

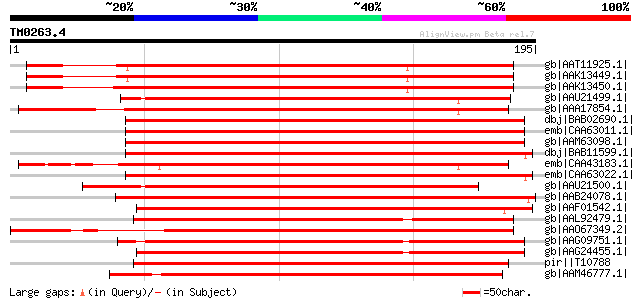
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0263.4
(195 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAT11925.1| oleosin isoform [Arachis hypogaea] 201 7e-51
gb|AAK13449.1| oleosin variant A [Arachis hypogaea] 199 5e-50
gb|AAK13450.1| oleosin variant B [Arachis hypogaea] 198 6e-50
gb|AAU21499.1| oleosin 1 [Arachis hypogaea] 189 3e-47
gb|AAA17854.1| p24 oleosin isoform A [Glycine max] gi|7442013|pi... 184 1e-45
dbj|BAB02690.1| oleosin-like protein [Arabidopsis thaliana] gi|9... 182 5e-45
emb|CAA63011.1| oleosin type4 [Arabidopsis thaliana] 182 5e-45
gb|AAM63098.1| oleosin isoform [Arabidopsis thaliana] 180 2e-44
dbj|BAB11599.1| oleosin, isoform 21K [Arabidopsis thaliana] gi|2... 180 2e-44
emb|CAA43183.1| soybean 24 kDa oleosin isoform [Glycine max] gi|... 178 7e-44
emb|CAA63022.1| oleosin type2 [Arabidopsis thaliana] 177 1e-43
gb|AAU21500.1| oleosin 2 [Arachis hypogaea] 177 2e-43
gb|AAB24078.1| lipid body membrane protein [Daucus carota] gi|82... 176 3e-43
gb|AAF01542.1| putative oleosin [Arabidopsis thaliana] gi|201486... 169 5e-41
gb|AAL92479.1| oleosin [Olea europaea subsp. europaea] 165 6e-40
gb|AAO67349.2| oleosin [Corylus avellana] 158 9e-38
gb|AAG09751.1| oleosin [Perilla frutescens] 155 5e-37
gb|AAG24455.1| 19 kDa oleosin [Perilla frutescens] 154 1e-36
pir||T10788 oleosin, 18.2K - upland cotton gi|266690|sp|P29527|O... 153 3e-36
gb|AAM46777.1| 16.9 kDa oleosin [Theobroma cacao] 150 2e-35
>gb|AAT11925.1| oleosin isoform [Arachis hypogaea]
Length = 176
Score = 201 bits (512), Expect = 7e-51
Identities = 108/183 (59%), Positives = 129/183 (70%), Gaps = 21/183 (11%)
Query: 7 QVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQR-YGGGGGGFSSLFPERGPSASQII 65
QV VHT T R + P+R Y GGG +L PERGPS SQII
Sbjct: 11 QVQVHTPTTQRVDV-------------------PRRGYDVSGGGIKTLLPERGPSTSQII 51
Query: 66 AVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGA 125
AV GVP GGTLLLL+G+SL+ ++IGLA+ TP+ I FSPVIVPAV+TIGLAV GILT+GA
Sbjct: 52 AVLVGVPTGGTLLLLSGLSLLGTIIGLAIATPVFIFFSPVIVPAVVTIGLAVTGILTAGA 111
Query: 126 FGLTGLMSFSWLMNYIRETRG-IVPEQLDYAKNRMADMADYVGQKTKEVGHDIQTKAHEA 184
GLTGLMS SW++N+IR+ G VP+QLD K RMADMADYVGQKTK+ G +IQTKA +
Sbjct: 112 CGLTGLMSLSWMINFIRQVHGTTVPDQLDSVKRRMADMADYVGQKTKDAGQEIQTKAQDV 171
Query: 185 KRT 187
KR+
Sbjct: 172 KRS 174
>gb|AAK13449.1| oleosin variant A [Arachis hypogaea]
Length = 176
Score = 199 bits (505), Expect = 5e-50
Identities = 107/183 (58%), Positives = 127/183 (68%), Gaps = 21/183 (11%)
Query: 7 QVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQR-YGGGGGGFSSLFPERGPSASQII 65
QV VHT T R + P+R Y GGG +L PERGPS SQII
Sbjct: 11 QVQVHTPTTQRVDV-------------------PRRGYDVSGGGIKTLLPERGPSTSQII 51
Query: 66 AVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGA 125
AV GVP GGTLLLL+G+SL+ ++IGLA+ TP+ FSPVIVPAV+TIGLAV GILT+GA
Sbjct: 52 AVLVGVPTGGTLLLLSGLSLLGTIIGLAIATPVFTFFSPVIVPAVVTIGLAVTGILTAGA 111
Query: 126 FGLTGLMSFSWLMNYIRETRG-IVPEQLDYAKNRMADMADYVGQKTKEVGHDIQTKAHEA 184
GLTGLMS SW++N+IR+ G VP+QLD K RMADMADYVGQKTK+ G IQTKA +
Sbjct: 112 CGLTGLMSLSWMINFIRQVHGTTVPDQLDSVKRRMADMADYVGQKTKDAGQQIQTKAQDV 171
Query: 185 KRT 187
KR+
Sbjct: 172 KRS 174
>gb|AAK13450.1| oleosin variant B [Arachis hypogaea]
Length = 176
Score = 198 bits (504), Expect = 6e-50
Identities = 105/182 (57%), Positives = 127/182 (69%), Gaps = 19/182 (10%)
Query: 7 QVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQIIA 66
QV VHT T R + Q + Y GGG + P+RGPS SQIIA
Sbjct: 11 QVQVHTPTTQRVDV------------------QRRGYDVSGGGVKTFLPDRGPSTSQIIA 52
Query: 67 VAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAF 126
V GVP GGTLLLL+G+SL+ ++IGLA+ TP+ FSPVIVPAV+TIGLAV GILT+GA
Sbjct: 53 VLVGVPTGGTLLLLSGLSLLGTIIGLAIATPVFTFFSPVIVPAVVTIGLAVIGILTAGAC 112
Query: 127 GLTGLMSFSWLMNYIRETRG-IVPEQLDYAKNRMADMADYVGQKTKEVGHDIQTKAHEAK 185
GLTGLMS SW++N+IR+ G VP+QLD AK RMADMADYVGQKTK+ G +IQTKA + K
Sbjct: 113 GLTGLMSLSWMINFIRQVHGTTVPDQLDSAKRRMADMADYVGQKTKDAGQEIQTKAQDVK 172
Query: 186 RT 187
R+
Sbjct: 173 RS 174
>gb|AAU21499.1| oleosin 1 [Arachis hypogaea]
Length = 169
Score = 189 bits (481), Expect = 3e-47
Identities = 97/152 (63%), Positives = 121/152 (78%), Gaps = 8/152 (5%)
Query: 42 RYGGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLIL 101
R+G G ++ +P+RGPS S++IAV G+PIGGTLLL AG++L +L+GLAV TPL IL
Sbjct: 18 RFGDTAAG-TNRYPDRGPSTSKVIAVITGLPIGGTLLLFAGLALAGTLLGLAVTTPLFIL 76
Query: 102 FSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMAD 161
FSPVIVPA++ +GL+VAG LTSGA GLTGL SFSW+MNYIR+T G VPEQL+ AK+RMAD
Sbjct: 77 FSPVIVPAIIVVGLSVAGFLTSGACGLTGLSSFSWVMNYIRQTHGSVPEQLEMAKHRMAD 136
Query: 162 MADY-------VGQKTKEVGHDIQTKAHEAKR 186
+A Y VGQKTKEVG +IQTKA ++KR
Sbjct: 137 VAGYVGQKTKDVGQKTKEVGQEIQTKAQDSKR 168
>gb|AAA17854.1| p24 oleosin isoform A [Glycine max] gi|7442013|pir||T06378 oleosin
p24, isoform A - soybean gi|1709459|sp|P29530|OLE1_SOYBN
P24 OLEOSIN ISOFORM A (P89)
Length = 226
Score = 184 bits (467), Expect = 1e-45
Identities = 100/189 (52%), Positives = 122/189 (63%), Gaps = 17/189 (8%)
Query: 4 PQQQVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQ 63
P V VHTT THRYE G P R+E Y G S ERGP+ SQ
Sbjct: 6 PPHSVQVHTTTTHRYEAGVVPPGARFETS----------YEAGVKAASIYHSERGPTTSQ 55
Query: 64 IIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTS 123
++AV AG+P+GG LLLLAG++L +L GLAV TPL +LFSPV+VPA + IGLAVAG LTS
Sbjct: 56 VLAVLAGLPVGGILLLLAGLTLAGTLTGLAVATPLFVLFSPVLVPATVAIGLAVAGFLTS 115
Query: 124 GAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADY-------VGQKTKEVGHD 176
GAFGLT L SFSW++NYIRET+ AK+ +A+ A+Y VGQKTKEVG D
Sbjct: 116 GAFGLTALSSFSWILNYIRETQPASENLAAAAKHHLAEAAEYVGQKTKEVGQKTKEVGQD 175
Query: 177 IQTKAHEAK 185
IQ+KA + +
Sbjct: 176 IQSKAQDTR 184
>dbj|BAB02690.1| oleosin-like protein [Arabidopsis thaliana]
gi|987014|emb|CAA90877.1| oleosin [Arabidopsis thaliana]
gi|17978745|gb|AAL47366.1| oleosin-like protein
[Arabidopsis thaliana] gi|15450920|gb|AAK96731.1|
oleosin-like protein [Arabidopsis thaliana]
gi|25090839|sp|Q42431|OLEO4_ARATH Oleosin 20.3 kDa
(Oleosin type 4) gi|15232227|ref|NP_189403.1|
glycine-rich protein / oleosin [Arabidopsis thaliana]
Length = 191
Score = 182 bits (462), Expect = 5e-45
Identities = 93/148 (62%), Positives = 108/148 (72%)
Query: 44 GGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFS 103
G GG G S + RGPS +QI+A+ AGVPIGGTLL LAG++L S+IGL V PL +LFS
Sbjct: 32 GYGGYGAGSDYKSRGPSTNQILALIAGVPIGGTLLTLAGLTLAGSVIGLLVSIPLFLLFS 91
Query: 104 PVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
PVIVPA +TIGLAV GIL SG FGLTGL S SW++NY+R T VPEQLDYAK RMAD
Sbjct: 92 PVIVPAALTIGLAVTGILASGLFGLTGLSSVSWVLNYLRGTSDTVPEQLDYAKRRMADAV 151
Query: 164 DYVGQKTKEVGHDIQTKAHEAKRTPITT 191
Y G K KE+G +Q KAHEA+ T T
Sbjct: 152 GYAGMKGKEMGQYVQDKAHEARETEFMT 179
>emb|CAA63011.1| oleosin type4 [Arabidopsis thaliana]
Length = 191
Score = 182 bits (462), Expect = 5e-45
Identities = 93/148 (62%), Positives = 108/148 (72%)
Query: 44 GGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFS 103
G GG G S + RGPS +QI+A+ AGVPIGGTLL LAG++L S+IGL V PL +LFS
Sbjct: 32 GYGGYGAGSDYKSRGPSTNQILALIAGVPIGGTLLTLAGLTLAGSVIGLLVSIPLFLLFS 91
Query: 104 PVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
PVIVPA +TIGLAV GIL SG FGLTGL S SW++NY+R T VPEQLDYAK RMAD
Sbjct: 92 PVIVPAALTIGLAVTGILASGLFGLTGLSSVSWVLNYLRGTSDTVPEQLDYAKRRMADAV 151
Query: 164 DYVGQKTKEVGHDIQTKAHEAKRTPITT 191
Y G K KE+G +Q KAHEA+ T T
Sbjct: 152 GYAGMKGKEMGQYVQDKAHEARETEFMT 179
>gb|AAM63098.1| oleosin isoform [Arabidopsis thaliana]
Length = 191
Score = 180 bits (457), Expect = 2e-44
Identities = 92/148 (62%), Positives = 107/148 (72%)
Query: 44 GGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFS 103
G GG G S + RGPS +QI+A+ AGVPIGGTLL LAG++L S+IGL V PL +LFS
Sbjct: 32 GYGGYGAGSDYKSRGPSTNQILALIAGVPIGGTLLTLAGLTLAGSVIGLLVSIPLFLLFS 91
Query: 104 PVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
PVIVPA +TIGLAV GIL SG FGLTGL S SW++NY+R T VPEQLDYAK RMAD
Sbjct: 92 PVIVPAALTIGLAVTGILASGLFGLTGLSSVSWVLNYLRGTSDTVPEQLDYAKRRMADAV 151
Query: 164 DYVGQKTKEVGHDIQTKAHEAKRTPITT 191
Y G K K +G +Q KAHEA+ T T
Sbjct: 152 SYAGMKGKXMGQYVQDKAHEARETEFMT 179
>dbj|BAB11599.1| oleosin, isoform 21K [Arabidopsis thaliana]
gi|21360407|gb|AAM47319.1| AT5g40420/MPO12_130
[Arabidopsis thaliana] gi|15242686|ref|NP_198858.1|
glycine-rich protein / oleosin [Arabidopsis thaliana]
gi|16209649|gb|AAL14385.1| AT5g40420/MPO12_130
[Arabidopsis thaliana] gi|20455173|sp|Q39165|OLEO2_ARATH
Oleosin 21.2 kDa (Oleosin type 2)
gi|725260|gb|AAA87295.1| oleosin
Length = 199
Score = 180 bits (456), Expect = 2e-44
Identities = 89/152 (58%), Positives = 112/152 (73%), Gaps = 1/152 (0%)
Query: 44 GGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFS 103
G GGGG+ S+ PE GPS++Q++++ GVP+ G+LL LAG+ L S+IGL V PL +LFS
Sbjct: 34 GYGGGGYKSMMPESGPSSTQVLSLLIGVPVVGSLLALAGLLLAGSVIGLMVALPLFLLFS 93
Query: 104 PVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
PVIVPA +TIGLA+ G L SG FGLTGL S SW+MNY+R TR VPEQL+YAK RMAD
Sbjct: 94 PVIVPAALTIGLAMTGFLASGMFGLTGLSSISWVMNYLRGTRRTVPEQLEYAKRRMADAV 153
Query: 164 DYVGQKTKEVGHDIQTKAHEAKRTPIT-THDT 194
Y GQK KE+G +Q KA + K+ I+ HDT
Sbjct: 154 GYAGQKGKEMGQHVQNKAQDVKQYDISKPHDT 185
>emb|CAA43183.1| soybean 24 kDa oleosin isoform [Glycine max]
gi|476216|gb|AAA17855.1| p24 oleosin isoform B [Glycine
max] gi|81806|pir||S17936 oleosin (clone P24/91) -
soybean gi|266689|sp|P29531|OLE2_SOYBN P24 OLEOSIN
ISOFORM B (P91)
Length = 223
Score = 178 bits (452), Expect = 7e-44
Identities = 101/190 (53%), Positives = 126/190 (66%), Gaps = 19/190 (10%)
Query: 4 PQQQVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLF-PERGPSAS 62
P V VHTT THRYE G PP R+EA RY G SS++ ERGP+ S
Sbjct: 5 PPHSVQVHTT-THRYEAGVV-PPARFEA---------PRYEAGIKAPSSIYHSERGPTTS 53
Query: 63 QIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILT 122
Q++AV AG+P+GG LLLLAG++L +L GL V TPL I+FSPV++PA + IGLAVAG LT
Sbjct: 54 QVLAVVAGLPVGGILLLLAGLTLAGTLTGLVVATPLFIIFSPVLIPATVAIGLAVAGFLT 113
Query: 123 SGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADY-------VGQKTKEVGH 175
SG FGLT L SFSW++NYIRET+ AK+ +A+ A+Y VGQKTKEVG
Sbjct: 114 SGVFGLTALSSFSWILNYIRETQPASENLAAAAKHHLAEAAEYVGQKTKEVGQKTKEVGQ 173
Query: 176 DIQTKAHEAK 185
DIQ+KA + +
Sbjct: 174 DIQSKAQDTR 183
>emb|CAA63022.1| oleosin type2 [Arabidopsis thaliana]
Length = 199
Score = 177 bits (450), Expect = 1e-43
Identities = 88/152 (57%), Positives = 112/152 (72%), Gaps = 1/152 (0%)
Query: 44 GGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFS 103
G GGGG+ S+ PE GPS++Q++++ GVP+ G+LL LAG+ L S+IGL V PL +LFS
Sbjct: 34 GYGGGGYKSMMPESGPSSTQVLSLLIGVPVVGSLLALAGLLLAGSVIGLMVALPLFLLFS 93
Query: 104 PVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
PVIVPA +TIGLA+ G L SG FGLTGL S SW+MNY+R T+ VPEQL+YAK RMAD
Sbjct: 94 PVIVPAGLTIGLAMTGFLASGMFGLTGLSSISWVMNYLRGTKRTVPEQLEYAKRRMADAV 153
Query: 164 DYVGQKTKEVGHDIQTKAHEAKRTPIT-THDT 194
Y GQK KE+G +Q KA + K+ I+ HDT
Sbjct: 154 GYAGQKGKEMGQHVQNKAQDVKQYDISKPHDT 185
>gb|AAU21500.1| oleosin 2 [Arachis hypogaea]
Length = 150
Score = 177 bits (448), Expect = 2e-43
Identities = 89/147 (60%), Positives = 113/147 (76%), Gaps = 1/147 (0%)
Query: 28 RYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIA 87
R + + + R+G G ++ + +RGPS S++IAV G+PIGGTLLL AG++L
Sbjct: 4 RTQPHAVQVHTTAGRFGDTAAG-TNRYADRGPSTSKVIAVITGLPIGGTLLLFAGLALAG 62
Query: 88 SLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGI 147
+L+GLAV TPL ILFSPVIVPA + +GL+VAG LTSGA GLTGL SFSW+MNYIR+T G
Sbjct: 63 TLLGLAVTTPLFILFSPVIVPATIVVGLSVAGFLTSGACGLTGLSSFSWVMNYIRQTHGS 122
Query: 148 VPEQLDYAKNRMADMADYVGQKTKEVG 174
VPEQL+ AK+RMAD+A YVGQKTK+VG
Sbjct: 123 VPEQLEMAKHRMADVAGYVGQKTKDVG 149
>gb|AAB24078.1| lipid body membrane protein [Daucus carota] gi|82051|pir||JQ0986
lipid body-associated membrane protein - carrot
Length = 180
Score = 176 bits (446), Expect = 3e-43
Identities = 90/157 (57%), Positives = 114/157 (72%), Gaps = 1/157 (0%)
Query: 40 PQRYGGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLL 99
PQ+ GG SL P+ PS SQ++AV +P+GGTLL LAGI+L+ +LIGLAV TPL
Sbjct: 15 PQQTANQPGGVKSLLPKNSPSTSQVLAVVTLLPVGGTLLFLAGITLVGTLIGLAVATPLF 74
Query: 100 ILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRM 159
+LFSPV+VPA +TIGLAV G L SGAFGLTGL S SW+++Y R+ VP+Q++ AK R
Sbjct: 75 LLFSPVLVPAALTIGLAVTGFLGSGAFGLTGLSSLSWVLSYFRQASQRVPDQIELAKKRA 134
Query: 160 ADMADYVGQKTKEVGHDIQTKAHEAKRTPITT-HDTK 195
+MA Y GQKTKEVG IQ+KA +A+ T TT DT+
Sbjct: 135 QEMAAYAGQKTKEVGDTIQSKAAQAQDTTATTGRDTR 171
>gb|AAF01542.1| putative oleosin [Arabidopsis thaliana] gi|20148653|gb|AAM10217.1|
putative oleosin [Arabidopsis thaliana]
gi|16649133|gb|AAL24418.1| putative oleosin [Arabidopsis
thaliana] gi|15232141|ref|NP_186806.1| glycine-rich
protein / oleosin [Arabidopsis thaliana]
Length = 183
Score = 169 bits (427), Expect = 5e-41
Identities = 83/150 (55%), Positives = 109/150 (72%), Gaps = 3/150 (2%)
Query: 48 GGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIV 107
GG L+P+ GPS++Q++AV GVPIGGTLL +AG++L S+IGL + PL ++FSPVIV
Sbjct: 21 GGIKVLYPQSGPSSTQVLAVFVGVPIGGTLLTIAGLTLAGSVIGLMLAFPLFLIFSPVIV 80
Query: 108 PAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVG 167
PA IGLA+ G L SGA GLTGL S SW++NYIR +PE+L+ AK+R+ADMA+YVG
Sbjct: 81 PAAFVIGLAMTGFLASGAIGLTGLSSMSWVLNYIRRAGQHIPEELEEAKHRLADMAEYVG 140
Query: 168 QKTKEVGHDIQTKAH---EAKRTPITTHDT 194
Q+TK+ G I+ KAH EAK + DT
Sbjct: 141 QRTKDAGQTIEDKAHDVREAKTFDVRDRDT 170
>gb|AAL92479.1| oleosin [Olea europaea subsp. europaea]
Length = 165
Score = 165 bits (418), Expect = 6e-40
Identities = 83/141 (58%), Positives = 104/141 (72%), Gaps = 3/141 (2%)
Query: 47 GGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVI 106
GGG S+ P++GPS SQ++AV +P+GGTLL LAG++L+ SLIGLAV TPL I+FSPV+
Sbjct: 23 GGGMKSVLPKKGPSTSQVLAVVTLLPVGGTLLALAGLTLVESLIGLAVTTPLFIIFSPVL 82
Query: 107 VPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYV 166
VPA + +GLAV LTSGAFGLTGL S SW++N++R+ G LD AK+RM D A V
Sbjct: 83 VPATILVGLAVTAFLTSGAFGLTGLSSLSWVVNFLRQVSG---SMLDLAKSRMGDAAIQV 139
Query: 167 GQKTKEVGHDIQTKAHEAKRT 187
GQKTKE G IQ KA E K +
Sbjct: 140 GQKTKETGQTIQQKAPEGKES 160
>gb|AAO67349.2| oleosin [Corylus avellana]
Length = 159
Score = 158 bits (399), Expect = 9e-38
Identities = 86/187 (45%), Positives = 116/187 (61%), Gaps = 28/187 (14%)
Query: 1 MAQPQQQVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPS 60
MA QQ+ VH H + GG + + GG GPS
Sbjct: 1 MADRPQQLQVHPQRGHGHYEGGI----KNQRGG------------------------GPS 32
Query: 61 ASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGI 120
A +++AV A +P+GGTLL LAG++L S+IGL V +PL I+FSPV+VPA + +GLAVA
Sbjct: 33 AVKVMAVLAALPVGGTLLALAGLTLAGSVIGLLVTSPLFIIFSPVLVPAAIVVGLAVASF 92
Query: 121 LTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQTK 180
L+SGA GLTGL S SW++NY+R +P ++D AK RM DMA +VGQKT+EVG +IQ++
Sbjct: 93 LSSGALGLTGLSSLSWVLNYLRCASQSLPREMDQAKRRMQDMAAFVGQKTREVGQEIQSR 152
Query: 181 AHEAKRT 187
A E +RT
Sbjct: 153 AQEGRRT 159
>gb|AAG09751.1| oleosin [Perilla frutescens]
Length = 175
Score = 155 bits (393), Expect = 5e-37
Identities = 84/151 (55%), Positives = 101/151 (66%), Gaps = 5/151 (3%)
Query: 41 QRYGGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLI 100
QRY GG SL P++GPS QI+A+ +PIGGTLL LAGI+L SLIGLA TPL +
Sbjct: 24 QRYEGGA---KSLLPQKGPSTGQILAIITLLPIGGTLLCLAGITLAGSLIGLAFATPLFV 80
Query: 101 LFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMA 160
+FSPV+VPA + LAV LTSGAFGLTGL S SW+ N R+ G E LDYAK RM
Sbjct: 81 IFSPVLVPAAFLLALAVTAFLTSGAFGLTGLSSLSWVFNSFRQATG--QEPLDYAKRRMQ 138
Query: 161 DMADYVGQKTKEVGHDIQTKAHEAKRTPITT 191
+ YVG+KTK+VG I++KA E TT
Sbjct: 139 EGTMYVGEKTKQVGETIKSKAQEGGHDDRTT 169
>gb|AAG24455.1| 19 kDa oleosin [Perilla frutescens]
Length = 177
Score = 154 bits (390), Expect = 1e-36
Identities = 81/144 (56%), Positives = 98/144 (67%), Gaps = 2/144 (1%)
Query: 48 GGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIV 107
GG SL P++GPS QI+A+ +PIGGTLL LAGI+L SLIGLA TPL ++FSPV+V
Sbjct: 30 GGAKSLLPQKGPSTGQILAIITLLPIGGTLLCLAGITLAGSLIGLAFATPLFVIFSPVLV 89
Query: 108 PAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVG 167
PA + LAV LTSGAFGLTGL S SW+ N R+ G E LDYAK RM + YVG
Sbjct: 90 PAAFLLALAVTAFLTSGAFGLTGLSSLSWVFNSFRQATG--QEPLDYAKRRMQEGTMYVG 147
Query: 168 QKTKEVGHDIQTKAHEAKRTPITT 191
+KTK+VG I++KA E TT
Sbjct: 148 EKTKQVGETIKSKAQEGGHDDRTT 171
>pir||T10788 oleosin, 18.2K - upland cotton gi|266690|sp|P29527|OLE6_GOSHI
Oleosin 18.2 kDa gi|167365|gb|AAA18525.1| 18.2 kDa
oleosin gi|167363|gb|AAA18524.1| 18.2 kDa oleosin
Length = 168
Score = 153 bits (386), Expect = 3e-36
Identities = 76/139 (54%), Positives = 100/139 (71%)
Query: 47 GGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVI 106
GGG+ + GPS SQ++AV +PIGGTLL LAG++L ++IGL + TPL I+FSPV+
Sbjct: 26 GGGYGAKNYHSGPSTSQVLAVLTLLPIGGTLLALAGLTLAGTVIGLMLATPLFIIFSPVL 85
Query: 107 VPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYV 166
VPA + I +AV G L+SGAFGLTGL S S+++N +R G LD+AK R+ DM +YV
Sbjct: 86 VPAAIAIAMAVTGFLSSGAFGLTGLSSLSYVLNRLRYATGTEQLDLDHAKRRVQDMTEYV 145
Query: 167 GQKTKEVGHDIQTKAHEAK 185
GQKTKEVG I+ KAHE +
Sbjct: 146 GQKTKEVGQKIENKAHEGQ 164
>gb|AAM46777.1| 16.9 kDa oleosin [Theobroma cacao]
Length = 160
Score = 150 bits (379), Expect = 2e-35
Identities = 78/146 (53%), Positives = 102/146 (69%), Gaps = 3/146 (2%)
Query: 38 NQPQRYGGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTP 97
+Q R+ GG + S GPSA+Q++AV +P+GG LL LAG++L ++IGL V TP
Sbjct: 13 HQHHRFDQGGKNYQSA---SGPSATQVLAVLTLLPVGGILLALAGLTLTGTVIGLCVATP 69
Query: 98 LLILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKN 157
L I+FSPV+VPA + +GLAVAG L+SGAFGLTGL S +++ N +R G +D AK
Sbjct: 70 LFIIFSPVLVPAAIAVGLAVAGFLSSGAFGLTGLSSLAYVFNRLRRATGTEQLDMDQAKR 129
Query: 158 RMADMADYVGQKTKEVGHDIQTKAHE 183
RM DMA YVGQKTKEVG I+ KA+E
Sbjct: 130 RMQDMAGYVGQKTKEVGQKIEGKANE 155
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 356,711,655
Number of Sequences: 2540612
Number of extensions: 16129395
Number of successful extensions: 68626
Number of sequences better than 10.0: 347
Number of HSP's better than 10.0 without gapping: 204
Number of HSP's successfully gapped in prelim test: 146
Number of HSP's that attempted gapping in prelim test: 68270
Number of HSP's gapped (non-prelim): 433
length of query: 195
length of database: 863,360,394
effective HSP length: 121
effective length of query: 74
effective length of database: 555,946,342
effective search space: 41140029308
effective search space used: 41140029308
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0263.4


