G + SSLQ LRGS+VDI+ EA I++
Sbjct: 212 LTVVGLVPCVFHVFCLFFIPESPRWLAKLGRDKECRSSLQRLRGSDVDISREANTIRDTI 271
Query: 244 YTVCRPQEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAA 294
E K+ F G+GL+ LQQL G +GV +YAS +F G S+
Sbjct: 272 DMTENGGETKMSELFQRRYAYPLIIGVGLMFLQQLCGSSGVTYYASSLFNKGGFPSA-IG 330
Query: 295 TFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHN 354
T + I V +AT LVDK GRR LL+ S S M +S LL++ ++ G S G L
Sbjct: 331 TSVIATIMVPKAMLATVLVDKMGRRTLLMASCSAMGLSALLLSVSY---GFQSFGI-LPE 386
Query: 355 ILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTA 414
+ I + +G++ I+ FA+G+G +PW+IM+EI P N+K AG++ T +NW +IT T
Sbjct: 387 LTPIFTCIGVLGHIVSFAMGMGGLPWIIMAEIFPMNVKVSAGTLVTVTNWLFGWIITYTF 446
Query: 415 NFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQA 457
NF+L W+++G F I+++ SA ++ F VPETK R+LEEIQA
Sbjct: 447 NFMLEWNASGMFLIFSMVSASSIVFIYFLVPETKGRSLEEIQA 489
>gb|AAK11720.1| sugar-porter family protein 1 [Arabidopsis thaliana]
Length = 474
Score = 330 bits (846), Expect = 6e-89
Identities = 193/467 (41%), Positives = 270/467 (57%), Gaps = 19/467 (4%)
Query: 8 EDSRNIRKPFINNNNAGSGS-----FFVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDL 62
E+ R+I + + N S V+L V G FG GY+S + +++DL
Sbjct: 4 EEGRSIEEGLLQLKNKNDDSECRITACVILSTFVAVCGSFSFGVATGYTSGAETGVMKDL 63
Query: 63 NLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSL 122
+LSI++FS FGS + +GA +GA G+LA GR+G++ V+ I GW++I+ AK+ L
Sbjct: 64 DLSIAQFSAFGSFATLGAAIGALFCGNLAMVIGRRGTMWVSDFLCITGWLSIAFAKEVML 123
Query: 123 LFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWR 182
L GR++ G G G+ SYVVPVYIAEI P+ +RG+ NQL G+ + Y G F NWR
Sbjct: 124 LNFGRIISGIGFGLTSYVVPVYIAEITPKHVRGTFTFSNQLLQNAGLAMIYFCGNFINWR 183
Query: 183 TLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNR 242
TLALLG PC I + GL+F+PESPRWLA++G + E+SL LRG + DI+ EA EIQ
Sbjct: 184 TLALLGALPCFIQVIGLFFVPESPRWLAKVGSDKELENSLFRLRGRDADISREASEIQVM 243
Query: 243 FYTVCRPQEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDA 293
V + F GIGL+++QQ SG V YAS IF AG S +
Sbjct: 244 TKMVENDSKSSFSDLFQRKYRYTLVVGIGLMLIQQFSGSAAVISYASTIFRKAGFSVAIG 303
Query: 294 ATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELH 353
T LG + + LVDK GRR LL+ S+ M+++ +L+ AF L+ K L
Sbjct: 304 TTM-LGIFVIPKAMIGLILVDKWGRRPLLMTSAFGMSMTCMLLGVAFTLQ----KMQLLS 358
Query: 354 NILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLT 413
+ ILS + ++ I +A+G+G +PW+IMSEI P NIK AGS+ T ++ ++S++T
Sbjct: 359 ELTPILSFICVMMYIATYAIGLGGLPWVIMSEIFPINIKVTAGSIVTLVSFSSSSIVTYA 418
Query: 414 ANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASFI 460
NFL WS+ GTF I+A + F L VPETK +LEEIQ S I
Sbjct: 419 FNFLFEWSTQGTFFIFAGIGGAALLFIWLLVPETKGLSLEEIQVSLI 465
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.141 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 726,677,064
Number of Sequences: 2540612
Number of extensions: 29413969
Number of successful extensions: 133550
Number of sequences better than 10.0: 4942
Number of HSP's better than 10.0 without gapping: 2433
Number of HSP's successfully gapped in prelim test: 2512
Number of HSP's that attempted gapping in prelim test: 120947
Number of HSP's gapped (non-prelim): 7540
length of query: 461
length of database: 863,360,394
effective HSP length: 131
effective length of query: 330
effective length of database: 530,540,222
effective search space: 175078273260
effective search space used: 175078273260
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0283.14


