

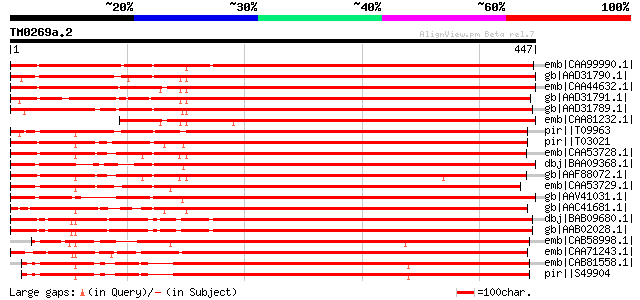
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269a.2
(447 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA99990.1| mitotic cyclin [Sesbania rostrata] 726 0.0
gb|AAD31790.1| mitotic cyclin B1-3 [Lupinus luteus] gi|3253137|g... 686 0.0
emb|CAA44632.1| mitotic cyclin [Glycine max] gi|99895|pir||S1652... 683 0.0
gb|AAD31791.1| mitotic cyclin B1-4 [Lupinus luteus] gi|3253103|g... 662 0.0
gb|AAD31789.1| mitotic cyclin B1-2 [Lupinus luteus] gi|3253135|g... 657 0.0
emb|CAA81232.1| cyclin [Glycine max] 552 e-156
pir||T09963 mitosis-specific cyclin B-type - Madagascar periwink... 550 e-155
pir||T03021 mitosis-specific cyclin CYM, B-type - common tobacco... 544 e-153
emb|CAA53728.1| mitotic-like cyclin [Antirrhinum majus] gi|54209... 517 e-145
dbj|BAA09368.1| B-type cyclin [Nicotiana tabacum] gi|7438486|pir... 515 e-145
gb|AAF88072.1| cyclin [Cicer arietinum] 515 e-144
emb|CAA53729.1| mitotic-like cyclin [Antirrhinum majus] gi|54209... 507 e-142
gb|AAV41031.1| cyclin B-like protein [Nicotiana tabacum] 507 e-142
gb|AAC41681.1| mitotic cyclin gi|7438499|pir||T14916 mitosis-spe... 498 e-139
dbj|BAB09680.1| mitosis-specific cyclin 1b [Arabidopsis thaliana... 476 e-133
gb|AAB02028.1| cyclin gi|2146728|pir||S65734 mitosis-specific cy... 474 e-132
emb|CAB58998.1| CYCB1-1 protein [Petunia x hybrida] 473 e-132
emb|CAA71243.1| mitotic cyclin [Chenopodium rubrum] gi|7438507|p... 468 e-130
emb|CAB81558.1| cyclin B1 [Nicotiana tabacum] 463 e-129
pir||S49904 cyclin - common tobacco 463 e-129
>emb|CAA99990.1| mitotic cyclin [Sesbania rostrata]
Length = 445
Score = 726 bits (1874), Expect = 0.0
Identities = 373/449 (83%), Positives = 402/449 (89%), Gaps = 8/449 (1%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITR 60
MASRP+VPQQPRGDA +G GKQQ KKNGAA GRNR+ALGDIGNL VRG+EVKPNRPITR
Sbjct: 1 MASRPIVPQQPRGDAALGAGKQQ-KKNGAADGRNRKALGDIGNLVTVRGVEVKPNRPITR 59
Query: 61 SFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKP 120
SFCAQLLANAQAA AAENNKKQACPNVAGPP V GVAVAKR APKP K+VT KPKP
Sbjct: 60 SFCAQLLANAQAAAAAENNKKQACPNVAGPPPVVE--GVAVAKRVAPKPGQKKVTTKPKP 117
Query: 121 VVEVIEISPDEQIKKEKSVQKKKEDSKKK---TLTSVLTARSKAACGLTNKPKEEIVDID 177
EVIEISPDE++ K+ + +K+ + + KK T +SVLTARSKAACGLTNKPKE I+DID
Sbjct: 118 E-EVIEISPDEEVHKDNNKKKEGDANTKKKSHTYSSVLTARSKAACGLTNKPKE-IIDID 175
Query: 178 ASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFE 237
A+D NELAAVEYIEDIYKFYKMVENESRPH YM SQPEINE+MRAILVDWLIDVH+KF+
Sbjct: 176 AADTANELAAVEYIEDIYKFYKMVENESRPHDYMDSQPEINERMRAILVDWLIDVHSKFD 235
Query: 238 LSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAY 297
LSLETLYLTINIVDRFLAVKTVPRRELQLVGIS+MLMA+KYEEIWPPEVNDFVCLSDRAY
Sbjct: 236 LSLETLYLTINIVDRFLAVKTVPRRELQLVGISAMLMASKYEEIWPPEVNDFVCLSDRAY 295
Query: 298 SHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTL 357
+HEQIL MEKIILG+LEWTLTVPTPFVFL RFIKASVPDE + NMAHFLSELGMMHY TL
Sbjct: 296 THEQILFMEKIILGKLEWTLTVPTPFVFLVRFIKASVPDEALENMAHFLSELGMMHYATL 355
Query: 358 MYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLK 417
MYC SM+AASAVYAARCTLNKSP WNETLK HT YSEEQLMDCARLLVS H TVGNGKLK
Sbjct: 356 MYCSSMVAASAVYAARCTLNKSPVWNETLKQHTGYSEEQLMDCARLLVSLHSTVGNGKLK 415
Query: 418 VVFRKYSDPERGAVAVLPPAKNLMPQEAS 446
VV+RKYSDPERG+VAVLPPAKNL+ + S
Sbjct: 416 VVYRKYSDPERGSVAVLPPAKNLLSEGKS 444
>gb|AAD31790.1| mitotic cyclin B1-3 [Lupinus luteus] gi|3253137|gb|AAC61889.1|
cyclin [Lupinus luteus] gi|7438501|pir||T10526 cyclin
B1c-ll - yellow lupine
Length = 459
Score = 686 bits (1769), Expect = 0.0
Identities = 354/457 (77%), Positives = 397/457 (86%), Gaps = 18/457 (3%)
Query: 1 MASRPVVP---QQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRP 57
MASRPVVP QQ RG+ V+GGGKQ KKN AA G+NRRALGDIGNLD V+G+EVKPNRP
Sbjct: 1 MASRPVVPIHQQQVRGEGVIGGGKQ--KKNVAADGKNRRALGDIGNLDRVKGVEVKPNRP 58
Query: 58 ITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGV---AVAKREAPKPVTKRV 114
ITRSFCAQLLANAQ A A ENNKK A PNV G A P V AVAKR APKP K+V
Sbjct: 59 ITRSFCAQLLANAQVAAAVENNKKLAIPNVGG-----AKPNVVEGAVAKRVAPKPAEKKV 113
Query: 115 TGKPKPVVEVIEISPDEQIKKEKSVQKKKE--DSKKK--TLTSVLTARSKAACGLTNKPK 170
KPKP E +EISP E+++K KSV KKKE ++KKK TL+SVLTARSKAACGLT KP+
Sbjct: 114 VEKPKPR-EAVEISPHEEVQKNKSVVKKKEGGENKKKPQTLSSVLTARSKAACGLTKKPQ 172
Query: 171 EEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLI 230
E+I+DIDA+D NELAA+EYIEDIYKFYK+ E+ESRPH Y+ SQPEINE+MRAILVDWLI
Sbjct: 173 EQIIDIDANDSGNELAALEYIEDIYKFYKLEESESRPHQYLDSQPEINERMRAILVDWLI 232
Query: 231 DVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFV 290
DV+ KF+LSLETLYLTINIVDRFLAVK VPRRELQL+GIS+ML+A+KYEEIWPPEVNDFV
Sbjct: 233 DVNNKFDLSLETLYLTINIVDRFLAVKVVPRRELQLLGISAMLLASKYEEIWPPEVNDFV 292
Query: 291 CLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELG 350
CLSDRAY+HEQILVMEKIILG+LEWTLTVPTP+VFL RFIKASVPD+ + NM+HFLSELG
Sbjct: 293 CLSDRAYTHEQILVMEKIILGKLEWTLTVPTPYVFLVRFIKASVPDQELENMSHFLSELG 352
Query: 351 MMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCT 410
MMHY TLMYCPSM+AASAV+AARCTLNK+P WNETLKLHT YSEEQLMDCA+LLVSFH T
Sbjct: 353 MMHYSTLMYCPSMVAASAVFAARCTLNKTPFWNETLKLHTSYSEEQLMDCAKLLVSFHST 412
Query: 411 VGNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQEASS 447
+G GKLKVV RKYSDP++GAVAVLPPAK LMP+ +SS
Sbjct: 413 IGGGKLKVVHRKYSDPQKGAVAVLPPAKYLMPESSSS 449
>emb|CAA44632.1| mitotic cyclin [Glycine max] gi|99895|pir||S16522 mitosis-specific
cyclin S13-6 - soybean gi|116157|sp|P25011|CCNB1_SOYBN
G2/mitotic-specific cyclin S13-6 (B-like cyclin)
Length = 454
Score = 683 bits (1762), Expect = 0.0
Identities = 358/456 (78%), Positives = 398/456 (86%), Gaps = 14/456 (3%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGI-EVKPNRPIT 59
MASR V QQ RG+AVVGGGKQQ KKNG A GRNR+ALGDIGNL VRG+ + KPNRPIT
Sbjct: 1 MASRIVQQQQARGEAVVGGGKQQ-KKNGVADGRNRKALGDIGNLANVRGVVDAKPNRPIT 59
Query: 60 RSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPK 119
RSF AQLLANAQAA AA+N+K+QAC NVAGPPA VA GVAVAKR APKPV+K+V KPK
Sbjct: 60 RSFGAQLLANAQAAAAADNSKRQACANVAGPPA-VANEGVAVAKRAAPKPVSKKVIVKPK 118
Query: 120 PVVEVIEI--SPDEQIKKEKSVQKKKE---DSKKK---TLTSVLTARSKAACGLTNKPKE 171
P +V +I SPD KKE KKKE + KKK TLTSVLTARSKAACG+TNKPKE
Sbjct: 119 PSEKVTDIDASPD---KKEVLKDKKKEGDANPKKKSQHTLTSVLTARSKAACGITNKPKE 175
Query: 172 EIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLID 231
+I+DIDASDVDNELAAVEYI+DIYKFYK+VENESRPH Y+ SQPEINE+MRAILVDWLID
Sbjct: 176 QIIDIDASDVDNELAAVEYIDDIYKFYKLVENESRPHDYIGSQPEINERMRAILVDWLID 235
Query: 232 VHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVC 291
VHTKFELSLETLYLTINI+DRFLAVKTVPRRELQLVGIS+MLMA+KYEEIWPPEVNDFVC
Sbjct: 236 VHTKFELSLETLYLTINIIDRFLAVKTVPRRELQLVGISAMLMASKYEEIWPPEVNDFVC 295
Query: 292 LSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGM 351
LSDRAY+HE IL MEK IL +LEWTLTVPTP VFL RFIKASVPD+ + NMAHFLSELGM
Sbjct: 296 LSDRAYTHEHILTMEKTILNKLEWTLTVPTPLVFLVRFIKASVPDQELDNMAHFLSELGM 355
Query: 352 MHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTV 411
M+Y TLMYCPSM+AASAV AARCTLNK+P WNETLKLHT YS+EQLMDCARLLV F+ T+
Sbjct: 356 MNYATLMYCPSMVAASAVLAARCTLNKAPFWNETLKLHTGYSQEQLMDCARLLVGFYSTL 415
Query: 412 GNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQEASS 447
NGKL+VV+RKYSDP++GAVAVLPPAK L+P+ ++S
Sbjct: 416 ENGKLRVVYRKYSDPQKGAVAVLPPAKFLLPEGSAS 451
>gb|AAD31791.1| mitotic cyclin B1-4 [Lupinus luteus] gi|3253103|gb|AAC24245.1|
cyclin CycB1d-ll [Lupinus luteus] gi|7438502|pir||T10527
cyclin B1d-ll - yellow lupine
Length = 452
Score = 662 bits (1707), Expect = 0.0
Identities = 346/449 (77%), Positives = 386/449 (85%), Gaps = 16/449 (3%)
Query: 1 MASRPVV--PQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPI 58
MASRP V QQ R + VVGGGKQQ KNGA G+NR+ALGDIGNLD VKPNRP+
Sbjct: 1 MASRPTVRLQQQARDEEVVGGGKQQ--KNGAGDGKNRKALGDIGNLD-----HVKPNRPV 53
Query: 59 TRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKP 118
TRSFCAQLLANAQAA AAEN KK A PNVA P P A GV VAKR APKP K+VT KP
Sbjct: 54 TRSFCAQLLANAQAAAAAENKKKLAIPNVADPK-PNVADGV-VAKRVAPKPAEKKVTAKP 111
Query: 119 KPVVEVIEISPDEQIKKEKSVQKKKE--DSKKK--TLTSVLTARSKAACGLTNKPKEEIV 174
K EVIEI+P E+ +K KSV KKKE ++KKK TLTSVLTARSKAACGLTNKPKE+I+
Sbjct: 112 K-AQEVIEINPAEEAQKNKSVNKKKEGGENKKKSRTLTSVLTARSKAACGLTNKPKEKII 170
Query: 175 DIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHT 234
DIDA D NELAAVEYIEDIYKFYK+ ENE+RPH YM SQP+INEKMRAILVDWLI+VHT
Sbjct: 171 DIDAGDSGNELAAVEYIEDIYKFYKLAENENRPHQYMDSQPDINEKMRAILVDWLINVHT 230
Query: 235 KFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSD 294
KF+LSLETLYLTINI+DRFLA+KTVPR+ELQLVGIS+MLMA+KYEEIWPPEV++FVCLSD
Sbjct: 231 KFDLSLETLYLTINIIDRFLALKTVPRKELQLVGISAMLMASKYEEIWPPEVDEFVCLSD 290
Query: 295 RAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHY 354
RA+ HE++L MEKIILG+LEWTLTVPTP+VFL RFIKASVPD+ + NMAHFLSELGMMHY
Sbjct: 291 RAFIHEEVLAMEKIILGKLEWTLTVPTPYVFLVRFIKASVPDQELENMAHFLSELGMMHY 350
Query: 355 DTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNG 414
TLMYCPSMIAASAV+AARCTLNK+P WNETLKLHT YS+EQLMDCA+LLVSFH ++
Sbjct: 351 GTLMYCPSMIAASAVFAARCTLNKTPIWNETLKLHTGYSKEQLMDCAKLLVSFHSSIRGE 410
Query: 415 KLKVVFRKYSDPERGAVAVLPPAKNLMPQ 443
KLKV++RKYSDPERGAVAVL PAKNLM +
Sbjct: 411 KLKVLYRKYSDPERGAVAVLSPAKNLMSE 439
>gb|AAD31789.1| mitotic cyclin B1-2 [Lupinus luteus] gi|3253135|gb|AAC61888.1|
cyclin [Lupinus luteus] gi|7438500|pir||T10525 cyclin
B1b-ll - yellow lupine
Length = 454
Score = 657 bits (1696), Expect = 0.0
Identities = 334/454 (73%), Positives = 383/454 (83%), Gaps = 14/454 (3%)
Query: 1 MASRPVVPQQP---RGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRP 57
MASRP+VP QP +G+ V+GG ++Q KNGAA G+NR LGDIGNLD V+G + NRP
Sbjct: 1 MASRPIVPLQPQQAKGEGVIGGRRKQENKNGAANGKNRVVLGDIGNLDRVKGANINLNRP 60
Query: 58 ITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGK 117
ITRS CAQLLA A+A EN+K A PNV GP P A GV +R APKP K+VT K
Sbjct: 61 ITRSLCAQLLAKAEAG---ENDKNLAIPNVTGPK-PQVADGVVAKRRVAPKPAEKKVTAK 116
Query: 118 PKPVVEVIEISPDEQIKKEKSVQKKKED----SKKK--TLTSVLTARSKAACGLTNKPKE 171
PKPV E++EIS ++++K+KS K KE SKKK TLTSVLTARSKAACGLT KPK+
Sbjct: 117 PKPV-EIVEISSGKEVQKDKSANKNKEQGDALSKKKSQTLTSVLTARSKAACGLTEKPKD 175
Query: 172 EIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLID 231
+I+DIDA D NELAAVEYIED+YKFYK+ ENE+RPH YM SQPEINE+MRAILVDWLID
Sbjct: 176 QIIDIDAGDSRNELAAVEYIEDMYKFYKLAENENRPHQYMDSQPEINERMRAILVDWLID 235
Query: 232 VHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVC 291
V TKF+LSLETLYLTINIVDRFLAVKTV RRELQLVG+S+MLMA+KYEEIWPPEVNDFVC
Sbjct: 236 VQTKFDLSLETLYLTINIVDRFLAVKTVLRRELQLVGVSAMLMASKYEEIWPPEVNDFVC 295
Query: 292 LSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGM 351
L+DRAY+HEQILVMEKIILG+LEWTLTVPT FVFLTRFIKASVPD+ + NM HFLSELGM
Sbjct: 296 LTDRAYTHEQILVMEKIILGKLEWTLTVPTTFVFLTRFIKASVPDQELENMGHFLSELGM 355
Query: 352 MHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTV 411
MHY TL+YCPSM+AASAV+AARCTLNK+P WNETL+LHT YSEEQLMDCARLLVSFH T+
Sbjct: 356 MHYATLVYCPSMVAASAVFAARCTLNKTPIWNETLQLHTGYSEEQLMDCARLLVSFHSTL 415
Query: 412 GNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQEA 445
NGKLKV++RKYSDP+RGAV++ PPAKNLMP+ +
Sbjct: 416 ANGKLKVLYRKYSDPQRGAVSMHPPAKNLMPESS 449
>emb|CAA81232.1| cyclin [Glycine max]
Length = 373
Score = 552 bits (1422), Expect = e-156
Identities = 286/367 (77%), Positives = 318/367 (85%), Gaps = 16/367 (4%)
Query: 94 VAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEI--SPDEQIKKEKSVQKKKE---DSKK 148
VA GVAVAKR APKPV+K+V KPKP +V +I SPD KK KKKE + KK
Sbjct: 7 VANEGVAVAKRAAPKPVSKKVIVKPKPSEKVTDIDASPD---KKRVLKDKKKEGDANPKK 63
Query: 149 K---TLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNELAAVE-----YIEDIYKFYKM 200
K TLTSVLTARSKAACG+TNKPKE+I+DIDASDVDNELAAVE YI+DIYKFYK+
Sbjct: 64 KSQHTLTSVLTARSKAACGITNKPKEQIIDIDASDVDNELAAVELAAVEYIDDIYKFYKL 123
Query: 201 VENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVP 260
VENESRPH Y+ SQPEINE+MRAILVDWLIDVHTKFELSLETLYLTINI+DRFLAVKTVP
Sbjct: 124 VENESRPHDYIGSQPEINERMRAILVDWLIDVHTKFELSLETLYLTINIIDRFLAVKTVP 183
Query: 261 RRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVP 320
RRELQLVGIS+MLMA+KYEEIWPPEVNDFVCLSDRAY+HE IL MEK IL +LEWTLTVP
Sbjct: 184 RRELQLVGISAMLMASKYEEIWPPEVNDFVCLSDRAYTHEHILTMEKTILNKLEWTLTVP 243
Query: 321 TPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSP 380
TP VFL RFIKASVPD+ + NMAHFLSELGMM+Y TLMYCPSM+AASAV AARCTLNK+P
Sbjct: 244 TPLVFLVRFIKASVPDQELDNMAHFLSELGMMNYATLMYCPSMVAASAVLAARCTLNKAP 303
Query: 381 AWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
WNETLKLHT YS+EQLMDCARLLV FH T+ NGKL+VV+RKYSDP++GAVAVLPPAK L
Sbjct: 304 FWNETLKLHTGYSQEQLMDCARLLVGFHSTLENGKLRVVYRKYSDPQKGAVAVLPPAKFL 363
Query: 441 MPQEASS 447
+P+ ++S
Sbjct: 364 LPEGSAS 370
>pir||T09963 mitosis-specific cyclin B-type - Madagascar periwinkle
gi|2190261|dbj|BAA20411.1| B-type cyclin [Catharanthus
roseus]
Length = 436
Score = 550 bits (1417), Expect = e-155
Identities = 299/449 (66%), Positives = 347/449 (76%), Gaps = 22/449 (4%)
Query: 1 MASRPVV--PQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKP---- 54
MA+R VV PQQ R DA G KQ KN A G+ RRALGDIGNL VRG+E KP
Sbjct: 1 MATRNVVQVPQQNR-DAAPAGMKQ---KNMAGEGKIRRALGDIGNLVTVRGVEGKPLPQV 56
Query: 55 NRPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRV 114
+RPITRSFCAQLLANAQAA AENNKK NV G AA G + K K
Sbjct: 57 SRPITRSFCAQLLANAQAAAVAENNKKCKAVNVDG-----AADGGIIPKGARKPAQKKAA 111
Query: 115 TGKPKPVVEVIEISPD-EQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEI 173
T KPKP VIEIS D EQ+KKEK KK TLTS LTARSKAACGL+ KPK++I
Sbjct: 112 TIKPKPEA-VIEISSDSEQVKKEKKPSKKDAP----TLTSTLTARSKAACGLSKKPKDQI 166
Query: 174 VDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVH 233
+DIDA+D DNELA VEY+EDIYKFYK+ ENESR H YM SQPEIN+KMRAIL+DWLI+VH
Sbjct: 167 IDIDAADADNELAVVEYVEDIYKFYKIAENESRVHNYMDSQPEINDKMRAILIDWLIEVH 226
Query: 234 TKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLS 293
KFEL+ ETLYLTINIVDR+LAV+T R+ELQLVG+S+ML+A+KYEEIW PEVNDFVC+S
Sbjct: 227 HKFELNPETLYLTINIVDRYLAVQTTLRKELQLVGMSAMLIASKYEEIWAPEVNDFVCIS 286
Query: 294 DRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMH 353
DRAYSH+Q+LVMEK ILG LEW LTVPTP+VFL RFIKASVPD + NM +F +ELGMM+
Sbjct: 287 DRAYSHQQVLVMEKRILGGLEWNLTVPTPYVFLVRFIKASVPDSNMENMVYFFAELGMMN 346
Query: 354 YD-TLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVG 412
Y +MYC SMIAASAVYAARCT NK+P+W++TLKLHT +SE QL+DCA++LV+ H
Sbjct: 347 YSVAMMYCSSMIAASAVYAARCTFNKTPSWDDTLKLHTGFSEYQLIDCAKVLVNLHAMAA 406
Query: 413 NGKLKVVFRKYSDPERGAVAVLPPAKNLM 441
+ KLKV+FRKYS ERG+VA+LPPAK+L+
Sbjct: 407 DNKLKVIFRKYSSLERGSVALLPPAKSLL 435
>pir||T03021 mitosis-specific cyclin CYM, B-type - common tobacco
gi|2196453|dbj|BAA20425.1| B-type cyclin [Nicotiana
tabacum]
Length = 446
Score = 544 bits (1401), Expect = e-153
Identities = 294/448 (65%), Positives = 349/448 (77%), Gaps = 15/448 (3%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGN-LDPVRGIEVKP--NRP 57
MASR V+ QQ G+AV G KQ+ AA GRNR+ALGDIGN + VRG+E KP RP
Sbjct: 1 MASRNVLQQQNIGEAVPGALKQK-NMAAAAQGRNRKALGDIGNNMVTVRGVEGKPLPQRP 59
Query: 58 ITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGK 117
ITR FCAQLLANAQAA AEN KK V AP+ A GV K A KPV K+ K
Sbjct: 60 ITRGFCAQLLANAQAA--AENQKKSM---VVNGDAPIVAKGVLPVKGAAKKPVQKKAAVK 114
Query: 118 PKPVVEVIEISPD--EQIKKEKSVQKKKEDS--KKKTLTSVLTARSKAACGLTNKPKEEI 173
PKP +VIEISPD EQ+K+ K +K +DS KK TLTS LTARSKAACGL++KPK +I
Sbjct: 115 PKP--DVIEISPDTEEQVKENKQKKKAGDDSSVKKATLTSTLTARSKAACGLSHKPKVQI 172
Query: 174 VDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVH 233
VDIDA+DV+NELA VEY+EDIY FYK+ ENESR H YM SQPEI +MRAIL+DWLI+VH
Sbjct: 173 VDIDAADVNNELAVVEYVEDIYNFYKIAENESRIHDYMDSQPEITARMRAILIDWLIEVH 232
Query: 234 TKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLS 293
KFELS ETLYLTINIVDR+LAV T RRELQLVG+S+ML+A+KYEEIW PEVNDFVC+S
Sbjct: 233 HKFELSQETLYLTINIVDRYLAVTTTSRRELQLVGMSAMLIASKYEEIWAPEVNDFVCIS 292
Query: 294 DRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMH 353
D+AYSHEQ+L MEK ILG+LEW LTVPTP+VFL R+IKA+V + + NM +FL+ELG+M+
Sbjct: 293 DKAYSHEQVLGMEKRILGQLEWYLTVPTPYVFLVRYIKAAVSNAQMENMVYFLAELGLMN 352
Query: 354 YDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGN 413
Y T +YCPSMIAASAVY A+ TLN +P WN+TLKLHT +SE QL+ CA+LLVS+H
Sbjct: 353 YATNIYCPSMIAASAVYVAQHTLNCTPFWNDTLKLHTGFSESQLLGCAKLLVSYHMEAPE 412
Query: 414 GKLKVVFRKYSDPERGAVAVLPPAKNLM 441
KLKV+++KYS PERGAVA+ PPAK+L+
Sbjct: 413 HKLKVIYKKYSKPERGAVALQPPAKSLL 440
>emb|CAA53728.1| mitotic-like cyclin [Antirrhinum majus] gi|542091|pir||S41709
mitosis-specific cyclin 1 - garden snapdragon
gi|461725|sp|P34800|CCN1_ANTMA G2/mitotic-specific
cyclin 1
Length = 473
Score = 517 bits (1332), Expect = e-145
Identities = 286/455 (62%), Positives = 342/455 (74%), Gaps = 26/455 (5%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKP------ 54
M SR +V QQ R +A V G +Q KN A +NRRALGDIGNL VRG++ K
Sbjct: 1 MGSRNIVQQQNRAEAAVPGAMKQ--KNIAGEKKNRRALGDIGNLVTVRGVDGKAKAIPQV 58
Query: 55 NRPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVT--- 111
+RP+TRSFCAQLLANAQ A AA+NNK A + GV +R A V
Sbjct: 59 SRPVTRSFCAQLLANAQTA-AADNNKINA-------KGAIVVDGVLPDRRVAAARVPAQK 110
Query: 112 KRVTGKPKPVVEVIEISPDEQI-KKEKSVQKKK---EDSKKK--TLTSVLTARSKAACGL 165
K KP+P E+I ISPD KKEK ++K+K + +KKK TLTS LTARSKAA G+
Sbjct: 111 KAAVVKPRPE-EIIVISPDSVAEKKEKPIEKEKAAEKSAKKKAPTLTSTLTARSKAASGV 169
Query: 166 TNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAIL 225
K KE+IVDIDA+DV+N+LA VEY+ED+YKFYK VENESRPH YM SQPEINEKMRAIL
Sbjct: 170 KTKTKEQIVDIDAADVNNDLAVVEYVEDMYKFYKSVENESRPHDYMGSQPEINEKMRAIL 229
Query: 226 VDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPE 285
+DWL+ VH KFELS ETLYLTINIVDR+LA +T RRELQLVGI +ML+A+KYEEIW PE
Sbjct: 230 IDWLVQVHHKFELSPETLYLTINIVDRYLASETTIRRELQLVGIGAMLIASKYEEIWAPE 289
Query: 286 VNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHF 345
V++ VC+SD YS +QILVMEK ILG LEW LTVPTP+VFL RFIKAS+ D V NM +F
Sbjct: 290 VHELVCISDNTYSDKQILVMEKKILGALEWYLTVPTPYVFLVRFIKASMTDSDVENMVYF 349
Query: 346 LSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLV 405
L+ELGMM+Y TL+YCPSMIAA++VYAARCTLNK+P WNETL+LHT +SE QLMDCA+LLV
Sbjct: 350 LAELGMMNYATLIYCPSMIAAASVYAARCTLNKAPFWNETLQLHTGFSEPQLMDCAKLLV 409
Query: 406 SFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
+F G+ KLK ++RKYS+ ERGAVA+L PAK++
Sbjct: 410 AFPKMAGDQKLKSIYRKYSNLERGAVALLSPAKSV 444
>dbj|BAA09368.1| B-type cyclin [Nicotiana tabacum] gi|7438486|pir||T03611 cyclin,
B-type - common tobacco
Length = 473
Score = 515 bits (1327), Expect = e-145
Identities = 273/451 (60%), Positives = 336/451 (73%), Gaps = 37/451 (8%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITR 60
MASR V+ QQ RG+AV G KQ KKN A GRNR+ALGDIGN+ RG+E K P
Sbjct: 1 MASRIVLQQQNRGEAVPGAVKQ--KKNMAPEGRNRKALGDIGNVATGRGLEGKKPLP--- 55
Query: 61 SFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKP 120
Q VA K NVA PA A KP K+ T KP P
Sbjct: 56 ----------QKPVAV----KVKGANVAKVPA-------------ARKPAQKKATVKPNP 88
Query: 121 VVEVIEISPDEQIKKEKSVQKKKEDS----KKKTLTSVLTARSKAACGLTNKPKEEIVDI 176
++IEISPD Q K ++ +Q+KK D +K TLTS LTARSKAACGL+ KPKE++VDI
Sbjct: 89 E-DIIEISPDTQEKLKEKMQRKKADKDSLKQKATLTSTLTARSKAACGLSKKPKEQVVDI 147
Query: 177 DASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKF 236
DA+DV+NELA VEY+EDIY FYK+ ENE+R H YM SQPEIN++MRA+L+DWL++VH KF
Sbjct: 148 DAADVNNELAVVEYVEDIYSFYKLAENETRVHDYMDSQPEINDRMRAVLIDWLVEVHQKF 207
Query: 237 ELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRA 296
EL+ ETLYLTINIVDR+LAVKT RRELQL+GIS+ML+A+KYEEIW PEVNDFVC+SD++
Sbjct: 208 ELNPETLYLTINIVDRYLAVKTTSRRELQLLGISAMLIASKYEEIWAPEVNDFVCISDKS 267
Query: 297 YSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDT 356
Y+H+Q+L MEK ILG+LEW LTVPTP+VFL RFIKAS+PD + NM +FL+ELG+M+Y T
Sbjct: 268 YTHDQVLAMEKEILGQLEWYLTVPTPYVFLARFIKASLPDSEIENMVYFLAELGLMNYAT 327
Query: 357 LMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKL 416
++YCPSMIAASAVYAAR TLN++P WNETLKLHT +SE QL++CARLLVS+ KL
Sbjct: 328 IIYCPSMIAASAVYAARHTLNRTPFWNETLKLHTGFSESQLIECARLLVSYQSAAATHKL 387
Query: 417 KVVFRKYSDPERGAVAVLPPAKNLMPQEASS 447
KV+++KYS PERG V++L PAK+L+ +SS
Sbjct: 388 KVIYKKYSSPERGVVSLLTPAKSLLAASSSS 418
>gb|AAF88072.1| cyclin [Cicer arietinum]
Length = 505
Score = 515 bits (1326), Expect = e-144
Identities = 287/457 (62%), Positives = 341/457 (73%), Gaps = 28/457 (6%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKP------ 54
M SR +V QQ R +A V G +Q KN A +NRRALGDIGNL VRG++ K
Sbjct: 1 MGSRNIVQQQNRAEAAVPGAMKQ--KNIAGEKKNRRALGDIGNLVTVRGVDGKAKAIPQV 58
Query: 55 NRPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVT--- 111
+RP+TRSFCAQLLANAQ A AA+NNK A + GV +R A V
Sbjct: 59 SRPVTRSFCAQLLANAQTA-AADNNKINA-------KGAIVVDGVLPDRRVAAARVPAQK 110
Query: 112 KRVTGKPKPVVEVIEISPDEQI-KKEKSVQKKK---EDSKKK--TLTSVLTARSKAACGL 165
K KP+P E+I ISPD KKEK ++K+K + +KKK TLTS LTARSK A G+
Sbjct: 111 KAAVVKPRPE-EIIVISPDSVAEKKEKPIEKEKAAEKSAKKKASTLTSTLTARSKVASGV 169
Query: 166 TNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAIL 225
K KE+IVDIDA+DV N+LA VEY+ED+YKFYK VENESRPH YM SQPEINEKMRAIL
Sbjct: 170 KTKTKEQIVDIDAADVTNDLAVVEYVEDMYKFYKSVENESRPHDYMGSQPEINEKMRAIL 229
Query: 226 VDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPE 285
+DWL+ VH KFELS ETLYLTINIVDR+LA +T RRELQLVGI +ML+A+KYEEIW PE
Sbjct: 230 IDWLVQVHHKFELSPETLYLTINIVDRYLASETTIRRELQLVGIGAMLIASKYEEIWAPE 289
Query: 286 VNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHF 345
V++ VC+SD YS +QILVMEK ILG LEW LTVPTP+VFL RFIKAS+ D V NM +F
Sbjct: 290 VHELVCISDNTYSDKQILVMEKKILGALEWYLTVPTPYVFLVRFIKASMTDSDVENMVYF 349
Query: 346 LSELGMMHYDTLMYCPSMIAASA--VYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARL 403
L+ELGMM+Y TL+YCPSMIAA++ VYAARCTLNK+P WNETL+LHT +SE QLMDCA+L
Sbjct: 350 LAELGMMNYATLIYCPSMIAAASHQVYAARCTLNKAPFWNETLQLHTGFSEPQLMDCAKL 409
Query: 404 LVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
LV+FH G+ KLK ++RKYS+ ERGAVA+L PAK+L
Sbjct: 410 LVAFHKMAGDQKLKSIYRKYSNLERGAVALLSPAKSL 446
>emb|CAA53729.1| mitotic-like cyclin [Antirrhinum majus] gi|542092|pir||S41710
mitosis-specific cyclin 2 - garden snapdragon
gi|461726|sp|P34801|CCN2_ANTMA G2/mitotic-specific
cyclin 2
Length = 441
Score = 507 bits (1306), Expect = e-142
Identities = 283/447 (63%), Positives = 334/447 (74%), Gaps = 23/447 (5%)
Query: 1 MASR-PVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKP----N 55
M SR VV QQ RGD V G KQ K+ A +NRRALGDIGN+ VRG+E K +
Sbjct: 1 MGSRHQVVQQQNRGDVVPGAIKQ---KSMAVEKKNRRALGDIGNVVTVRGVEGKALPQVS 57
Query: 56 RPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVT 115
RPITR FCAQL+ANA+AA AAENNK N G A G KR + ++ T
Sbjct: 58 RPITRGFCAQLIANAEAA-AAENNKNSLAVNAKG------ADGALPIKRAVARVPVQKKT 110
Query: 116 GKPKPVVEVIEISPDEQIKK----EKSVQKKKEDSKKK-TLTSVLTARSKAACGLTNKPK 170
K KP E+IEISPD + KK EK + +K KK TLTS LTARSKAA + KPK
Sbjct: 111 VKSKPQ-EIIEISPDTEKKKAPVLEKEITGEKSLKKKAPTLTSTLTARSKAASVVRTKPK 169
Query: 171 EEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLI 230
E+IVDIDA+DV+N+LA VEY+ED+YKFYK EN+SRPH YM SQPEINEKMRAIL+DWL+
Sbjct: 170 EQIVDIDAADVNNDLAVVEYVEDMYKFYKSAENDSRPHDYMDSQPEINEKMRAILIDWLV 229
Query: 231 DVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFV 290
VH KFELS ETLYLTINIVDR+LA KT RRELQL+G+SSML+A+KYEEIW PEVND V
Sbjct: 230 QVHYKFELSPETLYLTINIVDRYLASKTTSRRELQLLGMSSMLIASKYEEIWAPEVNDLV 289
Query: 291 CLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGV-TNMAHFLSEL 349
C+SD +YS+EQ+L MEK ILG LEW LTVPTP+VFL RFIKAS+PD V NM +FL+EL
Sbjct: 290 CISDGSYSNEQVLRMEKKILGALEWYLTVPTPYVFLVRFIKASLPDSDVEKNMVYFLAEL 349
Query: 350 GMMHYDT-LMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFH 408
GMM+Y T +MYCPSMIAA+AVYAARCTLNK P WNETL++HT +SE QLMDCA+LL+ FH
Sbjct: 350 GMMNYATIIMYCPSMIAAAAVYAARCTLNKMPIWNETLRMHTGFSEVQLMDCAKLLIDFH 409
Query: 409 CTVGNGKLKVVFRKYSDPERGAVAVLP 435
+ KL+ ++RKYS E+GAVA+LP
Sbjct: 410 GGSTDQKLQGIYRKYSRLEKGAVALLP 436
>gb|AAV41031.1| cyclin B-like protein [Nicotiana tabacum]
Length = 473
Score = 507 bits (1305), Expect = e-142
Identities = 265/452 (58%), Positives = 333/452 (73%), Gaps = 39/452 (8%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITR 60
MASR V+ QQ RG+AV G KQ K+N A GRNR+ALGDIGN+ RG+E K +P+ +
Sbjct: 1 MASRIVLQQQNRGEAVPGAVKQ--KRNMAPEGRNRKALGDIGNVATGRGVEGK--KPLPQ 56
Query: 61 SFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAK-REAPKPVTKRVTGKPK 119
P V G VAK KP K+ T KP
Sbjct: 57 K-----------------------------PVAVKVKGAIVAKVPPVRKPAQKKATVKPN 87
Query: 120 PVVEVIEISPDEQIKKEKSVQKKKED----SKKKTLTSVLTARSKAACGLTNKPKEEIVD 175
P ++IEISPD Q K ++++Q+ K D +K TLTS LTARSKAACGL+ KPKE+IVD
Sbjct: 88 P-EDIIEISPDTQEKLKENMQRTKADKDSLKQKATLTSTLTARSKAACGLSKKPKEQIVD 146
Query: 176 IDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTK 235
IDA+DV+NELA VEY+ED+Y FYK+ ENE+R H YM SQPEIN++MRA+L+DWL++VH K
Sbjct: 147 IDAADVNNELAVVEYVEDVYSFYKLAENETRVHDYMDSQPEINDRMRAVLIDWLVEVHQK 206
Query: 236 FELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR 295
FEL+ ETLYLTINIVDR+LAVKT RRELQLVGIS+ML+A+KYEEIW PEVNDFVC+S++
Sbjct: 207 FELNPETLYLTINIVDRYLAVKTTSRRELQLVGISAMLIASKYEEIWAPEVNDFVCISNK 266
Query: 296 AYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYD 355
+Y+ +Q+L MEK LG+LEW LTVPTP+VFL RFIKAS PD + NM +FL+ELG+M+Y
Sbjct: 267 SYTRDQVLAMEKEFLGQLEWYLTVPTPYVFLARFIKASPPDSEIKNMVYFLAELGLMNYA 326
Query: 356 TLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGK 415
T++YCPSMIAASAVYAAR T+N++P WNETLKLHT +SE QL++CARLLVS+H K
Sbjct: 327 TIIYCPSMIAASAVYAARHTINRTPFWNETLKLHTGFSESQLIECARLLVSYHSAAATHK 386
Query: 416 LKVVFRKYSDPERGAVAVLPPAKNLMPQEASS 447
LKV+++KYS PERG V++L PAK+L+ +SS
Sbjct: 387 LKVIYKKYSSPERGVVSLLTPAKSLLAASSSS 418
>gb|AAC41681.1| mitotic cyclin gi|7438499|pir||T14916 mitosis-specific cyclin -
parsley
Length = 443
Score = 498 bits (1282), Expect = e-139
Identities = 278/456 (60%), Positives = 334/456 (72%), Gaps = 35/456 (7%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKP------ 54
MAS PVV QQ R +A GGG + K A GRNRRALGDIGNL GIE K
Sbjct: 1 MASIPVV-QQNRVEAA-GGGAVKQKNMAAVEGRNRRALGDIGNLVTGHGIEGKQQQIPQV 58
Query: 55 NRPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRV 114
+RP+TR FCAQLLANAQA V ENNKKQ + GV ++ K+V
Sbjct: 59 SRPVTRGFCAQLLANAQAGVV-ENNKKQRA---------IVGDGVVAVRQ-------KKV 101
Query: 115 TGKPKPVVEVIEISPD----EQIKKEKSVQKKKEDSKKK---TLTSVLTARSKAAC-GLT 166
+ KPKP ++I ISPD +++ K + +K E S KK T TS LTARSKAA GLT
Sbjct: 102 SVKPKPE-DIIVISPDTEEADRVNKHLNRKKATEGSLKKKGQTFTSTLTARSKAAAFGLT 160
Query: 167 NKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILV 226
KPKE+IVDIDA+D +NELAAVEY+ED+YKFYK+ E+ESR Y+ QPEIN+KMRAILV
Sbjct: 161 RKPKEQIVDIDAADANNELAAVEYVEDMYKFYKLAEHESRVFDYIDFQPEINQKMRAILV 220
Query: 227 DWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEV 286
DWLI+VH KFEL ETLYL INI+DR+L+ ++V R+ELQLVGISSML A+KYEEIWPPEV
Sbjct: 221 DWLIEVHNKFELMPETLYLAINILDRYLSTESVARKELQLVGISSMLTASKYEEIWPPEV 280
Query: 287 NDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDE-GVTNMAHF 345
ND +SD AY+++Q+L+MEK ILG+LEW LTVPTP+VFL RFIKAS+P+E V NMA F
Sbjct: 281 NDLTKISDNAYTNQQVLIMEKKILGQLEWNLTVPTPYVFLVRFIKASIPNEPAVENMACF 340
Query: 346 LSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLV 405
L+ELGMM+Y T+ YCPSM+AASAVY ARCTL+K+P WNETLK HT +SEEQLM+C R LV
Sbjct: 341 LTELGMMNYATVTYCPSMVAASAVYGARCTLDKAPFWNETLKSHTGFSEEQLMECGRTLV 400
Query: 406 SFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLM 441
FH KL+V++RKYS ERG VA+LPPAK L+
Sbjct: 401 RFHSCATENKLRVIYRKYSLDERGCVAMLPPAKALL 436
>dbj|BAB09680.1| mitosis-specific cyclin 1b [Arabidopsis thaliana]
gi|15239938|ref|NP_196233.1| cyclin 1b (CYC1b)
[Arabidopsis thaliana]
Length = 445
Score = 476 bits (1224), Expect = e-133
Identities = 260/450 (57%), Positives = 324/450 (71%), Gaps = 18/450 (4%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIE---VKP--N 55
MA+R VP+Q RG +V G K Q KNGA ++RRALGDIGNL V G++ +P N
Sbjct: 1 MATRANVPEQVRGAPLVDGLKIQ-NKNGAV--KSRRALGDIGNLVSVPGVQGGKAQPPIN 57
Query: 56 RPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVT 115
RPITRSF AQLLANAQ N + P + P+AA + K + +
Sbjct: 58 RPITRSFRAQLLANAQLERKPINGDNKV-PALGPKRQPLAARNPEAQRAVQKKNLVVKQQ 116
Query: 116 GKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVD 175
KP VEVIE + E KKE ++ K +KK T +SVL+ARSKAACG+ NKPK I+D
Sbjct: 117 TKP---VEVIE-TKKEVTKKEVAMSPK---NKKVTYSSVLSARSKAACGIVNKPK--IID 167
Query: 176 IDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTK 235
ID SD DN LAAVEY++D+Y FYK VE ES+P YM Q E+NEKMRAIL+DWL++VH K
Sbjct: 168 IDESDKDNHLAAVEYVDDMYSFYKEVEKESQPKMYMHIQTEMNEKMRAILIDWLLEVHIK 227
Query: 236 FELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR 295
FEL+LETLYLT+NI+DRFL+VK VP+RELQLVGIS++L+A+KYEEIWPP+VND V ++D
Sbjct: 228 FELNLETLYLTVNIIDRFLSVKAVPKRELQLVGISALLIASKYEEIWPPQVNDLVYVTDN 287
Query: 296 AYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYD 355
AYS QILVMEK ILG LEW LTVPT +VFL RFIKAS+ D + NM HFL+ELGMMHYD
Sbjct: 288 AYSSRQILVMEKAILGNLEWYLTVPTQYVFLVRFIKASMSDPEMENMVHFLAELGMMHYD 347
Query: 356 TLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGK 415
TL +CPSM+AASAVY ARC+LNKSPAW +TL+ HT Y+E ++MDC++LL H G +
Sbjct: 348 TLTFCPSMLAASAVYTARCSLNKSPAWTDTLQFHTGYTESEIMDCSKLLAFLHSRCGESR 407
Query: 416 LKVVFRKYSDPERGAVAVLPPAKNLMPQEA 445
L+ V++KYS E G VA++ PAK+L+ A
Sbjct: 408 LRAVYKKYSKAENGGVAMVSPAKSLLSAAA 437
>gb|AAB02028.1| cyclin gi|2146728|pir||S65734 mitosis-specific cyclin 1b -
Arabidopsis thaliana
Length = 445
Score = 474 bits (1220), Expect = e-132
Identities = 260/450 (57%), Positives = 323/450 (71%), Gaps = 18/450 (4%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIE---VKP--N 55
MA+R VP+Q RG +V G K Q KNGA ++RRALGDIGNL V G++ +P N
Sbjct: 1 MATRANVPEQVRGAPLVDGLKIQ-NKNGAV--KSRRALGDIGNLVSVPGVQGGKAQPPIN 57
Query: 56 RPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVT 115
RPITRSF AQLLANAQ N + P + P+AA + K + +
Sbjct: 58 RPITRSFRAQLLANAQLERKPINGDNKV-PALGPKRQPLAARNPEAQRAVQKKNLVVKQQ 116
Query: 116 GKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVD 175
KP VEVIE + E KKE ++ K +KK T +SVL+ARSKAACG+ NKPK I+D
Sbjct: 117 TKP---VEVIE-TKKEVTKKEVAMSPK---NKKVTYSSVLSARSKAACGIVNKPK--IID 167
Query: 176 IDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTK 235
ID SD DN LAAVEY++D+Y FYK VE ES+P YM Q E+NEKMRAIL+DWL++VH K
Sbjct: 168 IDESDKDNHLAAVEYVDDMYSFYKEVEKESQPRMYMHIQTEMNEKMRAILIDWLLEVHIK 227
Query: 236 FELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR 295
FEL+LETLYLT+NI+DRFL+VK VP+RELQLVGIS++L+A+KYEEIWPP+VND V ++D
Sbjct: 228 FELNLETLYLTVNIIDRFLSVKAVPKRELQLVGISALLIASKYEEIWPPQVNDLVYVTDN 287
Query: 296 AYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYD 355
AYS QILVMEK ILG LEW LTVPT +VFL RFIKAS+ D + NM HFL+ELGMMHYD
Sbjct: 288 AYSSRQILVMEKAILGNLEWYLTVPTQYVFLVRFIKASMSDPEMENMVHFLAELGMMHYD 347
Query: 356 TLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGK 415
TL +CPSM AASAVY ARC+LNKSPAW +TL+ HT Y+E ++MDC++LL H G +
Sbjct: 348 TLTFCPSMQAASAVYTARCSLNKSPAWTDTLQFHTGYTESEIMDCSKLLAFLHSRCGESR 407
Query: 416 LKVVFRKYSDPERGAVAVLPPAKNLMPQEA 445
L+ V++KYS E G VA++ PAK+L+ A
Sbjct: 408 LRAVYKKYSKAENGGVAMVSPAKSLLSAAA 437
>emb|CAB58998.1| CYCB1-1 protein [Petunia x hybrida]
Length = 437
Score = 473 bits (1216), Expect = e-132
Identities = 263/439 (59%), Positives = 314/439 (70%), Gaps = 41/439 (9%)
Query: 19 GGKQQPKKNGAAAGRN-RRALGDIGNLDPVRG---IEVKP----NRPITRSFCAQLLANA 70
GGKQ KNG A GRN RR LGDIGNL V G IE KP +RP TRSFCAQLLANA
Sbjct: 16 GGKQ---KNGQADGRNNRRVLGDIGNL--VTGAPVIEGKPKAQISRPATRSFCAQLLANA 70
Query: 71 QAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEISPD 130
QA E NK + V P K+ + KP VI ISPD
Sbjct: 71 QA----EKNKVKPLAEVVNKV-----------------PAKKKASDKPAVQEAVIVISPD 109
Query: 131 EQIKK---EKS-VQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNELA 186
E++KK EKS + K+K KTLTS LTARSKAACGL+N+PK EI DIDA+D N LA
Sbjct: 110 EEVKKKTIEKSPLSKRKAKKTGKTLTSTLTARSKAACGLSNRPKNEIDDIDAADAANHLA 169
Query: 187 AVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLT 246
VEY+EDIY FYK+ E+ESR + YM QPE+N KMRAILVDWLI+VH KFEL E+LYLT
Sbjct: 170 VVEYVEDIYNFYKLTEDESRVNNYMEFQPELNHKMRAILVDWLIEVHRKFELMPESLYLT 229
Query: 247 INIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVME 306
INI+DRFL++KTVPR+ELQLVGIS+ML+A KYEEIW PEVNDF+ +SD Y+ + IL ME
Sbjct: 230 INILDRFLSMKTVPRKELQLVGISAMLIACKYEEIWAPEVNDFMHISDNVYTRDHILQME 289
Query: 307 KIILGRLEWTLTVPTPFVFLTRFIKASVP--DEGVTNMAHFLSELGMMHY-DTLMYCPSM 363
K ILG+LEW LTVPTP+VFL R+IKA++P D+ + NMA F +ELG+M+Y T+ YCPSM
Sbjct: 290 KAILGKLEWYLTVPTPYVFLVRYIKAAMPSDDQEIQNMAFFFAELGLMNYTTTISYCPSM 349
Query: 364 IAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKY 423
+AASAVYAAR TLNK P W TL+ HT YSEEQLM+C + LVS+H KLK ++RK+
Sbjct: 350 LAASAVYAARGTLNKGPLWTPTLQHHTGYSEEQLMECTKQLVSYHKGAAESKLKAIYRKF 409
Query: 424 SDPERGAVAVLPPAKNLMP 442
S P+RGAVA+ PPA+NL+P
Sbjct: 410 SSPDRGAVALFPPARNLLP 428
>emb|CAA71243.1| mitotic cyclin [Chenopodium rubrum] gi|7438507|pir||T09960
mitosis-specific cyclin 1 - red goosefoot
Length = 446
Score = 468 bits (1205), Expect = e-130
Identities = 268/458 (58%), Positives = 325/458 (70%), Gaps = 33/458 (7%)
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEV-------K 53
MA+R V+ +QPRG G QQ G RNR ALGDIGN+ RG + K
Sbjct: 1 MAARAVLQEQPRG------GVQQKNGQGGER-RNRTALGDIGNVVTARGGAIGAANGIAK 53
Query: 54 P---NRPITRSFCAQLLANAQAAVAAENNKKQACP----NVAGPPAPVAAPGVAVAKREA 106
P NRP+TRSFCAQL+ANAQAA AE KKQ NV G A A GVAVA A
Sbjct: 54 PQAINRPLTRSFCAQLVANAQAA--AEKKKKQGVKAVDVNVGG--ARKAGVGVAVAPAAA 109
Query: 107 PKPVTKRVTGKPKPVVEVIEI-SPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGL 165
+ K+V K K V V E +P K E+ KK+ K T TS LTARSK ACGL
Sbjct: 110 AQ---KKVQEKKKTEVVVAETKTPSGHGKVEQKPASKKK--KTPTFTSFLTARSKEACGL 164
Query: 166 TNKPKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAIL 225
+ KP+E IV+ID V++ELA VEY+EDIY FYK+ E+ESR YM SQP+INEKMR+IL
Sbjct: 165 SKKPQELIVNIDEGSVEDELAVVEYVEDIYSFYKIAEDESRVRDYMDSQPDINEKMRSIL 224
Query: 226 VDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPE 285
VDWLI+VH KFEL ETLYLTINI+DRFL++K VPR+ELQLVGI+SML+A KYEEIW PE
Sbjct: 225 VDWLIEVHYKFELRQETLYLTINIIDRFLSMKIVPRKELQLVGIASMLIACKYEEIWAPE 284
Query: 286 VNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVP-DEGVTNMAH 344
VNDFV +SD+AY EQ+L MEK ILG LEW LTVPTP++FLTR++KASV D + NM++
Sbjct: 285 VNDFVQISDKAYVREQVLCMEKTILGNLEWYLTVPTPYMFLTRYVKASVTLDSEMENMSY 344
Query: 345 FLSELGMMHYD-TLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARL 403
F SELGMM+Y T+ Y PS++AAS+VY ARCTLN SP+W ETLK +T YSE QL++CARL
Sbjct: 345 FFSELGMMNYSTTIKYPPSLLAASSVYTARCTLNNSPSWTETLKHYTGYSENQLLECARL 404
Query: 404 LVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLM 441
LVSFH G+L+ V++K+S P+ GAVA+ PPAK+L+
Sbjct: 405 LVSFHMAAPEGRLRAVYKKFSKPDNGAVALRPPAKSLL 442
>emb|CAB81558.1| cyclin B1 [Nicotiana tabacum]
Length = 425
Score = 463 bits (1192), Expect = e-129
Identities = 256/441 (58%), Positives = 307/441 (69%), Gaps = 48/441 (10%)
Query: 11 PRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKP-----NRPITRSFCAQ 65
PRG+ GGKQ KN A GRNRRALGDIGNL P E KP +RP+TRSFCAQ
Sbjct: 13 PRGEM---GGKQ---KNAQADGRNRRALGDIGNLVPAPAAEGKPKAAQISRPVTRSFCAQ 66
Query: 66 LLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVI 125
LLANAQ E NKK P A V V K+ + K + K V G P
Sbjct: 67 LLANAQE----EKNKK--------PLAEVVNKDVPAKKKASDKEM-KTVGGSP------- 106
Query: 126 EISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNEL 185
+ K+K KTLTS LTARSKAACGL+N+PK EI DID +D DN L
Sbjct: 107 -------------LSKRKAKKSGKTLTSTLTARSKAACGLSNRPKYEIEDIDVADADNHL 153
Query: 186 AAVEYIEDIYKFYKMVENESR-PHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLY 244
AAVEY+EDIY FYK+ E ESR YM QP++N KMRAILVDWLI+VH KFEL E+LY
Sbjct: 154 AAVEYVEDIYNFYKLTEGESRVDDDYMNFQPDLNHKMRAILVDWLIEVHRKFELMPESLY 213
Query: 245 LTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILV 304
LTI I+DRFL++KTVPR+ELQLVGISSML+A KYEEIW PEVNDF+ +SD AY+ EQIL
Sbjct: 214 LTITILDRFLSLKTVPRKELQLVGISSMLIACKYEEIWAPEVNDFIHISDNAYAREQILQ 273
Query: 305 MEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEG--VTNMAHFLSELGMMHYD-TLMYCP 361
MEK ILG+LEW LTVPTP+VFL R+IKA+ P + + NM F +ELG+M+Y T+ Y P
Sbjct: 274 MEKAILGKLEWYLTVPTPYVFLVRYIKAATPSDNQEMENMTFFFAELGLMNYKITISYRP 333
Query: 362 SMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFR 421
SM+AAS+VYAAR TLNK+P W +TL+ HT YSE+QLM+CA++LVS+H KLK ++R
Sbjct: 334 SMLAASSVYAARSTLNKTPLWTQTLQHHTGYSEDQLMECAKILVSYHLDAAESKLKAIYR 393
Query: 422 KYSDPERGAVAVLPPAKNLMP 442
K+S P+RGAVA PPA+NL+P
Sbjct: 394 KFSSPDRGAVAFFPPARNLLP 414
>pir||S49904 cyclin - common tobacco
Length = 449
Score = 463 bits (1192), Expect = e-129
Identities = 256/441 (58%), Positives = 307/441 (69%), Gaps = 48/441 (10%)
Query: 11 PRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKP-----NRPITRSFCAQ 65
PRG+ GGKQ KN A GRNRRALGDIGNL P E KP +RP+TRSFCAQ
Sbjct: 13 PRGEM---GGKQ---KNAQADGRNRRALGDIGNLVPAPAAEGKPKAAQISRPVTRSFCAQ 66
Query: 66 LLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVI 125
LLANAQ E NKK P A V V K+ + K + K V G P
Sbjct: 67 LLANAQE----EKNKK--------PLAEVVNKDVPAKKKASDKEM-KTVGGSP------- 106
Query: 126 EISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNEL 185
+ K+K KTLTS LTARSKAACGL+N+PK EI DID +D DN L
Sbjct: 107 -------------LSKRKAKKSGKTLTSTLTARSKAACGLSNRPKYEIEDIDVADADNHL 153
Query: 186 AAVEYIEDIYKFYKMVENESR-PHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLY 244
AAVEY+EDIY FYK+ E ESR YM QP++N KMRAILVDWLI+VH KFEL E+LY
Sbjct: 154 AAVEYVEDIYNFYKLTEGESRVDDDYMNFQPDLNHKMRAILVDWLIEVHRKFELMPESLY 213
Query: 245 LTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILV 304
LTI I+DRFL++KTVPR+ELQLVGISSML+A KYEEIW PEVNDF+ +SD AY+ EQIL
Sbjct: 214 LTITILDRFLSLKTVPRKELQLVGISSMLIACKYEEIWAPEVNDFIHISDNAYAREQILQ 273
Query: 305 MEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEG--VTNMAHFLSELGMMHYD-TLMYCP 361
MEK ILG+LEW LTVPTP+VFL R+IKA+ P + + NM F +ELG+M+Y T+ Y P
Sbjct: 274 MEKAILGKLEWYLTVPTPYVFLVRYIKAATPSDNQEMENMTFFFAELGLMNYKITISYRP 333
Query: 362 SMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFR 421
SM+AAS+VYAAR TLNK+P W +TL+ HT YSE+QLM+CA++LVS+H KLK ++R
Sbjct: 334 SMLAASSVYAARSTLNKTPLWTQTLQHHTGYSEDQLMECAKILVSYHLDAAESKLKAIYR 393
Query: 422 KYSDPERGAVAVLPPAKNLMP 442
K+S P+RGAVA PPA+NL+P
Sbjct: 394 KFSSPDRGAVAFFPPARNLLP 414
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 746,126,728
Number of Sequences: 2540612
Number of extensions: 31451691
Number of successful extensions: 150985
Number of sequences better than 10.0: 1210
Number of HSP's better than 10.0 without gapping: 831
Number of HSP's successfully gapped in prelim test: 393
Number of HSP's that attempted gapping in prelim test: 148311
Number of HSP's gapped (non-prelim): 1778
length of query: 447
length of database: 863,360,394
effective HSP length: 131
effective length of query: 316
effective length of database: 530,540,222
effective search space: 167650710152
effective search space used: 167650710152
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0269a.2


