

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.1
(346 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
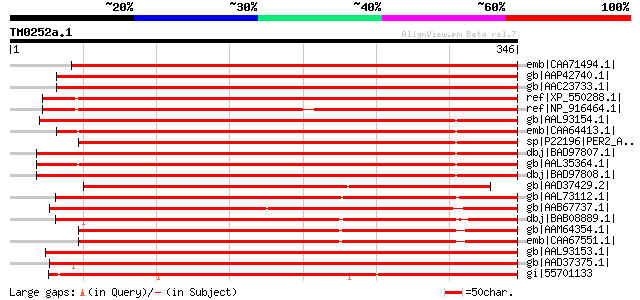
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA71494.1| peroxidase [Spinacia oleracea] gi|7433080|pir||T... 457 e-127
gb|AAP42740.1| At2g41480 [Arabidopsis thaliana] gi|22655091|gb|A... 456 e-127
gb|AAC23733.1| putative peroxidase [Arabidopsis thaliana] gi|743... 456 e-127
ref|XP_550288.1| putative peroxidase [Oryza sativa (japonica cul... 456 e-127
ref|NP_916464.1| putative peroxidase [Oryza sativa (japonica cul... 437 e-121
gb|AAL93154.1| bacterial-induced class III peroxidase [Gossypium... 410 e-113
emb|CAA64413.1| peroxidase precursor [Lycopersicon esculentum] g... 401 e-110
sp|P22196|PER2_ARAHY Cationic peroxidase 2 precursor (PNPC2) gi|... 390 e-107
dbj|BAD97807.1| peroxidase [Nicotiana tabacum] 388 e-106
gb|AAL35364.1| peroxidase [Capsicum annuum] 384 e-105
dbj|BAD97808.1| peroxidase [Nicotiana tabacum] gi|5381255|dbj|BA... 380 e-104
gb|AAD37429.2| peroxidase 4 precursor [Phaseolus vulgaris] 364 2e-99
gb|AAL73112.1| bacterial-induced peroxidase [Gossypium hirsutum] 360 3e-98
gb|AAB67737.1| cationic peroxidase [Stylosanthes humilis] 336 5e-91
dbj|BAB08889.1| peroxidase [Arabidopsis thaliana] gi|28972995|gb... 333 6e-90
gb|AAM64354.1| peroxidase [Arabidopsis thaliana] 328 1e-88
emb|CAA67551.1| peroxidase [Arabidopsis thaliana] gi|10176960|db... 328 2e-88
gb|AAL93153.1| class III peroxidase [Gossypium hirsutum] 313 5e-84
gb|AAD37375.1| peroxidase [Glycine max] 308 2e-82
gi|55701133 TPA: class III peroxidase 133 precursor [Oryza sativ... 302 8e-81
>emb|CAA71494.1| peroxidase [Spinacia oleracea] gi|7433080|pir||T09167 probable
peroxidase (EC 1.11.1.7) (clone PC36) - spinach
(fragment)
Length = 308
Score = 457 bits (1176), Expect = e-127
Identities = 209/304 (68%), Positives = 261/304 (85%)
Query: 43 AQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVG 102
A++QL +Y+SSCP AE VRST++S+FN DPTIAP L+RLHFHDCFVQGCD S+LI G
Sbjct: 5 AKSQLSIAYYASSCPQAEGIVRSTVQSHFNSDPTIAPGLLRLHFHDCFVQGCDASILISG 64
Query: 103 SSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPT 162
+S+ER A N+ L+GF+VI+DAK+Q+E++CPGVVSCADILA+AARD+V L+ GP+W VP
Sbjct: 65 TSSERTAFTNVGLKGFDVIDDAKAQVESVCPGVVSCADILALAARDSVDLTGGPNWGVPL 124
Query: 163 GRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYR 222
GR DG+ SS++ A NLPSPL+ ++V RQKFA KGL+DHDLVTLVGAHTIGQT+CRFF YR
Sbjct: 125 GRLDGKRSSASDAVNLPSPLESIAVHRQKFADKGLNDHDLVTLVGAHTIGQTDCRFFQYR 184
Query: 223 LYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGV 282
LYNFT TGN+DP+INQ +AQ Q LCPKNGNGL KV+LD S KFDV+FFKN+RDGN V
Sbjct: 185 LYNFTPTGNADPSINQPNIAQLQTLCPKNGNGLTKVALDRDSRTKFDVNFFKNIRDGNAV 244
Query: 283 LESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVC 342
LESDQRLWGD ATQ++VQ+YAGN+RGL G++F ++FPKAM+K+S I VK+ ++GE+RK+C
Sbjct: 245 LESDQRLWGDDATQAIVQNYAGNLRGLFGVRFNFDFPKAMVKMSGIGVKSGSDGEVRKMC 304
Query: 343 SKFN 346
SKFN
Sbjct: 305 SKFN 308
>gb|AAP42740.1| At2g41480 [Arabidopsis thaliana] gi|22655091|gb|AAM98136.1|
putative peroxidase [Arabidopsis thaliana]
gi|25453194|sp|O80822|PER25_ARATH Peroxidase 25
precursor (Atperox P25) gi|30688665|ref|NP_181679.2|
peroxidase, putative [Arabidopsis thaliana]
Length = 328
Score = 456 bits (1173), Expect = e-127
Identities = 215/314 (68%), Positives = 264/314 (83%)
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
+++L++ + LK G+YS+SCP AE+ VRST+ES+F+ DPTI+P L+RLHFHDCFVQ
Sbjct: 15 MLVLVLGKEVRSQLLKNGYYSTSCPKAESIVRSTVESHFDSDPTISPGLLRLHFHDCFVQ 74
Query: 93 GCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYL 152
GCDGSVLI G SAE+ AL NL LRG EVI+DAK++LEA+CPGVVSCADILA+AARD+V L
Sbjct: 75 GCDGSVLIKGKSAEQAALPNLGLRGLEVIDDAKARLEAVCPGVVSCADILALAARDSVDL 134
Query: 153 SNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIG 212
S+GPSW VPTGR+DGRIS + +ASNLPSPLD V+VQ+QKF KGLD HDLVTL+GAHTIG
Sbjct: 135 SDGPSWRVPTGRKDGRISLATEASNLPSPLDSVAVQKQKFQDKGLDTHDLVTLLGAHTIG 194
Query: 213 QTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSF 272
QT+C FF YRLYNFT TGNSDPTI+ + L Q + LCP NG+G ++V+LD GSP+KFD SF
Sbjct: 195 QTDCLFFRYRLYNFTVTGNSDPTISPSFLTQLKTLCPPNGDGSKRVALDIGSPSKFDESF 254
Query: 273 FKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKT 332
FKN+RDGN +LESDQRLW D+ T +VV+ YA +RGLLG +F YEF KAMIK+S+I+VKT
Sbjct: 255 FKNLRDGNAILESDQRLWSDAETNAVVKKYASRLRGLLGFRFDYEFGKAMIKMSSIDVKT 314
Query: 333 DTEGEIRKVCSKFN 346
D +GE+RKVCSK N
Sbjct: 315 DVDGEVRKVCSKVN 328
>gb|AAC23733.1| putative peroxidase [Arabidopsis thaliana] gi|7433068|pir||T02443
probable peroxidase (EC 1.11.1.7), cationic -
Arabidopsis thaliana
Length = 357
Score = 456 bits (1173), Expect = e-127
Identities = 215/314 (68%), Positives = 264/314 (83%)
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
+++L++ + LK G+YS+SCP AE+ VRST+ES+F+ DPTI+P L+RLHFHDCFVQ
Sbjct: 44 MLVLVLGKEVRSQLLKNGYYSTSCPKAESIVRSTVESHFDSDPTISPGLLRLHFHDCFVQ 103
Query: 93 GCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYL 152
GCDGSVLI G SAE+ AL NL LRG EVI+DAK++LEA+CPGVVSCADILA+AARD+V L
Sbjct: 104 GCDGSVLIKGKSAEQAALPNLGLRGLEVIDDAKARLEAVCPGVVSCADILALAARDSVDL 163
Query: 153 SNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIG 212
S+GPSW VPTGR+DGRIS + +ASNLPSPLD V+VQ+QKF KGLD HDLVTL+GAHTIG
Sbjct: 164 SDGPSWRVPTGRKDGRISLATEASNLPSPLDSVAVQKQKFQDKGLDTHDLVTLLGAHTIG 223
Query: 213 QTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSF 272
QT+C FF YRLYNFT TGNSDPTI+ + L Q + LCP NG+G ++V+LD GSP+KFD SF
Sbjct: 224 QTDCLFFRYRLYNFTVTGNSDPTISPSFLTQLKTLCPPNGDGSKRVALDIGSPSKFDESF 283
Query: 273 FKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKT 332
FKN+RDGN +LESDQRLW D+ T +VV+ YA +RGLLG +F YEF KAMIK+S+I+VKT
Sbjct: 284 FKNLRDGNAILESDQRLWSDAETNAVVKKYASRLRGLLGFRFDYEFGKAMIKMSSIDVKT 343
Query: 333 DTEGEIRKVCSKFN 346
D +GE+RKVCSK N
Sbjct: 344 DVDGEVRKVCSKVN 357
>ref|XP_550288.1| putative peroxidase [Oryza sativa (japonica cultivar-group)]
gi|55700869|tpe|CAH69244.1| TPA: class III peroxidase 1
precursor [Oryza sativa (japonica cultivar-group)]
gi|55296784|dbj|BAD68110.1| putative peroxidase [Oryza
sativa (japonica cultivar-group)]
Length = 326
Score = 456 bits (1172), Expect = e-127
Identities = 222/324 (68%), Positives = 265/324 (81%), Gaps = 1/324 (0%)
Query: 23 SQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLI 82
S+M A+ +L S + +Q L+ GFY ++CP AE VRST+E Y+N D TIAP L+
Sbjct: 4 SEMSALFFLFSALLRSSLVHSQG-LQIGFYDNNCPDAEDIVRSTVEKYYNNDATIAPGLL 62
Query: 83 RLHFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADIL 142
RLHFHDCFVQGCD SVLI G+S+ER A N +RGFEVI+DAKSQLEA+C GVVSCADIL
Sbjct: 63 RLHFHDCFVQGCDASVLISGASSERTAPQNFGIRGFEVIDDAKSQLEAVCSGVVSCADIL 122
Query: 143 AVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDL 202
A+AARDAV L+ GPSWSVP GRRDGRISS++ A LPSP DPVSVQRQKFAA+GL D +L
Sbjct: 123 ALAARDAVDLTGGPSWSVPLGRRDGRISSASDAKALPSPADPVSVQRQKFAAQGLTDREL 182
Query: 203 VTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDS 262
VTLVGAHTIGQT+C FF YRLYNFT TGN+DPTI+ + L Q +ALCP G+G R+V+LD
Sbjct: 183 VTLVGAHTIGQTDCIFFRYRLYNFTATGNADPTISPSALPQLRALCPPAGDGSRRVALDL 242
Query: 263 GSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAM 322
GSP FDVSFFKNVRDG VLESDQRLWGD+ATQ+ VQS+AGN+RGL GL+F+YEFPKAM
Sbjct: 243 GSPGAFDVSFFKNVRDGGAVLESDQRLWGDAATQAAVQSFAGNVRGLFGLRFSYEFPKAM 302
Query: 323 IKLSNIEVKTDTEGEIRKVCSKFN 346
+++S+I VKT ++GEIR+ CSKFN
Sbjct: 303 VRMSSIAVKTGSQGEIRRKCSKFN 326
>ref|NP_916464.1| putative peroxidase [Oryza sativa (japonica cultivar-group)]
Length = 319
Score = 437 bits (1125), Expect = e-121
Identities = 216/324 (66%), Positives = 258/324 (78%), Gaps = 8/324 (2%)
Query: 23 SQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLI 82
S+M A+ +L S + +Q L+ GFY ++CP AE VRST+E Y+N D TIAP L+
Sbjct: 4 SEMSALFFLFSALLRSSLVHSQG-LQIGFYDNNCPDAEDIVRSTVEKYYNNDATIAPGLL 62
Query: 83 RLHFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADIL 142
RLHFHDCFVQGCD SVLI G+S+ER A N +RGFEVI+DAKSQLEA+C GVVSCADIL
Sbjct: 63 RLHFHDCFVQGCDASVLISGASSERTAPQNFGIRGFEVIDDAKSQLEAVCSGVVSCADIL 122
Query: 143 AVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDL 202
A+AARDAV L+ GPSWSVP GRRDGRISS++ A LPSP DPVSVQRQKFAA+GL D
Sbjct: 123 ALAARDAVDLTGGPSWSVPLGRRDGRISSASDAKALPSPADPVSVQRQKFAAQGLTDR-- 180
Query: 203 VTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDS 262
AHTIGQT+C FF YRLYNFT TGN+DPTI+ + L Q +ALCP G+G R+V+LD
Sbjct: 181 -----AHTIGQTDCIFFRYRLYNFTATGNADPTISPSALPQLRALCPPAGDGSRRVALDL 235
Query: 263 GSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAM 322
GSP FDVSFFKNVRDG VLESDQRLWGD+ATQ+ VQS+AGN+RGL GL+F+YEFPKAM
Sbjct: 236 GSPGAFDVSFFKNVRDGGAVLESDQRLWGDAATQAAVQSFAGNVRGLFGLRFSYEFPKAM 295
Query: 323 IKLSNIEVKTDTEGEIRKVCSKFN 346
+++S+I VKT ++GEIR+ CSKFN
Sbjct: 296 VRMSSIAVKTGSQGEIRRKCSKFN 319
>gb|AAL93154.1| bacterial-induced class III peroxidase [Gossypium hirsutum]
Length = 328
Score = 410 bits (1055), Expect = e-113
Identities = 202/327 (61%), Positives = 255/327 (77%), Gaps = 2/327 (0%)
Query: 21 VSSQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPS 80
VS + +LL +L++ + ++ + GFYS+SCP E+ VRST++S+F DPTIAP
Sbjct: 3 VSCFSQNVLLVTLLLAIAVSLVESQGTRVGFYSTSCPRVESIVRSTVQSHFGSDPTIAPG 62
Query: 81 LIRLHFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCAD 140
L+R+HFHDCFV GCD S+LI G E+ A NL LRG+EVI+DAK+QLEA CPGVVSCAD
Sbjct: 63 LLRMHFHDCFVHGCDASILIDGPGTEKTAPPNLLLRGYEVIDDAKTQLEAACPGVVSCAD 122
Query: 141 ILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDH 200
ILA+AARD+V LS+G SW+VPTGRRDG +S ++ A+NLP D V VQ+QKFAAKGL+
Sbjct: 123 ILALAARDSVVLSSGASWAVPTGRRDGTVSQASDAANLPGFRDSVDVQKQKFAAKGLNTQ 182
Query: 201 DLVTLVGAHTIGQTECRFFSYRLYNFTTTGN-SDPTINQATLAQFQALCPKNGNGLRKVS 259
DLVTLVG HTIG T C+FF YRLYNFTTTGN +DP+I A ++Q QALCP+NG+G R++
Sbjct: 183 DLVTLVGGHTIGTTACQFFRYRLYNFTTTGNGADPSITAAFVSQLQALCPQNGDGSRRIG 242
Query: 260 LDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFP 319
LD+GS +FD SFF N+RDG G+LESDQRLW D++T++ VQ + G IRGLLGL F EF
Sbjct: 243 LDTGSVNRFDNSFFANLRDGKGILESDQRLWTDASTKTFVQRFLG-IRGLLGLTFNIEFG 301
Query: 320 KAMIKLSNIEVKTDTEGEIRKVCSKFN 346
++M+K+SNIEVKT T GEIRKVCSK N
Sbjct: 302 RSMVKMSNIEVKTGTVGEIRKVCSKVN 328
>emb|CAA64413.1| peroxidase precursor [Lycopersicon esculentum]
gi|7433065|pir||T07008 peroxidase (EC 1.11.1.7)
precursor, defense-related - tomato
Length = 332
Score = 401 bits (1031), Expect = e-110
Identities = 191/314 (60%), Positives = 247/314 (77%), Gaps = 2/314 (0%)
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
L+I+ ++ + Q + GFYSS+CP AE+ V+ST+ S+F DPT+AP L+R+HFHDCFVQ
Sbjct: 21 LVIVDVTMVFGQGT-RVGFYSSTCPRAESIVQSTVRSHFQSDPTVAPGLLRMHFHDCFVQ 79
Query: 93 GCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYL 152
GCDGS+LI G+ ER A N +LRGFEVI+DAK Q+EA+CPGVVSCADILA+AARD+V +
Sbjct: 80 GCDGSILISGTGTERTAPPNSNLRGFEVIDDAKQQIEAVCPGVVSCADILALAARDSVLV 139
Query: 153 SNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIG 212
+ G +WSVPTGR DGR+SS++ SNLP + V+ Q+QKFAAKGL+ DLVTLVG HTIG
Sbjct: 140 TKGLTWSVPTGRTDGRVSSASDTSNLPGFTESVAAQKQKFAAKGLNTQDLVTLVGGHTIG 199
Query: 213 QTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSF 272
+ C+FFSYRLYNF +TG DP+I+ L+Q QALCP+NG+G ++V+LD+GS FD S+
Sbjct: 200 TSACQFFSYRLYNFNSTGGPDPSIDATFLSQLQALCPQNGDGSKRVALDTGSVNNFDTSY 259
Query: 273 FKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKT 332
F N+R+G G+LESDQ LW D++T+ VQ Y G +RG LGL+F EF K+M+K+SNIEV T
Sbjct: 260 FSNLRNGRGILESDQILWTDASTKVFVQRYLG-LRGFLGLRFGLEFGKSMVKMSNIEVLT 318
Query: 333 DTEGEIRKVCSKFN 346
T GEIRKVCS FN
Sbjct: 319 GTNGEIRKVCSAFN 332
>sp|P22196|PER2_ARAHY Cationic peroxidase 2 precursor (PNPC2) gi|166475|gb|AAA32676.1|
cationic peroxidase
Length = 330
Score = 390 bits (1001), Expect = e-107
Identities = 185/299 (61%), Positives = 233/299 (77%), Gaps = 1/299 (0%)
Query: 48 KTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAER 107
+ GFYS +CP AE+ VRST+ S+ N DPT+A ++R+HFHDCFVQGCDGS+LI G + E+
Sbjct: 33 RVGFYSRTCPRAESIVRSTVRSHVNSDPTLAAKILRMHFHDCFVQGCDGSILISGPATEK 92
Query: 108 NALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDG 167
A ANL LRG+E+I+DAK+QLEA CPGVVSCADILA+AARD+V LS G SW VPTGRRDG
Sbjct: 93 TAFANLGLRGYEIIDDAKTQLEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRDG 152
Query: 168 RISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFT 227
R+S ++ SNLP+P D V VQ+QKFAAKGL+ DLVTLVG HTIG +EC+FFS RL+NF
Sbjct: 153 RVSQASDVSNLPAPSDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTSECQFFSNRLFNFN 212
Query: 228 TTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQ 287
T +DP I+ + ++ QALCP+N +V+LD+GS KFD S+F N+R+ GVL+SDQ
Sbjct: 213 GTAAADPAIDPSFVSNLQALCPQNTGAANRVALDTGSQFKFDTSYFSNLRNRRGVLQSDQ 272
Query: 288 RLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
LW D +T+S VQ Y G +RG LGL F EF K+M+K+SNI VKT T+GEIRK+CS FN
Sbjct: 273 ALWNDPSTKSFVQRYLG-LRGFLGLTFNVEFGKSMVKMSNIGVKTGTDGEIRKICSAFN 330
>dbj|BAD97807.1| peroxidase [Nicotiana tabacum]
Length = 330
Score = 388 bits (997), Expect = e-106
Identities = 190/328 (57%), Positives = 242/328 (72%), Gaps = 1/328 (0%)
Query: 19 YKVSSQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIA 78
Y S AM + L++ + ++ + GFYSS+CP AE+ V+ST+ ++F DPT+A
Sbjct: 4 YHHSINKMAMFMVILVLAIDVTMVLGQGTRVGFYSSTCPRAESIVQSTVRAHFQSDPTVA 63
Query: 79 PSLIRLHFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSC 138
P ++R+HFHDCFV GCDGS+LI GS AER A+ N +LRGF+VIEDAK Q+EAICPGVVSC
Sbjct: 64 PGILRMHFHDCFVLGCDGSILIEGSDAERTAIPNRNLRGFDVIEDAKKQIEAICPGVVSC 123
Query: 139 ADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLD 198
ADILA+AARD+V + G +WSVPTGRRDGR+S +A A NLP+ D V VQ+QKF AKGL+
Sbjct: 124 ADILALAARDSVVATRGLTWSVPTGRRDGRVSRAADAGNLPAFFDSVDVQKQKFTAKGLN 183
Query: 199 DHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKV 258
DLV L GAHTIG C RL+NF +TG DP+I+ L Q QALCP+NG+ R+V
Sbjct: 184 TQDLVALTGAHTIGTAGCAVIRGRLFNFNSTGGPDPSIDATFLPQLQALCPQNGDAARRV 243
Query: 259 SLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEF 318
+LD+GS FD S+F N+R+G GVLESDQ+LW D++T+ VQ + G IRGLLGL F EF
Sbjct: 244 ALDTGSANNFDTSYFSNLRNGRGVLESDQKLWTDASTKVFVQRFLG-IRGLLGLTFGVEF 302
Query: 319 PKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
++M+K+SNIEVKT T GEIRKVCS N
Sbjct: 303 GRSMVKMSNIEVKTGTNGEIRKVCSAIN 330
>gb|AAL35364.1| peroxidase [Capsicum annuum]
Length = 332
Score = 384 bits (987), Expect = e-105
Identities = 182/328 (55%), Positives = 247/328 (74%), Gaps = 2/328 (0%)
Query: 19 YKVSSQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIA 78
Y ++M +++ ++ + ++ ++ Q + GFYSS+CP AE+ V+ST+ S+F DPT+A
Sbjct: 7 YNSINKMVSIIFILVLAIDLTMVLGQGT-RVGFYSSTCPRAESIVQSTVRSHFQSDPTVA 65
Query: 79 PSLIRLHFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSC 138
P L+ +HFHDCFVQGCD S+LI GS ER A N LRG+EVI+DAK Q+EAICPGVVSC
Sbjct: 66 PGLLTMHFHDCFVQGCDASILISGSGTERTAPPNSLLRGYEVIDDAKQQIEAICPGVVSC 125
Query: 139 ADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLD 198
ADILA+AARD+V ++ G +WSVPTGRRDG +S ++ S+LP + V Q+QKF+AKGL+
Sbjct: 126 ADILALAARDSVLVTKGLTWSVPTGRRDGLVSRASDTSDLPGFTESVDSQKQKFSAKGLN 185
Query: 199 DHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKV 258
DLVTLVG HTIG + C+FFSYRLYNF +TG DP+I+ + L + LCP+NG+G ++V
Sbjct: 186 TQDLVTLVGGHTIGTSACQFFSYRLYNFNSTGGPDPSIDASFLPTLRGLCPQNGDGSKRV 245
Query: 259 SLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEF 318
+LD+GS FD S+F N+R+G G+LESDQ+LW D +T+ +Q Y G +RG LGL+F EF
Sbjct: 246 ALDTGSVNNFDTSYFSNLRNGRGILESDQKLWTDDSTKVFIQRYLG-LRGFLGLRFGVEF 304
Query: 319 PKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
++M+K+SNIEVKT T GEIRKVCS N
Sbjct: 305 GRSMVKMSNIEVKTGTNGEIRKVCSAIN 332
>dbj|BAD97808.1| peroxidase [Nicotiana tabacum] gi|5381255|dbj|BAA82307.1|
peroxidase [Nicotiana tabacum]
Length = 330
Score = 380 bits (977), Expect = e-104
Identities = 185/328 (56%), Positives = 241/328 (73%), Gaps = 1/328 (0%)
Query: 19 YKVSSQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIA 78
Y S AM + L++ + ++ + GFYSS+CP AE+ V+ST+ ++F DPT+A
Sbjct: 4 YHHSINKMAMFMVILVLAIDVTMVLGQGTRVGFYSSTCPRAESIVQSTVRAHFQSDPTVA 63
Query: 79 PSLIRLHFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSC 138
P ++R+HFHDCFV GCDGS+LI GS AER A+ N +L+GF+VIEDAK+Q+EAICPGVVSC
Sbjct: 64 PGILRMHFHDCFVLGCDGSILIEGSDAERTAIPNRNLKGFDVIEDAKTQIEAICPGVVSC 123
Query: 139 ADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLD 198
ADILA+AARD+V + G +WSVPTGRRDGR+S +A A +LP+ D V +Q++KF KGL+
Sbjct: 124 ADILALAARDSVVATRGLTWSVPTGRRDGRVSRAADAGDLPAFFDSVDIQKRKFLTKGLN 183
Query: 199 DHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKV 258
DLV L GAHTIG C RL+NF +TG DP+I+ L Q +ALCP+NG+ R+V
Sbjct: 184 TQDLVALTGAHTIGTAGCAVIRDRLFNFNSTGGPDPSIDATFLPQLRALCPQNGDASRRV 243
Query: 259 SLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEF 318
LD+GS FD S+F N+R+G GVLESDQ+LW D++TQ VQ + G IRGLLGL F EF
Sbjct: 244 GLDTGSVNNFDTSYFSNLRNGRGVLESDQKLWTDASTQVFVQRFLG-IRGLLGLTFGVEF 302
Query: 319 PKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
++M+K+SNIEVKT T GEIRKVCS N
Sbjct: 303 GRSMVKMSNIEVKTGTNGEIRKVCSAIN 330
>gb|AAD37429.2| peroxidase 4 precursor [Phaseolus vulgaris]
Length = 278
Score = 364 bits (934), Expect = 2e-99
Identities = 176/278 (63%), Positives = 218/278 (78%), Gaps = 1/278 (0%)
Query: 51 FYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAERNAL 110
FYSSSCP AE+ V+ST++S+ D T+A L+R+HFHDCFVQGCDGSVLI G++ E+ A
Sbjct: 1 FYSSSCPRAESIVKSTVQSHVKSDSTLAAGLLRMHFHDCFVQGCDGSVLISGANTEKTAF 60
Query: 111 ANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRIS 170
ANL LRGFEV++DAK+QLEA CPGVVSCADILA+AARD+V LS G S+ VPTGRRDGRIS
Sbjct: 61 ANLGLRGFEVVDDAKTQLEAACPGVVSCADILALAARDSVVLSGGLSYQVPTGRRDGRIS 120
Query: 171 SSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTG 230
++ SNLP+P D V VQ+QKF AKGL+ DLVTL+GAHTIG T C+FFS RLYNFT G
Sbjct: 121 QASDVSNLPAPFDSVDVQKQKFTAKGLNTQDLVTLLGAHTIGTTACQFFSNRLYNFTANG 180
Query: 231 NSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLW 290
D +I+ + L Q+LCP+NG+G +V+LD+GS FD+S++ N+R G G+L+SDQ LW
Sbjct: 181 -PDSSIDPSFLPTLQSLCPQNGDGSTRVALDTGSQKLFDLSYYNNLRKGRGILQSDQALW 239
Query: 291 GDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNI 328
D +TQ VVQ Y G IRGLLGLKF EF AM+K+ NI
Sbjct: 240 SDDSTQKVVQRYLGLIRGLLGLKFNVEFGNAMVKMGNI 277
>gb|AAL73112.1| bacterial-induced peroxidase [Gossypium hirsutum]
Length = 327
Score = 360 bits (924), Expect = 3e-98
Identities = 174/315 (55%), Positives = 231/315 (73%), Gaps = 2/315 (0%)
Query: 32 SLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFV 91
+ M+ M + ++ + GFY+ +CP AE+ VRST++S+F +P IAP L+R+HFHDCFV
Sbjct: 15 TFMLAMAAALVQAQGTRVGFYARTCPRAESIVRSTVQSHFRSNPNIAPGLLRMHFHDCFV 74
Query: 92 QGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVY 151
QGCD S+LI G + E+ A N LRG+EVI+DAK+QLEA CPGVVSCADIL +AARD+V+
Sbjct: 75 QGCDASILIDGPNTEKTAPPNRLLRGYEVIDDAKTQLEATCPGVVSCADILTLAARDSVF 134
Query: 152 LSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTI 211
L+ G +W+VPTGRRDGR+S ++ + LP + + Q+QKFAA GL+ DLV LVG HTI
Sbjct: 135 LTRGINWAVPTGRRDGRVSLASDTTILPGFRESIDSQKQKFAAFGLNTQDLVALVGGHTI 194
Query: 212 GQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVS 271
G + C+ FSYRLYNF T G DPTIN A + Q QALCP+NG+G R + LD+GS +FD S
Sbjct: 195 GTSACQLFSYRLYNF-TNGGPDPTINPAFVPQLQALCPQNGDGSRLIDLDTGSGNRFDTS 253
Query: 272 FFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVK 331
FF N+R+ G+LESDQ+LW D +T++ VQ + G RG L F EF ++M+K+SNI VK
Sbjct: 254 FFANLRNVRGILESDQKLWTDPSTRTFVQRFLGE-RGSRPLNFNVEFARSMVKMSNIGVK 312
Query: 332 TDTEGEIRKVCSKFN 346
T T GEIR++CS N
Sbjct: 313 TGTNGEIRRICSAIN 327
>gb|AAB67737.1| cationic peroxidase [Stylosanthes humilis]
Length = 319
Score = 336 bits (862), Expect = 5e-91
Identities = 170/320 (53%), Positives = 223/320 (69%), Gaps = 8/320 (2%)
Query: 28 MLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFH 87
++L + + +++ ++ + GFYSS+CP E+ VRST++S+ N D T+A L+R+HFH
Sbjct: 7 LVLALVSLGVVNSVVHGQGTRVGFYSSTCPGVESIVRSTVQSHLNSDLTLAAGLLRMHFH 66
Query: 88 DCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAAR 147
DCFV GCD S+LI G++ E+ A N+ LRGFEVI+ AK+QLEA CP VVSCADILA+AAR
Sbjct: 67 DCFVHGCDASLLIDGTNTEKTAPPNIGLRGFEVIDHAKTQLEAACPNVVSCADILALAAR 126
Query: 148 DAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVG 207
D+V LS G SW VPTGRRDG +SS+ LP P D V VQ+ KF+A GL+ DLVTLVG
Sbjct: 127 DSVVLSGGASWQVPTGRRDGLVSSAFDV-KLPGPGDSVDVQKHKFSALGLNTKDLVTLVG 185
Query: 208 AHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGN-GLRKVSLDSGSPA 266
HTIG T C+ S RL NF T DPTI+ + L Q +ALCP++G ++V LD+GS
Sbjct: 186 GHTIGTTSCQLLSSRLNNFNGTNGPDPTIDPSFLPQLKALCPQDGGASTKRVPLDNGSQT 245
Query: 267 KFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLS 326
KFD S+F NVR G G+L+SDQ LW D +T+ VQSY+ LG F +F +M+K+
Sbjct: 246 KFDTSYFNNVRRGRGILQSDQALWTDPSTKPFVQSYS------LGSTFNVDFGNSMVKMG 299
Query: 327 NIEVKTDTEGEIRKVCSKFN 346
NI VKT ++GEIRK CS FN
Sbjct: 300 NIGVKTGSDGEIRKKCSAFN 319
>dbj|BAB08889.1| peroxidase [Arabidopsis thaliana] gi|28972995|gb|AAO63822.1|
putative peroxidase [Arabidopsis thaliana]
gi|28393603|gb|AAO42221.1| putative peroxidase
[Arabidopsis thaliana] gi|15241812|ref|NP_198774.1|
peroxidase, putative [Arabidopsis thaliana]
gi|26397795|sp|Q9FKA4|PER62_ARATH Peroxidase 62
precursor (Atperox P62) (ATP24a)
Length = 319
Score = 333 bits (853), Expect = 6e-90
Identities = 172/319 (53%), Positives = 231/319 (71%), Gaps = 11/319 (3%)
Query: 32 SLMILMISFIIAQAQLKT--GFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDC 89
+L+I+ +S +IA T GFYS++CP AE VR+T+ S+F DP +AP L+R+H HDC
Sbjct: 8 ALVIVFLSCLIAVYGQGTRIGFYSTTCPNAETIVRTTVASHFGSDPKVAPGLLRMHNHDC 67
Query: 90 FVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDA 149
FVQGCDGSVL+ G ++ER A AN++L GFEVI+DAK QLEA CPGVVSCADILA+AARD+
Sbjct: 68 FVQGCDGSVLLSGPNSERTAGANVNLHGFEVIDDAKRQLEAACPGVVSCADILALAARDS 127
Query: 150 VYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLV-GA 208
V L+NG SW VPTGRRDGR+S ++ +NLPSP D +++Q++KF+A L+ DLVTLV G
Sbjct: 128 VSLTNGQSWQVPTGRRDGRVSLASNVNNLPSPSDSLAIQQRKFSAFRLNTRDLVTLVGGG 187
Query: 209 HTIGQTECRFFSYRLYNFTTTGN-SDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAK 267
HTIG C F + R++N ++GN +DPT++Q + Q Q LCP+NG+G +V LD+GS
Sbjct: 188 HTIGTAACGFITNRIFN--SSGNTADPTMDQTFVPQLQRLCPQNGDGSARVDLDTGSGNT 245
Query: 268 FDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSN 327
FD S+F N+ G+L+SD LW AT+S+VQ + RG F +F ++M+K+SN
Sbjct: 246 FDTSYFINLSRNRGILQSDHVLWTSPATRSIVQEFMAP-RG----NFNVQFARSMVKMSN 300
Query: 328 IEVKTDTEGEIRKVCSKFN 346
I VKT T GEIR+VCS N
Sbjct: 301 IGVKTGTNGEIRRVCSAVN 319
>gb|AAM64354.1| peroxidase [Arabidopsis thaliana]
Length = 328
Score = 328 bits (842), Expect = 1e-88
Identities = 163/299 (54%), Positives = 212/299 (70%), Gaps = 6/299 (2%)
Query: 48 KTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAER 107
+ GFY ++CP AE VR+ + + F+ DP IAP ++R+HFHDCFVQGCDGS+LI G++ ER
Sbjct: 36 RIGFYLTTCPRAETIVRNAVNAGFSSDPRIAPGILRMHFHDCFVQGCDGSILISGANTER 95
Query: 108 NALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDG 167
A NL+L+GFEVI++AK+QLEA CPGVVSCADILA+AARD V L+ G W VPTGRRDG
Sbjct: 96 TASPNLNLQGFEVIDNAKTQLEAACPGVVSCADILALAARDTVILTQGTGWQVPTGRRDG 155
Query: 168 RISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFT 227
R+S ++ A+NLP P D V+VQ+QKF+A GL+ DLV LVG HTIG C F RL+N T
Sbjct: 156 RVSLASNANNLPGPRDSVAVQQQKFSALGLNTRDLVVLVGGHTIGTAGCGVFRNRLFN-T 214
Query: 228 TTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQ 287
T +DPTI+ LAQ Q CP+NG+G +V LD+GS + +D S++ N+ G GVL+SDQ
Sbjct: 215 TGQTADPTIDPTFLAQLQTQCPQNGDGSVRVDLDTGSGSTWDTSYYNNLSRGRGVLQSDQ 274
Query: 288 RLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
LW D AT+ +VQ F EF ++M+++SNI V T GEIR+VCS N
Sbjct: 275 VLWTDPATRPIVQQLMAP-----RSTFNVEFARSMVRMSNIGVVTGANGEIRRVCSAVN 328
>emb|CAA67551.1| peroxidase [Arabidopsis thaliana] gi|10176960|dbj|BAB10280.1|
peroxidase [Arabidopsis thaliana]
gi|27363238|gb|AAO11538.1| At5g64120/MHJ24_10
[Arabidopsis thaliana] gi|15237615|ref|NP_201217.1|
peroxidase, putative [Arabidopsis thaliana]
gi|16226219|gb|AAL16106.1| AT5g64120/MHJ24_10
[Arabidopsis thaliana] gi|26397643|sp|Q43387|PER71_ARATH
Peroxidase 71 precursor (Atperox P71) (ATP15a) (ATPO2)
Length = 328
Score = 328 bits (840), Expect = 2e-88
Identities = 163/299 (54%), Positives = 212/299 (70%), Gaps = 6/299 (2%)
Query: 48 KTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAER 107
+ GFY ++CP AE VR+ + + F+ DP IAP ++R+HFHDCFVQGCDGS+LI G++ ER
Sbjct: 36 RIGFYLTTCPRAETIVRNAVNAGFSSDPRIAPGILRMHFHDCFVQGCDGSILISGANTER 95
Query: 108 NALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDG 167
A NL+L+GFEVI++AK+QLEA CPGVVSCADILA+AARD V L+ G W VPTGRRDG
Sbjct: 96 TAGPNLNLQGFEVIDNAKTQLEAACPGVVSCADILALAARDTVILTQGTGWQVPTGRRDG 155
Query: 168 RISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFT 227
R+S ++ A+NLP P D V+VQ+QKF+A GL+ DLV LVG HTIG C F RL+N T
Sbjct: 156 RVSLASNANNLPGPRDSVAVQQQKFSALGLNTRDLVVLVGGHTIGTAGCGVFRNRLFN-T 214
Query: 228 TTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQ 287
T +DPTI+ LAQ Q CP+NG+G +V LD+GS + +D S++ N+ G GVL+SDQ
Sbjct: 215 TGQTADPTIDPTFLAQLQTQCPQNGDGSVRVDLDTGSGSTWDTSYYNNLSRGRGVLQSDQ 274
Query: 288 RLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
LW D AT+ +VQ F EF ++M+++SNI V T GEIR+VCS N
Sbjct: 275 VLWTDPATRPIVQQLMAP-----RSTFNVEFARSMVRMSNIGVVTGANGEIRRVCSAVN 328
>gb|AAL93153.1| class III peroxidase [Gossypium hirsutum]
Length = 323
Score = 313 bits (802), Expect = 5e-84
Identities = 158/323 (48%), Positives = 213/323 (65%), Gaps = 1/323 (0%)
Query: 25 MEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRL 84
M+ M L++ + + Q QL+ GFYS++CP AE+ V S + + I P L+RL
Sbjct: 1 MKTMGFVFLLLPFFAIGVVQGQLRVGFYSNTCPDAESIVSSVVRNAAQSISNIPPVLLRL 60
Query: 85 HFHDCFVQGCDGSVLIV-GSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILA 143
HFHDCFV+GCDGS+LI G AER+A + + GFEVIE AK+QLEA CPGVVSCADI+A
Sbjct: 61 HFHDCFVEGCDGSILIENGPKAERHAFGHQGVGGFEVIEQAKAQLEATCPGVVSCADIVA 120
Query: 144 VAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLV 203
+AARDA+ L+NGPS+ VPTGRRDGR+S + A+N+P D + + KF KGL + DLV
Sbjct: 121 LAARDAIALANGPSYEVPTGRRDGRVSDVSLAANMPDVSDSIQQLKAKFLQKGLSEKDLV 180
Query: 204 TLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSG 263
L AHTIG T C F + RLY F+ G SDP I+ L Q Q++CP+NG+ ++ +D G
Sbjct: 181 LLSAAHTIGTTACFFMTKRLYKFSPAGGSDPAISPDFLPQLQSICPQNGDVNVRLPMDRG 240
Query: 264 SPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMI 323
S FD N+R+G VLESD RL+ D T+ VV SY G + + G F +F +++
Sbjct: 241 SERTFDKQILDNIRNGFAVLESDARLYDDETTRMVVDSYFGILTPIFGPSFESDFVDSIV 300
Query: 324 KLSNIEVKTDTEGEIRKVCSKFN 346
K+ I VKT ++GEIR+VC+ FN
Sbjct: 301 KMGQIGVKTGSKGEIRRVCTAFN 323
>gb|AAD37375.1| peroxidase [Glycine max]
Length = 341
Score = 308 bits (788), Expect = 2e-82
Identities = 157/323 (48%), Positives = 219/323 (67%), Gaps = 4/323 (1%)
Query: 28 MLLGSLMILMISFII--AQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLH 85
M L L +L SF++ +++QL+ GFYS++CP ++ +R+ + DP +A L+RLH
Sbjct: 19 MALFVLSLLFFSFLMGSSESQLQVGFYSNTCPQVDSIIRAVVRDAVLSDPNMAAVLLRLH 78
Query: 86 FHDCFVQGCDGSVLIV-GSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAV 144
FHDCF QGCDGS+LI G +ER+A + +RGFEVIE AK+QLE CPG+VSCADI+A+
Sbjct: 79 FHDCFAQGCDGSILIENGPQSERHAFGHQGVRGFEVIERAKAQLEGSCPGLVSCADIVAL 138
Query: 145 AARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVT 204
AARDAV ++NGP++ VPTGRRDG +S+ + A ++P D + + + KF KGL DLV
Sbjct: 139 AARDAVVMANGPAYQVPTGRRDGLVSNLSLADDMPDVSDSIELLKTKFLNKGLTVKDLVL 198
Query: 205 LVGAHTIGQTECRFFSYRLYNFTTTG-NSDPTINQATLAQFQALCPKNGNGLRKVSLDSG 263
L GAHTIG T C F + RLYNF +G SDP I Q L + +A CP+NG+ ++++D G
Sbjct: 199 LSGAHTIGTTACFFMTRRLYNFFPSGEGSDPAIRQNFLPRLKARCPQNGDVNIRLAIDEG 258
Query: 264 SPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMI 323
S KFD++ KN+R+G VLESD RL D AT++V+ SY + G F +F ++++
Sbjct: 259 SEQKFDINILKNIREGFAVLESDARLNDDIATKNVIDSYVSPFSPMFGPSFEADFVESVV 318
Query: 324 KLSNIEVKTDTEGEIRKVCSKFN 346
K+ I VKT GEIR+VCS FN
Sbjct: 319 KMGQIGVKTGFLGEIRRVCSAFN 341
>gi|55701133 TPA: class III peroxidase 133 precursor [Oryza sativa (japonica
cultivar-group)]
Length = 334
Score = 302 bits (774), Expect = 8e-81
Identities = 160/325 (49%), Positives = 212/325 (65%), Gaps = 7/325 (2%)
Query: 27 AMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHF 86
AMLL + +L+ ++ AQLK GFYS SCPTAE+TV S + + + D TI P+L+RL F
Sbjct: 12 AMLLVAA-VLVAGAAVSNAQLKVGFYSKSCPTAESTVASAVRQFADADSTILPALVRLQF 70
Query: 87 HDCFVQGCDGSVLI--VGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAV 144
HDCF +GCDGSVLI VG++AE N + LRG +V++ K QLE+ CPGVVSCADI+ +
Sbjct: 71 HDCFAKGCDGSVLIKGVGNNAEVNNNKHQGLRGLDVVDSIKQQLESECPGVVSCADIVVL 130
Query: 145 AARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVT 204
A+RDA+ + GPS+ VPTGRRDGR SS A LP D + V R KFAA GLDD DLV
Sbjct: 131 ASRDAIAFTGGPSFDVPTGRRDGRTSSLRDADVLPDVKDSIDVLRSKFAANGLDDKDLVL 190
Query: 205 LVGAHTIGQTECRFFSYRLYNFTTTG---NSDPTINQATLAQFQALCPKNGNGLRKVSLD 261
L AHT+G T C F RLYNF G +DP+I +A L++ Q+ C G+ ++ LD
Sbjct: 191 LSAAHTVGTTACFFLQDRLYNFPLAGGGRGADPSIPEAFLSELQSRCAP-GDFNTRLPLD 249
Query: 262 SGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKA 321
GS A+FD S +N+R+G V+ SD L+ +AT VV +Y+ + G F +F A
Sbjct: 250 RGSEAEFDTSILRNIRNGFAVIASDAALYNATATVGVVDTYSSMLSAFFGPYFRQDFADA 309
Query: 322 MIKLSNIEVKTDTEGEIRKVCSKFN 346
M+K+ ++ V T GE+RKVCSKFN
Sbjct: 310 MVKMGSVGVLTGAAGEVRKVCSKFN 334
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 542,125,615
Number of Sequences: 2540612
Number of extensions: 21609460
Number of successful extensions: 54144
Number of sequences better than 10.0: 859
Number of HSP's better than 10.0 without gapping: 720
Number of HSP's successfully gapped in prelim test: 139
Number of HSP's that attempted gapping in prelim test: 51163
Number of HSP's gapped (non-prelim): 903
length of query: 346
length of database: 863,360,394
effective HSP length: 129
effective length of query: 217
effective length of database: 535,621,446
effective search space: 116229853782
effective search space used: 116229853782
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0252a.1


