

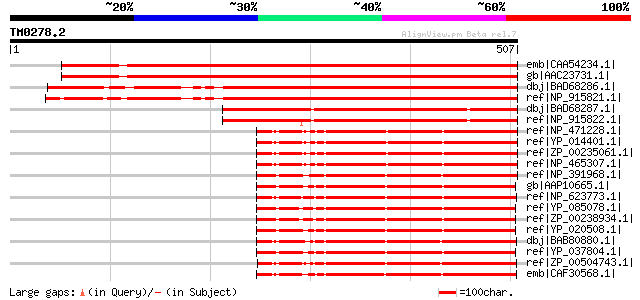
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0278.2
(507 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA54234.1| ARP protein [Arabidopsis thaliana] 584 e-165
gb|AAC23731.1| DNA-(apurinic or apyrimidinic site) lyase (ARP) [... 584 e-165
dbj|BAD68286.1| putative DNA-(apurinic or apyrimidinic site) lya... 534 e-150
ref|NP_915821.1| putative exodeoxyribonuclease [Oryza sativa (ja... 521 e-146
dbj|BAD68287.1| putative DNA-(apurinic or apyrimidinic site) lya... 408 e-112
ref|NP_915822.1| putative exodeoxyribonuclease [Oryza sativa (ja... 399 e-109
ref|NP_471228.1| hypothetical protein lin1894 [Listeria innocua ... 270 6e-71
ref|YP_014401.1| exodeoxyribonuclease [Listeria monocytogenes st... 269 2e-70
ref|ZP_00235061.1| exodeoxyribonuclease [Listeria monocytogenes ... 268 3e-70
ref|NP_465307.1| hypothetical protein lmo1782 [Listeria monocyto... 267 5e-70
ref|NP_391968.1| multifunctional DNA-repair enzyme [Bacillus sub... 263 7e-69
gb|AAP10665.1| Exodeoxyribonuclease III [Bacillus cereus ATCC 14... 258 2e-67
ref|NP_623773.1| Exonuclease III [Thermoanaerobacter tengcongens... 258 3e-67
ref|YP_085078.1| exodeoxyribonuclease III [Bacillus cereus E33L]... 256 9e-67
ref|ZP_00238934.1| exodeoxyribonuclease III [Bacillus cereus G92... 254 3e-66
ref|YP_020508.1| exodeoxyribonuclease iii [Bacillus anthracis st... 254 6e-66
dbj|BAB80880.1| 3'-exo-deoxyribonuclease [Clostridium perfringen... 253 8e-66
ref|YP_037804.1| exodeoxyribonuclease III [Bacillus thuringiensi... 253 1e-65
ref|ZP_00504743.1| AP endonuclease, family 1:Exodeoxyribonucleas... 250 8e-65
emb|CAF30568.1| exonuclease III [Methanococcus maripaludis S2] g... 249 1e-64
>emb|CAA54234.1| ARP protein [Arabidopsis thaliana]
Length = 527
Score = 584 bits (1506), Expect = e-165
Identities = 288/458 (62%), Positives = 358/458 (77%), Gaps = 9/458 (1%)
Query: 52 EIERLRNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKSFMDNNMGEQHPQIEEE 111
E+ L++D ++ MTVQELR TL+ + VP KGRK++L+STL+ MD+N+ +Q
Sbjct: 77 EMGTLQDDRKEIEAMTVQELRSTLRKLGVPVKGRKQELISTLRLHMDSNLPDQKETSS-- 134
Query: 112 HGLLISSENTSVEVKTKKVD-EDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTTKK 170
S+ + SV +K K + E+ +D N E ++ G++R+KQS ++ KV+ K
Sbjct: 135 -----STRSDSVTIKRKISNREEPTEDECTNSEAYDIEHGEKRVKQSTEKNLKAKVSAKA 189
Query: 171 KLLVESDEVSDFKPS-RAKRRVSSDIVRVVSQSDEISTTTIPTEPWTVLAHKKPQKDWIA 229
+ + K ++K SS I + +++EI ++ +EPWTVLAHKKPQKDW A
Sbjct: 190 IAKEQKSLMRTGKQQIQSKEETSSTISSELLKTEEIISSPSQSEPWTVLAHKKPQKDWKA 249
Query: 230 YNPKTMRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQE 289
YNPKTMRP PL TK VK+++WNVNGLR LLK E FSAL+LAQRE+FD+LCLQETKLQ
Sbjct: 250 YNPKTMRPPPLPEGTKCVKVMTWNVNGLRGLLKFESFSALQLAQRENFDILCLQETKLQV 309
Query: 290 KDINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTA 349
KD+ EIK+ LIDGY++SFW+CSVSKLGYSGTAIISRIKPLSVRYG G+S HDTEGR+VTA
Sbjct: 310 KDVEEIKKTLIDGYDHSFWSCSVSKLGYSGTAIISRIKPLSVRYGTGLSGHDTEGRIVTA 369
Query: 350 EFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEID 409
EFD+FYLI Y+PNSGDGLKRLSYR+ +WD +LSN++KELEK+KPVVLTGDLNCAHEEID
Sbjct: 370 EFDSFYLINTYVPNSGDGLKRLSYRIEEWDRTLSNHIKELEKSKPVVLTGDLNCAHEEID 429
Query: 410 IYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVGYTYWGYRHGGRKTNRGW 469
I+NPAGNK+SAGFT EER+SF AN L +GFVDTFR+QHP VVGYTYWGYRHGGRKTN+GW
Sbjct: 430 IFNPAGNKRSAGFTIEERQSFGANLLDKGFVDTFRKQHPGVVGYTYWGYRHGGRKTNKGW 489
Query: 470 RLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
RLDYFLVS+SIA VHDSYILPD+ GSDHCP+GLI+KL
Sbjct: 490 RLDYFLVSQSIAANVHDSYILPDINGSDHCPIGLILKL 527
>gb|AAC23731.1| DNA-(apurinic or apyrimidinic site) lyase (ARP) [Arabidopsis
thaliana] gi|8488963|sp|P45951|ARP_ARATH Apurinic
endonuclease-redox protein (DNA-(apurinic or
apyrimidinic site) lyase) gi|15227354|ref|NP_181677.1|
apurinic endonuclease-redox protein / DNA-(apurinic or
apyrimidinic site) lyase [Arabidopsis thaliana]
Length = 536
Score = 584 bits (1506), Expect = e-165
Identities = 288/458 (62%), Positives = 358/458 (77%), Gaps = 9/458 (1%)
Query: 52 EIERLRNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKSFMDNNMGEQHPQIEEE 111
E+ L++D ++ MTVQELR TL+ + VP KGRK++L+STL+ MD+N+ +Q
Sbjct: 86 EMGTLQDDRKEIEAMTVQELRSTLRKLGVPVKGRKQELISTLRLHMDSNLPDQKETSS-- 143
Query: 112 HGLLISSENTSVEVKTKKVD-EDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTTKK 170
S+ + SV +K K + E+ +D N E ++ G++R+KQS ++ KV+ K
Sbjct: 144 -----STRSDSVTIKRKISNREEPTEDECTNSEAYDIEHGEKRVKQSTEKNLKAKVSAKA 198
Query: 171 KLLVESDEVSDFKPS-RAKRRVSSDIVRVVSQSDEISTTTIPTEPWTVLAHKKPQKDWIA 229
+ + K ++K SS I + +++EI ++ +EPWTVLAHKKPQKDW A
Sbjct: 199 IAKEQKSLMRTGKQQIQSKEETSSTISSELLKTEEIISSPSQSEPWTVLAHKKPQKDWKA 258
Query: 230 YNPKTMRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQE 289
YNPKTMRP PL TK VK+++WNVNGLR LLK E FSAL+LAQRE+FD+LCLQETKLQ
Sbjct: 259 YNPKTMRPPPLPEGTKCVKVMTWNVNGLRGLLKFESFSALQLAQRENFDILCLQETKLQV 318
Query: 290 KDINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTA 349
KD+ EIK+ LIDGY++SFW+CSVSKLGYSGTAIISRIKPLSVRYG G+S HDTEGR+VTA
Sbjct: 319 KDVEEIKKTLIDGYDHSFWSCSVSKLGYSGTAIISRIKPLSVRYGTGLSGHDTEGRIVTA 378
Query: 350 EFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEID 409
EFD+FYLI Y+PNSGDGLKRLSYR+ +WD +LSN++KELEK+KPVVLTGDLNCAHEEID
Sbjct: 379 EFDSFYLINTYVPNSGDGLKRLSYRIEEWDRTLSNHIKELEKSKPVVLTGDLNCAHEEID 438
Query: 410 IYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVGYTYWGYRHGGRKTNRGW 469
I+NPAGNK+SAGFT EER+SF AN L +GFVDTFR+QHP VVGYTYWGYRHGGRKTN+GW
Sbjct: 439 IFNPAGNKRSAGFTIEERQSFGANLLDKGFVDTFRKQHPGVVGYTYWGYRHGGRKTNKGW 498
Query: 470 RLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
RLDYFLVS+SIA VHDSYILPD+ GSDHCP+GLI+KL
Sbjct: 499 RLDYFLVSQSIAANVHDSYILPDINGSDHCPIGLILKL 536
>dbj|BAD68286.1| putative DNA-(apurinic or apyrimidinic site) lyase (ARP) [Oryza
sativa (japonica cultivar-group)]
gi|55296319|dbj|BAD68137.1| putative DNA-(apurinic or
apyrimidinic site) lyase (ARP) [Oryza sativa (japonica
cultivar-group)]
Length = 499
Score = 534 bits (1376), Expect = e-150
Identities = 268/472 (56%), Positives = 345/472 (72%), Gaps = 36/472 (7%)
Query: 38 KHVIKGNDANSYSIEIERLRNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKSFM 97
+HV++ D +EIE+LRNDP + +MTV+ELR+ + + +P KG K+DL+S L
Sbjct: 62 EHVLEKEDVAESKLEIEQLRNDPDRLQSMTVKELREITRMMGIPVKGNKKDLVSALM--- 118
Query: 98 DNNMGEQHPQIEEEHGLLISSENTSVEVKTKKVDEDHVDDIN-DNPEVFEHSRGKR-RLK 155
+++G+ E+ G+ EV +K+ V + N D+ EV + KR R K
Sbjct: 119 -DSLGKVGTSSVEKIGVS--------EVPSKRKGASVVVEQNIDSSEVISETPSKRSRAK 169
Query: 156 QSGSESETVKVTTKKKLLVESDEVSDFKPSRAKRRVSSDIVRVVSQSDEISTTTIPTEPW 215
G+ E+ K+ K S K+++ +V+ S E EPW
Sbjct: 170 NKGTAEESSGANVKQS-----------KTSVQKKKL---VVQGASVDHE--------EPW 207
Query: 216 TVLAHKKPQKDWIAYNPKTMRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELAQRE 275
TVL HKKPQ WI YNPK MR L++DTK +K+LSWNVNGL+ALLK GFS +LAQRE
Sbjct: 208 TVLVHKKPQPAWIPYNPKVMRSPSLSKDTKALKILSWNVNGLKALLKSRGFSIHQLAQRE 267
Query: 276 DFDVLCLQETKLQEKDINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLSVRYGL 335
DFD+LCLQETK+QEKD+ IK L++GY +SFWTCSVSKLGYSGTAIISR+KPLS++YGL
Sbjct: 268 DFDILCLQETKMQEKDVEVIKEGLLEGYTHSFWTCSVSKLGYSGTAIISRVKPLSIKYGL 327
Query: 336 GISDHDTEGRLVTAEFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPV 395
G+ DHDTEGR+VT EF+ FYL+ Y+PNSGDGLKRL+YRVT+WDPSL NY+K+LEK+KPV
Sbjct: 328 GVPDHDTEGRVVTVEFNDFYLLTAYVPNSGDGLKRLTYRVTEWDPSLGNYMKDLEKSKPV 387
Query: 396 VLTGDLNCAHEEIDIYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVGYTY 455
+LTGDLNCAH+EIDI++PAGN++SAGFT EER+SF NFLS+GFVDTFR+QHP+VVGY+Y
Sbjct: 388 ILTGDLNCAHQEIDIHDPAGNRRSAGFTIEERESFETNFLSKGFVDTFRKQHPNVVGYSY 447
Query: 456 WGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
WGYRH RKTN+GWRLDYFLVSESIA++VHDSYI+PD+ SDH P+GL++KL
Sbjct: 448 WGYRHNARKTNKGWRLDYFLVSESIAERVHDSYIIPDISASDHSPLGLVLKL 499
>ref|NP_915821.1| putative exodeoxyribonuclease [Oryza sativa (japonica
cultivar-group)]
Length = 493
Score = 521 bits (1343), Expect = e-146
Identities = 269/475 (56%), Positives = 344/475 (71%), Gaps = 36/475 (7%)
Query: 36 SKKHVIKGNDANSYSIEIERLRNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKS 95
SK IK +D + ++LRNDP + +MTV+ELR+ + + +P KG K+DL+S L
Sbjct: 52 SKNTDIKKDDEHVLE---KQLRNDPDRLQSMTVKELREITRMMGIPVKGNKKDLVSAL-- 106
Query: 96 FMDNNMGEQHPQIEEEHGLLISSENTSV-EVKTKKVDEDHVDDIN-DNPEVFEHSRGKR- 152
MD+ E++ ++ S E V EV +K+ V + N D+ EV + KR
Sbjct: 107 -MDSLGKERNGKVGTS-----SVEKIGVSEVPSKRKGASVVVEQNIDSSEVISETPSKRS 160
Query: 153 RLKQSGSESETVKVTTKKKLLVESDEVSDFKPSRAKRRVSSDIVRVVSQSDEISTTTIPT 212
R K G+ E+ K+ K S K+++ +V+ S E
Sbjct: 161 RAKNKGTAEESSGANVKQS-----------KTSVQKKKL---VVQGASVDHE-------- 198
Query: 213 EPWTVLAHKKPQKDWIAYNPKTMRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELA 272
EPWTVL HKKPQ WI YNPK MR L++DTK +K+LSWNVNGL+ALLK GFS +LA
Sbjct: 199 EPWTVLVHKKPQPAWIPYNPKVMRSPSLSKDTKALKILSWNVNGLKALLKSRGFSIHQLA 258
Query: 273 QREDFDVLCLQETKLQEKDINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLSVR 332
QREDFD+LCLQETK+Q KD+ IK L++GY +SFWTCSVSKLGYSGTAIISR+KPLS++
Sbjct: 259 QREDFDILCLQETKMQAKDVEVIKEGLLEGYTHSFWTCSVSKLGYSGTAIISRVKPLSIK 318
Query: 333 YGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKT 392
YGLG+ DHDTEGR+VT EF+ FYL+ Y+PNSGDGLKRL+YRVT+WDPSL NY+K+LEK+
Sbjct: 319 YGLGVPDHDTEGRVVTVEFNDFYLLTAYVPNSGDGLKRLTYRVTEWDPSLGNYMKDLEKS 378
Query: 393 KPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVG 452
KPV+LTGDLNCAH+EIDI++PAGN++SAGFT EER+SF NFLS+GFVDTFR+QHP+VVG
Sbjct: 379 KPVILTGDLNCAHQEIDIHDPAGNRRSAGFTIEERESFETNFLSKGFVDTFRKQHPNVVG 438
Query: 453 YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
Y+YWGYRH RKTN+GWRLDYFLVSESIA++VHDSYI+PD+ SDH P+GL++KL
Sbjct: 439 YSYWGYRHNARKTNKGWRLDYFLVSESIAERVHDSYIIPDISASDHSPLGLVLKL 493
>dbj|BAD68287.1| putative DNA-(apurinic or apyrimidinic site) lyase (ARP) [Oryza
sativa (japonica cultivar-group)]
gi|55296320|dbj|BAD68138.1| putative DNA-(apurinic or
apyrimidinic site) lyase (ARP) [Oryza sativa (japonica
cultivar-group)]
Length = 307
Score = 408 bits (1048), Expect = e-112
Identities = 188/295 (63%), Positives = 235/295 (78%), Gaps = 4/295 (1%)
Query: 213 EPWTVLAHKKPQKDWIAYNPKTMRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELA 272
EPWT L H++ +W AYNPKTMRP PL+ DTK +K+LSWN+NGL ++ +GFSA +LA
Sbjct: 17 EPWTKLVHRERLPEWFAYNPKTMRPPPLSHDTKCMKILSWNINGLHDVVTTKGFSARDLA 76
Query: 273 QREDFDVLCLQETKLQEKDINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLSVR 332
QRE+FDVLCLQET L+EKD+ + K + D +S+W+CSVS+LGYSGTA+ISR+KP+SV+
Sbjct: 77 QRENFDVLCLQETHLEEKDVEKFKNLIAD--YDSYWSCSVSRLGYSGTAVISRVKPISVQ 134
Query: 333 YGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKT 392
YG+GI +HD EGR++T EFD FYL+ Y+PNSG L+RL+YRV WDP SNY+K LEK+
Sbjct: 135 YGIGIREHDHEGRVITLEFDGFYLVNAYVPNSGRFLRRLNYRVNNWDPCFSNYVKILEKS 194
Query: 393 KPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVG 452
KPV++ GDLNCA + IDI+NP KSAGFT EER+SF NF S+G VDTFR+QHP+ VG
Sbjct: 195 KPVIVAGDLNCARQSIDIHNPPAKTKSAGFTIEERESFETNFSSKGLVDTFRKQHPNAVG 254
Query: 453 YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
YT+WG R TN+GWRLDYFL SESI DKVHDSYILPDV SDH P+GL++KL
Sbjct: 255 YTFWG--ENQRITNKGWRLDYFLASESITDKVHDSYILPDVSFSDHSPIGLVLKL 307
>ref|NP_915822.1| putative exodeoxyribonuclease [Oryza sativa (japonica
cultivar-group)]
Length = 309
Score = 399 bits (1024), Expect = e-109
Identities = 186/297 (62%), Positives = 233/297 (77%), Gaps = 6/297 (2%)
Query: 213 EPWTVLAHKKPQKDWIAYNPKTMRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELA 272
EPWT L H++ +W AYNPKTMRP PL+ DTK +K+LSWN+NGL ++ +GFSA +LA
Sbjct: 17 EPWTKLVHRERLPEWFAYNPKTMRPPPLSHDTKCMKILSWNINGLHDVVTTKGFSARDLA 76
Query: 273 QREDFDVLCLQETKLQEK--DINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLS 330
QRE+FDVLCLQET L+ D+ + K + D +S+W+CSVS+LGYSGTA+ISR+KP+S
Sbjct: 77 QRENFDVLCLQETHLEASLGDVEKFKNLIAD--YDSYWSCSVSRLGYSGTAVISRVKPIS 134
Query: 331 VRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELE 390
V+YG+GI +HD EGR++T EFD FYL+ Y+PNSG L+RL+YRV WDP SNY+K LE
Sbjct: 135 VQYGIGIREHDHEGRVITLEFDGFYLVNAYVPNSGRFLRRLNYRVNNWDPCFSNYVKILE 194
Query: 391 KTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDV 450
K+KPV++ GDLNCA + IDI+NP KSAGFT EER+SF NF S+G VDTFR+QHP+
Sbjct: 195 KSKPVIVAGDLNCARQSIDIHNPPAKTKSAGFTIEERESFETNFSSKGLVDTFRKQHPNA 254
Query: 451 VGYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
VGYT+WG R TN+GWRLDYFL SESI DKVHDSYILPDV SDH P+GL++KL
Sbjct: 255 VGYTFWG--ENQRITNKGWRLDYFLASESITDKVHDSYILPDVSFSDHSPIGLVLKL 309
>ref|NP_471228.1| hypothetical protein lin1894 [Listeria innocua Clip11262]
gi|16414395|emb|CAC97124.1| lin1894 [Listeria innocua]
gi|25289128|pir||AD1669 3'-exo-deoxyribonuclease exoA
homolog lin1894 [imported] - Listeria innocua (strain
Clip11262)
Length = 251
Score = 270 bits (691), Expect = 6e-71
Identities = 142/262 (54%), Positives = 191/262 (72%), Gaps = 12/262 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA +K +GF LE + D D+ CLQETKLQE I E+ + Y++
Sbjct: 1 MKLISWNVNGLRAAVK-KGF--LEYFEEVDADIFCLQETKLQEGQI-ELD---LPAYKD- 52
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI ++++PLSV+YGLG+ +HDTEGR++T EF+ FY++ Y PNS
Sbjct: 53 YWNYAVKK-GYSGTAIFTKVEPLSVQYGLGVPEHDTEGRVITLEFEDFYMVTVYTPNSQA 111
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
LKRL YR+T ++ ++ Y+K L+KTKPVVL GDLN AHEEID+ NP N+K+AGF+DEE
Sbjct: 112 ELKRLDYRMT-FEDAILEYVKNLDKTKPVVLCGDLNVAHEEIDLKNPKTNRKNAGFSDEE 170
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVV-GYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R F+A FL GF+D+FR +PD+ Y++W YR R N GWR+DYF+VSE + DK+
Sbjct: 171 RAKFSA-FLDAGFIDSFRYFYPDLTDAYSWWSYRMNARARNTGWRIDYFVVSERLKDKLV 229
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
D+ I DV+GSDHCPV L + L
Sbjct: 230 DAKIHADVLGSDHCPVELELNL 251
>ref|YP_014401.1| exodeoxyribonuclease [Listeria monocytogenes str. 4b F2365]
gi|47094151|ref|ZP_00231871.1| exodeoxyribonuclease
[Listeria monocytogenes str. 4b H7858]
gi|47017476|gb|EAL08289.1| exodeoxyribonuclease
[Listeria monocytogenes str. 4b H7858]
gi|46881282|gb|AAT04578.1| exodeoxyribonuclease
[Listeria monocytogenes str. 4b F2365]
Length = 251
Score = 269 bits (687), Expect = 2e-70
Identities = 141/262 (53%), Positives = 191/262 (72%), Gaps = 12/262 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA +K +GF LE + D D+ CLQETKLQE I E+ + Y++
Sbjct: 1 MKLISWNVNGLRAAVK-KGF--LEYFEEVDADIFCLQETKLQEGQI-ELD---LPAYKD- 52
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI ++++PLSV+YGLG+ +HDTEGR++T EF+ F+++ Y PNS
Sbjct: 53 YWNYAVKK-GYSGTAIFTKVEPLSVQYGLGVPEHDTEGRVITLEFEEFFMVTVYTPNSQA 111
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
LKRL YR+T ++ ++ Y+K L+KTKPVVL GDLN AHEEID+ NP N+K+AGF+DEE
Sbjct: 112 ELKRLDYRMT-FEDAILEYVKNLDKTKPVVLCGDLNVAHEEIDLKNPKTNRKNAGFSDEE 170
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVV-GYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R F+A FL GF+D+FR +PD+ Y++W YR R N GWR+DYF+VSE + DK+
Sbjct: 171 RAKFSA-FLDAGFIDSFRYFYPDLTDAYSWWSYRMNARARNTGWRIDYFVVSERLKDKLV 229
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
D+ I DV+GSDHCPV L + L
Sbjct: 230 DAKIHADVLGSDHCPVELELNL 251
>ref|ZP_00235061.1| exodeoxyribonuclease [Listeria monocytogenes str. 1/2a F6854]
gi|47014105|gb|EAL05099.1| exodeoxyribonuclease
[Listeria monocytogenes str. 1/2a F6854]
Length = 251
Score = 268 bits (685), Expect = 3e-70
Identities = 141/262 (53%), Positives = 191/262 (72%), Gaps = 12/262 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
++L+SWNVNGLRA +K +GF LE + D D+ CLQETKLQE I E+ + Y++
Sbjct: 1 MRLISWNVNGLRAAVK-KGF--LEYFEEVDADIFCLQETKLQEGQI-ELD---LPAYKD- 52
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI ++++PLSV+YGLGI +HDTEGR++T EF+ F+++ Y PNS
Sbjct: 53 YWNYAVKK-GYSGTAIFTKVEPLSVQYGLGIPEHDTEGRVITLEFEEFFMVTVYTPNSQA 111
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
LKRL YR+T ++ ++ Y+K L+KTKPVVL GDLN AHEEID+ NP N+K+AGF+DEE
Sbjct: 112 ELKRLDYRMT-FEDAILEYVKNLDKTKPVVLCGDLNVAHEEIDLKNPKTNRKNAGFSDEE 170
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVV-GYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R F+A FL GF+D+FR +PD+ Y++W YR R N GWR+DYF+VSE + DK+
Sbjct: 171 RAKFSA-FLDAGFIDSFRYFYPDLTDAYSWWSYRMNARARNTGWRIDYFVVSERLKDKLV 229
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
D+ I DV+GSDHCPV L + L
Sbjct: 230 DAKIHADVLGSDHCPVELELNL 251
>ref|NP_465307.1| hypothetical protein lmo1782 [Listeria monocytogenes EGD-e]
gi|16411236|emb|CAC99860.1| lmo1782 [Listeria
monocytogenes] gi|25289129|pir||AF1297
3'-exo-deoxyribonuclease exoA homolog lmo1782 [imported]
- Listeria monocytogenes (strain EGD-e)
Length = 251
Score = 267 bits (683), Expect = 5e-70
Identities = 141/262 (53%), Positives = 190/262 (71%), Gaps = 12/262 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA +K +GF LE + D D+ CLQETKLQE I E+ + Y++
Sbjct: 1 MKLISWNVNGLRAAVK-KGF--LEYFEEVDADIFCLQETKLQEGQI-ELD---LPAYKD- 52
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI ++++PLSV+YGLGI +HDTEGR++T EF+ F+++ Y PNS
Sbjct: 53 YWNYAVKK-GYSGTAIFTKVEPLSVQYGLGIPEHDTEGRVITLEFEEFFMVTVYTPNSQA 111
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
LKRL YR+T ++ ++ Y+K L+ TKPVVL GDLN AHEEID+ NP N+K+AGF+DEE
Sbjct: 112 ELKRLDYRMT-FEDAILEYVKNLDNTKPVVLCGDLNVAHEEIDLKNPKTNRKNAGFSDEE 170
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVV-GYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R F+A FL GF+D+FR +PD+ Y++W YR R N GWR+DYF+VSE + DK+
Sbjct: 171 RAKFSA-FLDAGFIDSFRYFYPDLTDAYSWWSYRMNARARNTGWRIDYFVVSERLKDKLV 229
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
D+ I DV+GSDHCPV L + L
Sbjct: 230 DAKIHADVLGSDHCPVELELNL 251
>ref|NP_391968.1| multifunctional DNA-repair enzyme [Bacillus subtilis subsp.
subtilis str. 168] gi|2636635|emb|CAB16125.1| exoA
[Bacillus subtilis subsp. subtilis str. 168]
gi|585113|sp|P37454|EXOA_BACSU Exodeoxyribonuclease
gi|467372|dbj|BAA05218.1| 3'-exo-deoxyribonuclease
[Bacillus subtilis]
Length = 252
Score = 263 bits (673), Expect = 7e-69
Identities = 129/262 (49%), Positives = 187/262 (71%), Gaps = 11/262 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA+++ F L + ED D++CLQETK+Q+ ++ L +
Sbjct: 1 MKLISWNVNGLRAVMRKMDF--LSYLKEEDADIICLQETKIQDGQVD-----LQPEDYHV 53
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTA+ S+ +PL V YG+G+ +HD EGR++T EF+ +++ Y PNS
Sbjct: 54 YWNYAVKK-GYSGTAVFSKQEPLQVIYGIGVEEHDQEGRVITLEFENVFVMTVYTPNSRR 112
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL+R+ YR+ QW+ +L +Y+ EL++ KPV+L GDLN AH+EID+ NP N+ +AGF+D+E
Sbjct: 113 GLERIDYRM-QWEEALLSYILELDQKKPVILCGDLNVAHQEIDLKNPKANRNNAGFSDQE 171
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R++F FL GFVD+FR +PD+ G Y++W YR G R N GWR+DYF+VSES+ +++
Sbjct: 172 REAFT-RFLEAGFVDSFRHVYPDLEGAYSWWSYRAGARDRNIGWRIDYFVVSESLKEQIE 230
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
D+ I DVMGSDHCPV LII +
Sbjct: 231 DASISADVMGSDHCPVELIINI 252
>gb|AAP10665.1| Exodeoxyribonuclease III [Bacillus cereus ATCC 14579]
gi|30021833|ref|NP_833464.1| Exodeoxyribonuclease III
[Bacillus cereus ATCC 14579]
Length = 252
Score = 258 bits (660), Expect = 2e-67
Identities = 133/260 (51%), Positives = 181/260 (69%), Gaps = 11/260 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA++ GF LE + D+ CLQE KLQE I+ ++GY +
Sbjct: 1 MKLISWNVNGLRAVIAKGGF--LEYLEESSADIFCLQEIKLQEGQID----LNLEGYY-T 53
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI S+ +PLSV YGLGI +HD EGRL+T EF+ FY+I Y PNS
Sbjct: 54 YWNYAVKK-GYSGTAIFSKKEPLSVTYGLGIEEHDQEGRLITLEFEDFYMITLYTPNSKR 112
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL+RL YR+ +W+ +Y+K L++ KPV+ GDLN AH+EID+ NP N+++ GF+DEE
Sbjct: 113 GLERLDYRM-KWEDDFRSYIKRLDEKKPVIFCGDLNVAHKEIDLKNPKSNRENPGFSDEE 171
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ F+ L GF+DT+R +P+ G Y++W YR G R N GWRLDYF+VSE + D++
Sbjct: 172 REKFSC-ILEEGFIDTYRYLYPNQEGAYSWWSYRMGARAKNIGWRLDYFVVSEGMKDQIT 230
Query: 486 DSYILPDVMGSDHCPVGLII 505
++ I +VMGSDHCPV L I
Sbjct: 231 EAKINSEVMGSDHCPVELHI 250
>ref|NP_623773.1| Exonuclease III [Thermoanaerobacter tengcongensis MB4]
gi|20517232|gb|AAM25377.1| Exonuclease III
[Thermoanaerobacter tengcongensis MB4]
Length = 258
Score = 258 bits (659), Expect = 3e-67
Identities = 134/261 (51%), Positives = 182/261 (69%), Gaps = 9/261 (3%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA L+ +GF ++ + D D+ C+QETKLQE +I+ ++GY +
Sbjct: 1 MKLVSWNVNGLRACLQ-KGF--MDYFKAIDADIFCIQETKLQENQ-KDIEGLDLNGYY-A 55
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
FW + K GYSGTA+ ++ KPLSV YG+G HD EGR++T E+ F+L+ Y PNS
Sbjct: 56 FWNFAEKK-GYSGTAVFTKYKPLSVSYGIGTPHHDKEGRVITLEYKKFFLVNAYTPNSQR 114
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL RL+YR+ +W+ +YL +L+ KPV+L GDLN AH+EIDI NPA N+++AGFTDEE
Sbjct: 115 GLTRLNYRM-EWEEDFRSYLLKLDSVKPVILCGDLNVAHQEIDIKNPAANRRNAGFTDEE 173
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVV-GYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ L+ GF+DTFR +PD YT+W Y H R+ N GWR+DYF+VSE + D +
Sbjct: 174 REKMTI-LLNSGFIDTFRYFYPDKKDAYTWWSYMHNAREKNIGWRVDYFIVSERLKDYLI 232
Query: 486 DSYILPDVMGSDHCPVGLIIK 506
DS I +VMGSDHCPV LI+K
Sbjct: 233 DSQIHSEVMGSDHCPVVLIVK 253
>ref|YP_085078.1| exodeoxyribonuclease III [Bacillus cereus E33L]
gi|51975221|gb|AAU16771.1| exodeoxyribonuclease III
[Bacillus cereus E33L]
Length = 252
Score = 256 bits (655), Expect = 9e-67
Identities = 133/260 (51%), Positives = 178/260 (68%), Gaps = 11/260 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+K +SWNVNGLRA++ GF LE + + D+ CLQE KLQE +I + D Y +
Sbjct: 1 MKFISWNVNGLRAVIAKGGF--LEYLEESNADIFCLQEIKLQE---GQIDLNVEDYY--T 53
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI S+ +PLSV YGLGI +HD EGR++T EF+ FY+I Y PN+
Sbjct: 54 YWNYAVKK-GYSGTAIFSKKEPLSVTYGLGIEEHDQEGRVITLEFEGFYIITLYTPNAKR 112
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL+RL YR+ +W+ Y+K L++ KPV+ GDLN AH+EID+ NP N+K+ GF+DEE
Sbjct: 113 GLERLDYRM-KWEDDFRAYIKRLDEKKPVIFCGDLNVAHKEIDLKNPKSNRKNPGFSDEE 171
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ F L GF+DT+R +PD G Y++W YR G R N GWRLDYF+VSE I +++
Sbjct: 172 REKFTC-ILEEGFIDTYRYLYPDQEGAYSWWSYRMGARAKNIGWRLDYFVVSERIKNQIK 230
Query: 486 DSYILPDVMGSDHCPVGLII 505
D+ I +VMGSDHCPV L I
Sbjct: 231 DAKINSEVMGSDHCPVELHI 250
>ref|ZP_00238934.1| exodeoxyribonuclease III [Bacillus cereus G9241]
gi|47555059|gb|EAL13407.1| exodeoxyribonuclease III
[Bacillus cereus G9241]
Length = 252
Score = 254 bits (650), Expect = 3e-66
Identities = 132/260 (50%), Positives = 179/260 (68%), Gaps = 11/260 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA++ GF LE + + D+ CLQE KLQ+ +I L + Y +
Sbjct: 1 MKLISWNVNGLRAVIAKGGF--LEYLEESNADIFCLQEIKLQD---GQIDLNLEEYY--T 53
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI S+ +PLSV YGLGI +HD EGR++T EF+ FY+I Y PNS
Sbjct: 54 YWNYAVKK-GYSGTAIFSKKEPLSVTYGLGIEEHDQEGRVITLEFEDFYIITLYTPNSKR 112
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL+RL YR+ +W+ Y+K L++ KPV+ GDLN AH+EID+ NP N+K+ GF+DEE
Sbjct: 113 GLERLDYRM-KWEDDFRVYIKRLDEKKPVIFCGDLNVAHKEIDLKNPKSNRKNPGFSDEE 171
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ F L GF+DT+R +P+ G Y++W YR G R N GWRLDYF+VSE + D++
Sbjct: 172 REKFTC-ILEEGFIDTYRYLYPNQEGAYSWWSYRMGARAKNIGWRLDYFVVSERMKDQIT 230
Query: 486 DSYILPDVMGSDHCPVGLII 505
++ I +VMGSDHCPV L I
Sbjct: 231 EAKINSEVMGSDHCPVELHI 250
>ref|YP_020508.1| exodeoxyribonuclease iii [Bacillus anthracis str. 'Ames Ancestor']
gi|30263740|ref|NP_846117.1| exodeoxyribonuclease III
[Bacillus anthracis str. Ames]
gi|49186585|ref|YP_029837.1| exodeoxyribonuclease III
[Bacillus anthracis str. Sterne]
gi|30258384|gb|AAP27603.1| exodeoxyribonuclease III
[Bacillus anthracis str. Ames]
gi|47504307|gb|AAT32983.1| exodeoxyribonuclease III
[Bacillus anthracis str. 'Ames Ancestor']
gi|65321063|ref|ZP_00394022.1| COG0708: Exonuclease III
[Bacillus anthracis str. A2012]
gi|49180512|gb|AAT55888.1| exodeoxyribonuclease III
[Bacillus anthracis str. Sterne]
Length = 252
Score = 254 bits (648), Expect = 6e-66
Identities = 133/260 (51%), Positives = 177/260 (67%), Gaps = 11/260 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+K +SWNVNGLRA++ GF LE + + D+ CLQE KLQE I+ ++GY +
Sbjct: 1 MKFISWNVNGLRAVIAKGGF--LEYLEESNADIFCLQEIKLQEGQID----LNVEGYY-T 53
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI S+ +PLSV YGLGI +HD EGR++T EF+ FY+I Y PNS
Sbjct: 54 YWNYAVKK-GYSGTAIFSKKEPLSVTYGLGIEEHDQEGRVITLEFEDFYIITLYTPNSKR 112
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL+RL YR+ +W+ Y+K L++ K VV GDLN AH+EID+ NP N+K+ GF+DEE
Sbjct: 113 GLERLEYRM-KWEDDFRAYIKRLDEKKSVVFCGDLNVAHKEIDLKNPKSNRKNPGFSDEE 171
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ F L GF+DT+R +PD G Y++W YR G R N GWRLDYF+VSE + D++
Sbjct: 172 REKFTC-ILEEGFIDTYRYLYPDQEGAYSWWSYRMGARAKNIGWRLDYFVVSERMKDQIT 230
Query: 486 DSYILPDVMGSDHCPVGLII 505
+ I +VMGSDHCPV L I
Sbjct: 231 AAKINSEVMGSDHCPVELHI 250
>dbj|BAB80880.1| 3'-exo-deoxyribonuclease [Clostridium perfringens str. 13]
gi|18310156|ref|NP_562090.1| 3'-exo-deoxyribonuclease
[Clostridium perfringens str. 13]
Length = 250
Score = 253 bits (647), Expect = 8e-66
Identities = 131/261 (50%), Positives = 182/261 (69%), Gaps = 12/261 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA +K +GF L+ + ED D+ CLQETKLQE I+ ++GY +
Sbjct: 1 MKLISWNVNGLRACVK-KGF--LDYFKSEDADIFCLQETKLQEGQIDLD----LEGY-HQ 52
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W + K GYSGTAI ++ +PL+V YG+ + HD EGR++T EF+ F+++ Y PNS
Sbjct: 53 YWNYAEKK-GYSGTAIFTKKEPLNVYYGINMEHHDKEGRVITLEFEDFFMVTVYTPNSQS 111
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
L RL YR+ +W+ NYL EL K VV+ GDLN AH+EID+ NP N+K+AGFTDEE
Sbjct: 112 ELARLEYRM-EWEDDFRNYLLELSSKKGVVVCGDLNVAHKEIDLKNPKTNRKNAGFTDEE 170
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R F + LS GF+DTFR +PD+ G Y++W YR RK N GWR+DYFLVS ++ D++
Sbjct: 171 RDKF-STLLSSGFIDTFRYFNPDLEGIYSWWSYRFNARKNNAGWRIDYFLVSNNLEDRIK 229
Query: 486 DSYILPDVMGSDHCPVGLIIK 506
++ I +++GSDHCPV LI++
Sbjct: 230 EASIDTEILGSDHCPVKLIME 250
>ref|YP_037804.1| exodeoxyribonuclease III [Bacillus thuringiensis serovar konkukian
str. 97-27] gi|49329910|gb|AAT60556.1|
exodeoxyribonuclease III [Bacillus thuringiensis serovar
konkukian str. 97-27]
Length = 252
Score = 253 bits (645), Expect = 1e-65
Identities = 132/260 (50%), Positives = 177/260 (67%), Gaps = 11/260 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+K +SWNVNGLRA++ GF LE + + D+ C+QE KLQE I+ ++GY +
Sbjct: 1 MKFISWNVNGLRAVIAKGGF--LEYLEESNADIFCVQEIKLQEGQID----LNVEGYY-T 53
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTAI S+ +PLSV YGLGI +HD EGR++T EF+ FY+I Y PNS
Sbjct: 54 YWNYAVKK-GYSGTAIFSKKEPLSVTYGLGIEEHDQEGRVITLEFEDFYIITLYTPNSKR 112
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL+RL YR+ +W+ Y+K L++ K VV GDLN AH+EID+ NP N+K+ GF+DEE
Sbjct: 113 GLERLEYRM-KWEDDFRAYIKRLDEKKSVVFCGDLNVAHKEIDLKNPKSNRKNPGFSDEE 171
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ F L GF+DT+R +PD G Y++W YR G R N GWRLDYF+VSE + D++
Sbjct: 172 REKFTC-ILEEGFIDTYRYLYPDQEGAYSWWSYRMGARAKNIGWRLDYFVVSERMKDQIT 230
Query: 486 DSYILPDVMGSDHCPVGLII 505
+ I +VMGSDHCPV L I
Sbjct: 231 AAKINSEVMGSDHCPVELHI 250
>ref|ZP_00504743.1| AP endonuclease, family 1:Exodeoxyribonuclease III xth [Clostridium
thermocellum ATCC 27405] gi|67850592|gb|EAM46167.1| AP
endonuclease, family 1:Exodeoxyribonuclease III xth
[Clostridium thermocellum ATCC 27405]
Length = 251
Score = 250 bits (638), Expect = 8e-65
Identities = 131/260 (50%), Positives = 178/260 (68%), Gaps = 12/260 (4%)
Query: 248 KLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENSF 307
KL+SWNVNGLRA + +GF L+ ++ D D+ C+QE+K+Q I E++ +DGY + +
Sbjct: 3 KLISWNVNGLRACIN-KGF--LDYFKKADADIFCIQESKVQPGQI-ELE---LDGY-HQY 54
Query: 308 WTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGDG 367
W + K GYSGTA+ +RIKPL V+ G+GI +HD EGR++T EFD +YL+ Y PN+
Sbjct: 55 WNYAERK-GYSGTAVFTRIKPLCVQNGIGIDEHDREGRVITLEFDNYYLVNVYTPNAKKE 113
Query: 368 LKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEER 427
L+RL YR+ +W+ NYL L+ KPV++ GD+N AH+EID+ NP N++SAGFTDEER
Sbjct: 114 LERLDYRM-KWEDDFRNYLVGLKAKKPVIVCGDMNVAHKEIDLKNPESNRRSAGFTDEER 172
Query: 428 KSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHD 486
F L+ GF+DTFR +PD G YT+W Y R N GWR+DYF VSE + D++
Sbjct: 173 AKF-TELLNAGFIDTFRFFYPDKTGAYTWWSYMFNARARNAGWRIDYFCVSEELKDRLVS 231
Query: 487 SYILPDVMGSDHCPVGLIIK 506
+ I DVMGSDHCPV L IK
Sbjct: 232 ASIHDDVMGSDHCPVELQIK 251
>emb|CAF30568.1| exonuclease III [Methanococcus maripaludis S2]
gi|45358575|ref|NP_988132.1| exonuclease III
[Methanococcus maripaludis S2]
Length = 249
Score = 249 bits (636), Expect = 1e-64
Identities = 130/261 (49%), Positives = 177/261 (67%), Gaps = 13/261 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQL-IDGYEN 305
+K+LSWNVNG+RA LK GF + +RE D++C+QETK+Q + QL +DGY
Sbjct: 1 MKMLSWNVNGIRACLK-NGF--MNFLERESPDIMCIQETKVQSGQV-----QLGLDGYFQ 52
Query: 306 SFWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSG 365
+W + K GYSGTAI ++IKP +V G+ S+H+ EGR++T EFD +YL+ Y PNS
Sbjct: 53 -YWNYAERK-GYSGTAIFTKIKPNNVILGMENSEHNNEGRVITLEFDEYYLVNVYTPNSQ 110
Query: 366 DGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDE 425
GL RL YR +WD +Y+K LE KPVV GDLN AH+EID+ NP N K+AGFT E
Sbjct: 111 RGLTRLEYR-QKWDEDFLSYIKTLETKKPVVFCGDLNVAHKEIDLKNPKTNVKNAGFTPE 169
Query: 426 ERKSFAANFLSRGFVDTFRRQHPDVVGYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
ER F N ++ GF+DTFR + + Y++W YR R N GWR+DYF +S+S+ + +
Sbjct: 170 ERNGFD-NIVNSGFLDTFREFNNEPDNYSWWSYRFNARARNIGWRIDYFCISKSLRNNLK 228
Query: 486 DSYILPDVMGSDHCPVGLIIK 506
D+YI+P+VMGSDHCPVG+I +
Sbjct: 229 DAYIMPEVMGSDHCPVGIIFE 249
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 879,118,196
Number of Sequences: 2540612
Number of extensions: 38468995
Number of successful extensions: 114684
Number of sequences better than 10.0: 797
Number of HSP's better than 10.0 without gapping: 375
Number of HSP's successfully gapped in prelim test: 424
Number of HSP's that attempted gapping in prelim test: 112787
Number of HSP's gapped (non-prelim): 984
length of query: 507
length of database: 863,360,394
effective HSP length: 132
effective length of query: 375
effective length of database: 527,999,610
effective search space: 197999853750
effective search space used: 197999853750
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0278.2


