

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281b.2
(124 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
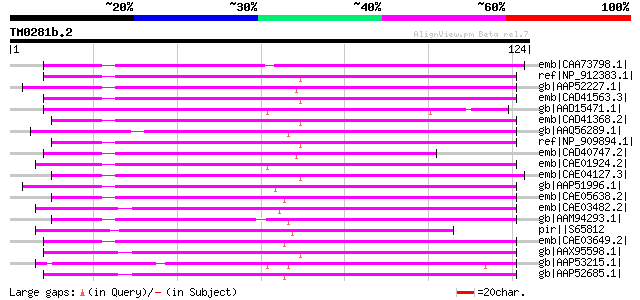
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulg... 92 4e-18
ref|NP_912383.1| putative reverse transcriptase [Oryza sativa (j... 61 5e-09
gb|AAP52227.1| putative reverse transcriptase [Oryza sativa (jap... 59 3e-08
emb|CAD41563.3| OSJNBa0006A01.18 [Oryza sativa (japonica cultiva... 57 1e-07
gb|AAD15471.1| putative non-LTR retroelement reverse transcripta... 56 2e-07
emb|CAD41368.2| OSJNBa0088A01.7 [Oryza sativa (japonica cultivar... 56 2e-07
gb|AAQ56289.1| putative reverse transcriptase [Oryza sativa (jap... 55 3e-07
ref|NP_909894.1| putative reverse transcriptase [Oryza sativa (j... 55 5e-07
emb|CAD40747.2| OSJNBa0072D21.1 [Oryza sativa (japonica cultivar... 54 9e-07
emb|CAE01924.2| OSJNBb0078D11.8 [Oryza sativa (japonica cultivar... 53 1e-06
emb|CAE04127.3| OSJNBa0009P12.14 [Oryza sativa (japonica cultiva... 53 2e-06
gb|AAP51996.1| putative non-LTR retroelement reverse transcripta... 52 3e-06
emb|CAE05638.2| OSJNBa0038O10.4 [Oryza sativa (japonica cultivar... 52 3e-06
emb|CAE03482.2| OSJNBa0065O17.7 [Oryza sativa (japonica cultivar... 52 3e-06
gb|AAM94293.1| putative reverse transcriptase [Sorghum bicolor] 52 3e-06
pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW1... 52 3e-06
emb|CAE03649.2| OSJNBa0060N03.14 [Oryza sativa (japonica cultiva... 51 6e-06
gb|AAX95598.1| hypothetical protein [Oryza sativa (japonica cult... 51 6e-06
gb|AAP53215.1| putative retroelement [Oryza sativa (japonica cul... 51 7e-06
gb|AAP52685.1| putative reverse transcriptase [Oryza sativa (jap... 51 7e-06
>emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulgaris]
gi|7484649|pir||T14619 reverse transcriptase - beet
retrotransposon (fragment)
Length = 507
Score = 91.7 bits (226), Expect = 4e-18
Identities = 44/115 (38%), Positives = 65/115 (56%), Gaps = 5/115 (4%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAAS 68
I GD+N ER + DF+ F++N+ LIE + +TWFR GQ+ S
Sbjct: 8 IMGDYNETLIPAER---GSHLISSQGARDFQNFINNMGLIEISPIKGFFTWFR--GQSKS 62
Query: 69 RLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCWFS 123
+LDR + +W + +P ++ RS SDHCP++ S+ NWGP+PFR LNCW +
Sbjct: 63 KLDRLFVQPEWLTSFPSLKLNILKRSPSDHCPLLAFSSHQNWGPKPFRFLNCWLT 117
>ref|NP_912383.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|29893671|gb|AAP06925.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 870
Score = 61.2 bits (147), Expect = 5e-09
Identities = 34/114 (29%), Positives = 56/114 (48%), Gaps = 4/114 (3%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAAS 68
I GDFN++ R+ ++ ++S + M F+A + L E P +K+TW +A
Sbjct: 142 ILGDFNMIYRARDK---NNSNLNLARMRRFRAVIDRCELHEIPLQNRKFTWSNERRKATL 198
Query: 69 -RLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+LDR + +W +PH V + SDHCP++ + PR F+ N W
Sbjct: 199 VKLDRCFCNEEWDLAFPHHVLHALPTGPSDHCPLVLSNPQGPHRPRTFKFENFW 252
>gb|AAP52227.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37531276|ref|NP_919940.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|22655786|gb|AAN04203.1| Putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|22655726|gb|AAN04143.1| Putative
reverse transcriptase [Oryza sativa]
Length = 773
Score = 58.5 bits (140), Expect = 3e-08
Identities = 30/119 (25%), Positives = 59/119 (49%), Gaps = 4/119 (3%)
Query: 4 ISCFCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPN 63
+ +C+ GDFN++A ++ ++ R ++ F+ + L L E G++YTW
Sbjct: 590 VGAWCVVGDFNLLATDGDK---NNGNVNRRLISKFRHTIDALELREVYLFGRRYTWSNEQ 646
Query: 64 GQAA-SRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+ +++D+ L++ DW + ++ S SDHCP++ M PR F+ + W
Sbjct: 647 ERPTLTKIDKVLVNGDWEEIFRDAHLQALSSSASDHCPLLLACDQMMHRPRKFKFESYW 705
>emb|CAD41563.3| OSJNBa0006A01.18 [Oryza sativa (japonica cultivar-group)]
Length = 1189
Score = 57.0 bits (136), Expect = 1e-07
Identities = 33/114 (28%), Positives = 55/114 (47%), Gaps = 4/114 (3%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAAS 68
I GDFN++ R+ ++ ++S + M F+A + L E P +K+TW +A
Sbjct: 142 ILGDFNMIYRARDK---NNSNLNLARMRRFRAVIDRCELHEIPLQNRKFTWSNERRKATL 198
Query: 69 -RLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+LDR + +W +PH V + SDH P++ + PR F+ N W
Sbjct: 199 VKLDRCFCNEEWDLAFPHHVLHALPTGPSDHYPLVLSNPQGPHRPRTFKFENFW 252
>gb|AAD15471.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411565|pir||B84517 hypothetical protein
At2g14430 [imported] - Arabidopsis thaliana
Length = 1277
Score = 56.2 bits (134), Expect = 2e-07
Identities = 34/115 (29%), Positives = 52/115 (44%), Gaps = 5/115 (4%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWF--RPNGQA 66
+ GDFN + E S + M DF+ + SL + G ++TW R NG
Sbjct: 385 LCGDFNEILDGSEHSNYDTSPFTPLGMRDFQEVVRYSSLTDLGYHGPRFTWCNKRENGLI 444
Query: 67 ASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHC--PVMFRSTDMNWGPRPFRVLN 119
+LD+ L++ WC +PH A SDH +M + + G RPF+ +N
Sbjct: 445 CKKLDKVLVNDSWCITFPHSYSVFEAGGCSDHTRGRIMLEAAAIG-GRRPFKFVN 498
>emb|CAD41368.2| OSJNBa0088A01.7 [Oryza sativa (japonica cultivar-group)]
gi|50928243|ref|XP_473649.1| OSJNBa0088A01.7 [Oryza
sativa (japonica cultivar-group)]
Length = 1324
Score = 55.8 bits (133), Expect = 2e-07
Identities = 32/112 (28%), Positives = 54/112 (47%), Gaps = 4/112 (3%)
Query: 11 GDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAAS-R 69
GDFN++ R+ ++ S+S M F+A +++ L E +K+TW R
Sbjct: 596 GDFNMIYRACDK---SNSNINLRRMRRFRAAINHCELKEIHLQNRKFTWTNERRHPTMVR 652
Query: 70 LDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
LDRF + +W + H + ++ SDHCP++ + PR F+ N W
Sbjct: 653 LDRFFCNENWDLAFGHHILHTLSSGTSDHCPLLLSNPQGPQRPRTFKFENFW 704
>gb|AAQ56289.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 854
Score = 55.5 bits (132), Expect = 3e-07
Identities = 34/117 (29%), Positives = 51/117 (43%), Gaps = 4/117 (3%)
Query: 6 CFCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQ 65
C+ + GDFN + E R I +D+ F +SN L+E P +G+ +TW Q
Sbjct: 677 CWILMGDFNFIRSVENRNIPGGDI---NDILIFNEIISNAGLVEIPLIGRNFTWSNMQEQ 733
Query: 66 -AASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+LD S+ W +PH + T ++ +SDH P FR N W
Sbjct: 734 PLLQQLDWVFTSTAWTLKFPHTMVTATSKYISDHAPCQISIETSVPKSSIFRFENFW 790
>ref|NP_909894.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|12957722|gb|AAK09240.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1135
Score = 54.7 bits (130), Expect = 5e-07
Identities = 31/112 (27%), Positives = 52/112 (45%), Gaps = 4/112 (3%)
Query: 11 GDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAAS-R 69
GDFN++ R+ ++ ++ + M F+A + L E P K++TW + +
Sbjct: 247 GDFNMIYRARDK---NNGNLNLARMRRFRATIDRCELREIPLQNKRFTWSNERQRPTLVK 303
Query: 70 LDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
LD + +W + H V + SDHCP++ + PRPFR N W
Sbjct: 304 LDHCFCNENWDLAFHHHVLHALPTGPSDHCPLVLSNPAAPCKPRPFRFENFW 355
>emb|CAD40747.2| OSJNBa0072D21.1 [Oryza sativa (japonica cultivar-group)]
gi|50923755|ref|XP_472238.1| OSJNBa0072D21.1 [Oryza
sativa (japonica cultivar-group)]
Length = 710
Score = 53.9 bits (128), Expect = 9e-07
Identities = 28/95 (29%), Positives = 49/95 (51%), Gaps = 4/95 (4%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAA- 67
+ GDFN++ E+ ++ Y F A + +L L E G+K+TW + +
Sbjct: 234 LGGDFNIIRNPTEK---NNDNYSEKWPFLFNAVIDSLDLRELELTGRKFTWANSSSKPTF 290
Query: 68 SRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVM 102
+LDR L+S++W +P ++R LSDH P++
Sbjct: 291 EKLDRILMSTEWEQKYPLSTARALSRELSDHTPLL 325
>emb|CAE01924.2| OSJNBb0078D11.8 [Oryza sativa (japonica cultivar-group)]
gi|50927957|ref|XP_473506.1| OSJNBb0078D11.8 [Oryza
sativa (japonica cultivar-group)]
Length = 1636
Score = 53.1 bits (126), Expect = 1e-06
Identities = 30/116 (25%), Positives = 57/116 (48%), Gaps = 4/116 (3%)
Query: 7 FCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWF-RPNGQ 65
+CI GDFN++A ++ ++ R + F+ L+ L L E G+++TW
Sbjct: 974 WCIVGDFNLLAAESDK---NNQNVDRRMINRFRCLLNLLELRELYLFGRRFTWSSEQESP 1030
Query: 66 AASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+++D+ L++++W + ++ S SDHCP++ M PR F+ W
Sbjct: 1031 TLTKIDKVLVNAEWEDFFRDAHLQALSSSASDHCPLLLACDAMVHRPRRFKFEAFW 1086
>emb|CAE04127.3| OSJNBa0009P12.14 [Oryza sativa (japonica cultivar-group)]
Length = 1206
Score = 52.8 bits (125), Expect = 2e-06
Identities = 31/114 (27%), Positives = 55/114 (48%), Gaps = 4/114 (3%)
Query: 11 GDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAAS-R 69
GDFN++ R+ ++ +++ M F+A L + L E +++TW + R
Sbjct: 409 GDFNMIYRACDK---NNNNLNLRRMGHFRAALEHCELKEVHLQNRRFTWSNERRKPTLVR 465
Query: 70 LDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCWFS 123
LDRF + +W + H + ++ SDHCP++ + PR F+ N W S
Sbjct: 466 LDRFYCNENWDLGFGHHILHALSSGTSDHCPLLLTNPQGPRRPRSFKFENFWTS 519
>gb|AAP51996.1| putative non-LTR retroelement reverse transcriptase [Oryza sativa
(japonica cultivar-group)] gi|37530814|ref|NP_919709.1|
putative non-LTR retroelement reverse transcriptase
[Oryza sativa (japonica cultivar-group)]
gi|21326501|gb|AAM47629.1| Putative non-LTR retroelement
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1345
Score = 52.0 bits (123), Expect = 3e-06
Identities = 32/119 (26%), Positives = 55/119 (45%), Gaps = 4/119 (3%)
Query: 4 ISCFCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRP- 62
+S + + GDFN++ ++ ++ ++S RS M FK L L + E P G+K+TW +
Sbjct: 520 MSPWLLIGDFNLIYKTSDK---NNSRLNRSMMQRFKGTLDRLEVKELPLSGRKFTWTQEC 576
Query: 63 NGQAASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+ S++DR DW + ++ S SDH P+ FR + W
Sbjct: 577 DNPTQSKIDRAFCCPDWDVLFDSAQMYALSSSCSDHAPLFITGCVEREQNNSFRFESFW 635
>emb|CAE05638.2| OSJNBa0038O10.4 [Oryza sativa (japonica cultivar-group)]
gi|50926570|ref|XP_473232.1| OSJNBa0038O10.4 [Oryza
sativa (japonica cultivar-group)]
Length = 1045
Score = 52.0 bits (123), Expect = 3e-06
Identities = 30/112 (26%), Positives = 51/112 (44%), Gaps = 4/112 (3%)
Query: 11 GDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNG-QAASR 69
GDFN++ R+ ++ ++ + M F+A + L E P +++TW +
Sbjct: 144 GDFNIIYRARDK---NNGNLNLARMRRFRATIDRCELHEIPLQNRRFTWSNERQWPTLVK 200
Query: 70 LDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
LDR + +W + H V + SDHCP++ + PR FR N W
Sbjct: 201 LDRCFCNENWDLAFHHHVLHALPTGPSDHCPLVLSNPAAPRKPRTFRFENFW 252
>emb|CAE03482.2| OSJNBa0065O17.7 [Oryza sativa (japonica cultivar-group)]
gi|50927885|ref|XP_473470.1| OSJNBa0065O17.7 [Oryza
sativa (japonica cultivar-group)]
Length = 3331
Score = 52.0 bits (123), Expect = 3e-06
Identities = 29/116 (25%), Positives = 53/116 (45%), Gaps = 4/116 (3%)
Query: 7 FCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPN-GQ 65
+ + GDFN++ ++++ ++ R M F+ + L L E P G+++TW
Sbjct: 807 WAVVGDFNLILEAQDKNNLN---LNRRMMGRFRRLIDELELRELPLDGRRFTWSNERESP 863
Query: 66 AASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+LDR L S+DW +P + + SDHC ++ + + FR W
Sbjct: 864 TLVKLDRVLFSADWEELFPSCLLQAASSIASDHCALLLHTCVTSPKAHRFRFEAFW 919
>gb|AAM94293.1| putative reverse transcriptase [Sorghum bicolor]
Length = 1323
Score = 52.0 bits (123), Expect = 3e-06
Identities = 31/114 (27%), Positives = 53/114 (46%), Gaps = 8/114 (7%)
Query: 11 GDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQ---AA 67
GDFN++ ++++ +++ R+ M F+ L L+E +KYTW NG+
Sbjct: 336 GDFNLICEAQDK---NNNNINRAHMRKFRQALDASELLEIKLQNRKYTW--SNGRKNPTL 390
Query: 68 SRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
LDR + DW +P ++ SLSDHCP+ + F+ + W
Sbjct: 391 VHLDRAFCNLDWDEIFPSATLIALSSSLSDHCPLFLCNQQQPHRRATFKFESFW 444
>pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW15) -
Arabidopsis thaliana retrotransposon Ta11-1
gi|976278|gb|AAA75254.1| reverse transcriptase
Length = 1333
Score = 52.0 bits (123), Expect = 3e-06
Identities = 31/102 (30%), Positives = 49/102 (47%), Gaps = 4/102 (3%)
Query: 7 FCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQA 66
+C+ GDFN + + E+ G G S F L + ++E P++G +TW +
Sbjct: 133 WCMLGDFNPILHNGEKRG--GPRRGDSSFLPFTDMLDSCDMLELPSIGNPFTWGGKTNEM 190
Query: 67 --ASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRST 106
SRLDR + +W +P Q + + SDH PV+ R T
Sbjct: 191 WIQSRLDRCFGNKNWFRFFPISNQEFLDKRGSDHRPVLVRLT 232
>emb|CAE03649.2| OSJNBa0060N03.14 [Oryza sativa (japonica cultivar-group)]
gi|50928607|ref|XP_473831.1| OSJNBa0060N03.14 [Oryza
sativa (japonica cultivar-group)]
Length = 1784
Score = 51.2 bits (121), Expect = 6e-06
Identities = 32/114 (28%), Positives = 55/114 (48%), Gaps = 4/114 (3%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNG-QAA 67
+AGDFN++A + ++ ++S R M F+ ++ L L E G++YTW
Sbjct: 1133 VAGDFNLIAAAADK---NNSRVNRRLMNAFRNKINELELKELYLFGRRYTWSSEQQFPTL 1189
Query: 68 SRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+ DR +S++W +P ++ S SDHC ++ + G R FR W
Sbjct: 1190 VKSDRVPVSTEWEDAFPDAHLQALSSSSSDHCALLLSCGETPRGRRRFRFETFW 1243
>gb|AAX95598.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 1090
Score = 51.2 bits (121), Expect = 6e-06
Identities = 34/114 (29%), Positives = 52/114 (44%), Gaps = 4/114 (3%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAAS 68
I GDFN++ + ++ ++ RS M F+A L+ L E +KYTW QA
Sbjct: 100 ILGDFNLIYEARDKNNLN---LNRSLMGQFRAALNRNGLKEIRLQNRKYTWGNEREQATL 156
Query: 69 -RLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+LDR S + + + M+ S SDHCP++ + D F N W
Sbjct: 157 VKLDRVFCSCQGDLLFTNFLLQAMSTSFSDHCPLLLLNADRPRRKAHFNFENFW 210
>gb|AAP53215.1| putative retroelement [Oryza sativa (japonica cultivar-group)]
gi|37533252|ref|NP_920928.1| putative retroelement [Oryza
sativa (japonica cultivar-group)]
gi|19920129|gb|AAM08561.1| Putative retroelement [Oryza
sativa]
Length = 1739
Score = 50.8 bits (120), Expect = 7e-06
Identities = 36/119 (30%), Positives = 57/119 (47%), Gaps = 7/119 (5%)
Query: 7 FCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWF-RPNGQ 65
FC+ GD N + + E+ G++ R ++ FK ++NL L++ G YTW + G+
Sbjct: 904 FCM-GDLNEIMHANEKYGLAPPNQNRINI--FKHHVNNLGLMDMGYNGPAYTWSNKQQGK 960
Query: 66 --AASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGP-RPFRVLNCW 121
RLDR L + +WC +P+ + SDH P++ N P R F N W
Sbjct: 961 DLVLERLDRCLANVEWCFNYPNTTVYHLPMLYSDHAPIIAILNPKNRRPKRSFMFENWW 1019
>gb|AAP52685.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37532192|ref|NP_920398.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|20451038|gb|AAM22009.1| Putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1792
Score = 50.8 bits (120), Expect = 7e-06
Identities = 32/114 (28%), Positives = 51/114 (44%), Gaps = 4/114 (3%)
Query: 9 IAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFRPNG-QAA 67
I GDFN++ + ++ + R M F++ ++ L L E G++YTW
Sbjct: 1084 ITGDFNLITAAADKNNLR---INRRLMNAFRSKVNELELKEVYLFGRRYTWSSEQAVPTL 1140
Query: 68 SRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
+LDR S+ W +P ++ S SDHCP + D R FR + W
Sbjct: 1141 VKLDRLFASTQWEDMFPEAHLQALSSSGSDHCPQLLTCGDRVPRQRRFRFESYW 1194
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.137 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 209,543,522
Number of Sequences: 2540612
Number of extensions: 7293581
Number of successful extensions: 16287
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 16193
Number of HSP's gapped (non-prelim): 97
length of query: 124
length of database: 863,360,394
effective HSP length: 100
effective length of query: 24
effective length of database: 609,299,194
effective search space: 14623180656
effective search space used: 14623180656
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0281b.2


