

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0280a.7
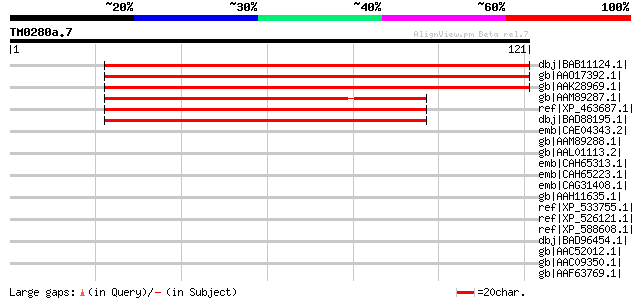
(121 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB11124.1| unnamed protein product [Arabidopsis thaliana] g... 73 2e-12
gb|AAO17392.1| SET domain histone methyltransferase SUVH4 [Arabi... 73 2e-12
gb|AAK28969.1| SUVH4 [Arabidopsis thaliana] 73 2e-12
gb|AAM89287.1| SET domain-containing protein SET118 [Zea mays] 63 2e-09
ref|XP_463687.1| putative SUVH4 [Oryza sativa (japonica cultivar... 62 3e-09
dbj|BAD88195.1| putative SET domain-containing protein [Oryza sa... 62 3e-09
emb|CAE04343.2| OSJNBb0038F03.7 [Oryza sativa (japonica cultivar... 42 0.004
gb|AAM89288.1| SET domain-containing protein SET104 [Zea mays] 40 0.010
gb|AAL01113.2| Su(VAR)3-9-related protein 4 [Arabidopsis thaliana] 40 0.017
emb|CAH65313.1| hypothetical protein [Gallus gallus] gi|60302716... 39 0.022
emb|CAH65223.1| hypothetical protein [Gallus gallus] 39 0.022
emb|CAG31408.1| hypothetical protein [Gallus gallus] 39 0.022
gb|AAH11635.1| SETMAR protein [Homo sapiens] 39 0.038
ref|XP_533755.1| PREDICTED: similar to SETMAR protein [Canis fam... 39 0.038
ref|XP_526121.1| PREDICTED: similar to SETMAR protein [Pan trogl... 39 0.038
ref|XP_588608.1| PREDICTED: similar to SETMAR protein, partial [... 39 0.038
dbj|BAD96454.1| SET domain and mariner transposase fusion gene v... 39 0.038
gb|AAC52012.1| orf; encodes putative chimeric protein with SET d... 39 0.038
gb|AAC09350.1| unknown [Homo sapiens] 39 0.038
gb|AAF63769.1| hypothetical protein [Arabidopsis thaliana] 38 0.050
>dbj|BAB11124.1| unnamed protein product [Arabidopsis thaliana]
gi|26983788|gb|AAN86146.1| unknown protein [Arabidopsis
thaliana] gi|15240758|ref|NP_196900.1| SET
domain-containing protein (SUVH4) [Arabidopsis thaliana]
gi|30580520|sp|Q8GZB6|SUV4_ARATH Histone-lysine
N-methyltransferase, H3 lysine-9 specific 4 (Histone
H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (Suppressor
of variegation 3-9 homolog 4) (Su(var)3-9 homolog 4)
(KRYPTONITE protein)
Length = 624
Score = 72.8 bits (177), Expect = 2e-12
Identities = 38/99 (38%), Positives = 61/99 (61%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR SQK L++ L+V+ KGWAV + ++I G+PV EY+ +R+ ++ S +E FE
Sbjct: 437 NRTSQKRLRFNLEVFRSAKKGWAVRSWEYIPAGSPVCEYIGVVRRTADVDTISDNEYIFE 496
Query: 83 IDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETME 121
IDC + + + GR R L +V++P + V +S +DE E
Sbjct: 497 IDCQQTMQGLGGRQRRLRDVAVPMNNGVSQSSEDENAPE 535
>gb|AAO17392.1| SET domain histone methyltransferase SUVH4 [Arabidopsis thaliana]
Length = 624
Score = 72.8 bits (177), Expect = 2e-12
Identities = 38/99 (38%), Positives = 61/99 (61%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR SQK L++ L+V+ KGWAV + ++I G+PV EY+ +R+ ++ S +E FE
Sbjct: 437 NRTSQKRLRFNLEVFRSAKKGWAVRSWEYIPAGSPVCEYIGVVRRTADVDTISDNEYIFE 496
Query: 83 IDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETME 121
IDC + + + GR R L +V++P + V +S +DE E
Sbjct: 497 IDCQQTMQGLGGRQRRLRDVAVPMNNGVSQSSEDENAPE 535
>gb|AAK28969.1| SUVH4 [Arabidopsis thaliana]
Length = 624
Score = 72.8 bits (177), Expect = 2e-12
Identities = 38/99 (38%), Positives = 61/99 (61%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR SQK L++ L+V+ KGWAV + ++I G+PV EY+ +R+ ++ S +E FE
Sbjct: 437 NRTSQKRLRFNLEVFRSAKKGWAVRSWEYIPAGSPVCEYIGVVRRTADVDTISDNEYIFE 496
Query: 83 IDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETME 121
IDC + + + GR R L +V++P + V +S +DE E
Sbjct: 497 IDCQQTMQGLGGRQRRLRDVAVPMNNGVSQSSEDENAPE 535
>gb|AAM89287.1| SET domain-containing protein SET118 [Zea mays]
Length = 696
Score = 62.8 bits (151), Expect = 2e-09
Identities = 33/75 (44%), Positives = 46/75 (61%), Gaps = 1/75 (1%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR SQ+ LQY L+V+ SKGW V T D IL G P+ EY LR+ +L + C F+
Sbjct: 510 NRTSQQGLQYHLEVFKTASKGWGVRTWDTILPGAPICEYTGVLRRTEDLDGSQNNYC-FD 568
Query: 83 IDCLKIINEVEGRNR 97
IDCL+ + ++GR +
Sbjct: 569 IDCLQTMKGLDGREK 583
>ref|XP_463687.1| putative SUVH4 [Oryza sativa (japonica cultivar-group)]
gi|20160732|dbj|BAB89674.1| putative SUVH4 [Oryza sativa
(japonica cultivar-group)]
Length = 676
Score = 62.0 bits (149), Expect = 3e-09
Identities = 33/75 (44%), Positives = 45/75 (60%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR SQK LQY L+V+ SKGW V T D IL G P+ EY LR+ E+ + F+
Sbjct: 490 NRTSQKGLQYRLEVFKTASKGWGVRTWDTILPGAPICEYTGVLRRTEEVDGLLQNNYIFD 549
Query: 83 IDCLKIINEVEGRNR 97
IDCL+ + ++GR +
Sbjct: 550 IDCLQTMKGLDGREK 564
>dbj|BAD88195.1| putative SET domain-containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 663
Score = 62.0 bits (149), Expect = 3e-09
Identities = 33/75 (44%), Positives = 45/75 (60%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR SQK LQY L+V+ SKGW V T D IL G P+ EY LR+ E+ + F+
Sbjct: 477 NRTSQKGLQYRLEVFKTASKGWGVRTWDTILPGAPICEYTGVLRRTEEVDGLLQNNYIFD 536
Query: 83 IDCLKIINEVEGRNR 97
IDCL+ + ++GR +
Sbjct: 537 IDCLQTMKGLDGREK 551
>emb|CAE04343.2| OSJNBb0038F03.7 [Oryza sativa (japonica cultivar-group)]
gi|50927315|ref|XP_473383.1| OSJNBb0038F03.7 [Oryza
sativa (japonica cultivar-group)]
Length = 841
Score = 41.6 bits (96), Expect = 0.004
Identities = 24/61 (39%), Positives = 33/61 (53%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGFE 82
NR+ Q L++ LQV+ GW V T DFI G+ V EY+ E+ ++ E S E F
Sbjct: 661 NRVGQHGLRFRLQVFKTKLMGWGVRTLDFIPSGSFVCEYIGEVLEDEEAQKRSTDEYLFA 720
Query: 83 I 83
I
Sbjct: 721 I 721
>gb|AAM89288.1| SET domain-containing protein SET104 [Zea mays]
Length = 886
Score = 40.4 bits (93), Expect = 0.010
Identities = 19/48 (39%), Positives = 30/48 (61%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVE 70
NR+ Q L++ LQ++ S GW V T +FI G+ V EY+ E+ ++ E
Sbjct: 704 NRVGQHGLKFRLQIFKTKSMGWGVRTLEFIPSGSFVCEYIGEVLEDEE 751
>gb|AAL01113.2| Su(VAR)3-9-related protein 4 [Arabidopsis thaliana]
Length = 488
Score = 39.7 bits (91), Expect = 0.017
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 16/99 (16%)
Query: 23 NRISQKDLQYCLQVYL-KISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGF 81
NR+ Q+ +++ LQVY + KGW + T + GT + EY+ E+ N EL +V
Sbjct: 293 NRVVQRGIRWQLQVYFTQEGKGWGLRTLQDLPKGTFICEYIGEILTNTELYDRNVR---- 348
Query: 82 EIDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETM 120
R + V+L A + E+ + DEE +
Sbjct: 349 -----------SSSERHTYPVTLDADWGSEKDLKDEEAL 376
>emb|CAH65313.1| hypothetical protein [Gallus gallus] gi|60302716|ref|NP_001012550.1|
euchromatic histone methyltransferase 1 [Gallus gallus]
Length = 1249
Score = 39.3 bits (90), Expect = 0.022
Identities = 24/67 (35%), Positives = 31/67 (45%)
Query: 18 WT*FDNRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVS 77
W NR+ Q L+ LQ+Y GW V T I GT V EYV EL + E +
Sbjct: 1062 WRTCRNRVVQNGLRTRLQLYRTQKMGWGVRTMQDIPLGTFVCEYVGELISDSEADVREED 1121
Query: 78 ECGFEID 84
F++D
Sbjct: 1122 SYLFDLD 1128
>emb|CAH65223.1| hypothetical protein [Gallus gallus]
Length = 904
Score = 39.3 bits (90), Expect = 0.022
Identities = 24/67 (35%), Positives = 31/67 (45%)
Query: 18 WT*FDNRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVS 77
W NR+ Q L+ LQ+Y GW V T I GT V EYV EL + E +
Sbjct: 717 WRTCRNRVVQNGLRTRLQLYRTQKMGWGVRTMQDIPLGTFVCEYVGELISDSEADVREED 776
Query: 78 ECGFEID 84
F++D
Sbjct: 777 SYLFDLD 783
>emb|CAG31408.1| hypothetical protein [Gallus gallus]
Length = 856
Score = 39.3 bits (90), Expect = 0.022
Identities = 24/67 (35%), Positives = 31/67 (45%)
Query: 18 WT*FDNRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVS 77
W NR+ Q L+ LQ+Y GW V T I GT V EYV EL + E +
Sbjct: 669 WRTCRNRVVQNGLRTRLQLYRTQKMGWGVRTMQDIPLGTFVCEYVGELISDSEADVREED 728
Query: 78 ECGFEID 84
F++D
Sbjct: 729 SYLFDLD 735
>gb|AAH11635.1| SETMAR protein [Homo sapiens]
Length = 352
Score = 38.5 bits (88), Expect = 0.038
Identities = 19/43 (44%), Positives = 25/43 (57%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR+ QK LQ+ QV+ KGW + T +FI G V EY E+
Sbjct: 117 NRVVQKGLQFHFQVFKTHKKGWGLRTLEFIPKGRFVCEYAGEV 159
>ref|XP_533755.1| PREDICTED: similar to SETMAR protein [Canis familiaris]
Length = 342
Score = 38.5 bits (88), Expect = 0.038
Identities = 19/43 (44%), Positives = 26/43 (60%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR+ Q+ LQ+ LQV+ KGW + T +FI G V EY E+
Sbjct: 158 NRVVQQGLQFQLQVFKTDKKGWGLRTLEFIPKGRFVCEYAGEV 200
>ref|XP_526121.1| PREDICTED: similar to SETMAR protein [Pan troglodytes]
Length = 365
Score = 38.5 bits (88), Expect = 0.038
Identities = 19/43 (44%), Positives = 25/43 (57%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR+ QK LQ+ QV+ KGW + T +FI G V EY E+
Sbjct: 130 NRVVQKGLQFHFQVFKTHKKGWGLRTLEFIPKGRFVCEYAGEV 172
>ref|XP_588608.1| PREDICTED: similar to SETMAR protein, partial [Bos taurus]
Length = 300
Score = 38.5 bits (88), Expect = 0.038
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 6/55 (10%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEY------VKELRKNVEL 71
NR+ Q LQ+ LQV+ KGW + T DFI G V EY + E+++ V+L
Sbjct: 117 NRVVQWGLQFHLQVFKTDHKGWGLRTLDFIPKGRFVCEYAGEVLGISEVQRRVQL 171
>dbj|BAD96454.1| SET domain and mariner transposase fusion gene variant [Homo
sapiens] gi|63079013|gb|AAY29570.1| metnase [Homo
sapiens]
Length = 671
Score = 38.5 bits (88), Expect = 0.038
Identities = 19/43 (44%), Positives = 25/43 (57%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR+ QK LQ+ QV+ KGW + T +FI G V EY E+
Sbjct: 117 NRVVQKGLQFHFQVFKTHKKGWGLRTLEFIPKGRFVCEYAGEV 159
>gb|AAC52012.1| orf; encodes putative chimeric protein with SET domain in
N-terminus with similarity to several other human,
Drosophila, nematode and yeast proteins [Homo sapiens]
gi|5730039|ref|NP_006506.1| SET domain and mariner
transposase fusion gene [Homo sapiens]
Length = 671
Score = 38.5 bits (88), Expect = 0.038
Identities = 19/43 (44%), Positives = 25/43 (57%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR+ QK LQ+ QV+ KGW + T +FI G V EY E+
Sbjct: 117 NRVVQKGLQFHFQVFKTHKKGWGLRTLEFIPKGRFVCEYAGEV 159
>gb|AAC09350.1| unknown [Homo sapiens]
Length = 671
Score = 38.5 bits (88), Expect = 0.038
Identities = 19/43 (44%), Positives = 25/43 (57%)
Query: 23 NRISQKDLQYCLQVYLKISKGWAVNTRDFILFGTPVLEYVKEL 65
NR+ QK LQ+ QV+ KGW + T +FI G V EY E+
Sbjct: 117 NRVVQKGLQFHFQVFKTHKKGWGLRTLEFIPKGRFVCEYAGEV 159
>gb|AAF63769.1| hypothetical protein [Arabidopsis thaliana]
Length = 424
Score = 38.1 bits (87), Expect = 0.050
Identities = 28/99 (28%), Positives = 45/99 (45%), Gaps = 16/99 (16%)
Query: 23 NRISQKDLQYCLQVYL-KISKGWAVNTRDFILFGTPVLEYVKELRKNVELVIDSVSECGF 81
NR+ Q+ ++ LQVY + KGW + T + GT + EY+ E+ N EL +V
Sbjct: 277 NRVVQRGIRCQLQVYFTQEGKGWGLRTLQDLPKGTFICEYIGEILTNTELYDRNVR---- 332
Query: 82 EIDCLKIINEVEGRNRLLHNVSLPASFCVERSVDDEETM 120
R + V+L A + E+ + DEE +
Sbjct: 333 -----------SSSERHTYPVTLDADWGSEKDLKDEEAL 360
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.337 0.148 0.478
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 185,407,748
Number of Sequences: 2540612
Number of extensions: 6539031
Number of successful extensions: 23636
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 84
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 23545
Number of HSP's gapped (non-prelim): 101
length of query: 121
length of database: 863,360,394
effective HSP length: 97
effective length of query: 24
effective length of database: 616,921,030
effective search space: 14806104720
effective search space used: 14806104720
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0280a.7


