

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0250.7
(423 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
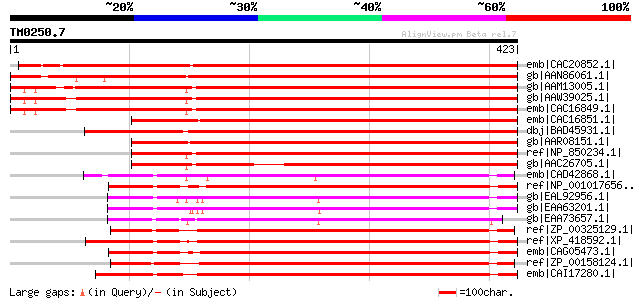
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC20852.1| geranyl diphosphat synthase [Quercus robur] 599 e-170
gb|AAN86061.1| geranylgeranyl diphosphate synthase [Citrus unshiu] 596 e-169
gb|AAM13005.1| putative trans-prenyltransferase [Arabidopsis tha... 533 e-150
gb|AAW39025.1| At2g34630 [Arabidopsis thaliana] 533 e-150
emb|CAC16849.1| geranyl diphosphate synthase [Arabidopsis thaliana] 531 e-149
emb|CAC16851.1| geranyl diphosphate synthase [Citrus sinensis] 528 e-148
dbj|BAD45931.1| putative geranyl diphosphat synthase [Oryza sati... 518 e-145
gb|AAR08151.1| geranyl diphosphate synthase [Vitis vinifera] 511 e-143
ref|NP_850234.1| geranyl diphosphate synthase, putative / GPPS, ... 479 e-134
gb|AAC26705.1| putative trans-prenyltransferase [Arabidopsis tha... 428 e-118
emb|CAD42868.1| solanesyl pyrophosphate synthase [Mucor circinel... 259 8e-68
ref|NP_001017656.1| hypothetical protein LOC550349 [Danio rerio]... 258 2e-67
gb|EAL92956.1| solanesyl pyrophosphate synthase [Aspergillus fum... 258 2e-67
gb|EAA63201.1| hypothetical protein AN2767.2 [Aspergillus nidula... 257 5e-67
gb|EAA73657.1| hypothetical protein FG10933.1 [Gibberella zeae P... 254 3e-66
ref|ZP_00325129.1| COG0142: Geranylgeranyl pyrophosphate synthas... 252 2e-65
ref|XP_418592.1| PREDICTED: similar to trans-prenyltransferase; ... 251 2e-65
emb|CAG05473.1| unnamed protein product [Tetraodon nigroviridis] 251 4e-65
ref|ZP_00158124.1| COG0142: Geranylgeranyl pyrophosphate synthas... 250 7e-65
emb|CAI17280.1| trans-prenyltransferase (TPT) [Homo sapiens] gi|... 248 3e-64
>emb|CAC20852.1| geranyl diphosphat synthase [Quercus robur]
Length = 416
Score = 599 bits (1544), Expect = e-170
Identities = 324/417 (77%), Positives = 356/417 (84%), Gaps = 5/417 (1%)
Query: 8 SRISRNLRGSLNGTRRLLSIQEQNHQSLQSHHYHTPTGSTAKVMRSLA-FSKGLPALHSS 66
SRISR R NG R LS + + Q L Y ST KV+ FS GLPALH
Sbjct: 4 SRISRIRRPGSNGFRWFLS-HKTHLQFLNPPAYSY--SSTHKVLGCREIFSWGLPALHGF 60
Query: 67 RYQIHDQSSSISEEELDPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKR 126
R+ IH QSSSI EE+ DPFSLVADELS +AN LR+MV+ EVPKLAS AEYFFKMGVEGKR
Sbjct: 61 RHNIHHQSSSIVEEQNDPFSLVADELSMVANRLRSMVVTEVPKLASAAEYFFKMGVEGKR 120
Query: 127 FRPTVLLLMSTALNLPIPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLD 186
FRPTVLLLM+TA+N+ I +P G T +LR+RQQRIAEITEMIHVASLLHDDVLD
Sbjct: 121 FRPTVLLLMATAMNISILEPSLRGP-GDALTTELRARQQRIAEITEMIHVASLLHDDVLD 179
Query: 187 DADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETM 246
DADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETM
Sbjct: 180 DADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETM 239
Query: 247 QMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQ 306
QM+TT +QRCSMEYYMQKTYYKTASLISNSCKAIA+L GQT++VAMLA++YGKNLGLA+Q
Sbjct: 240 QMTTTCEQRCSMEYYMQKTYYKTASLISNSCKAIALLGGQTSEVAMLAYEYGKNLGLAYQ 299
Query: 307 LIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIAL 366
LIDDVLDFTGTSASLGKGSLSDIRHGI+TAPILFAMEEFPQLR VVD GFD+P NVD+AL
Sbjct: 300 LIDDVLDFTGTSASLGKGSLSDIRHGIITAPILFAMEEFPQLREVVDRGFDDPANVDVAL 359
Query: 367 EYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITRTK 423
+YLGKSRGIQR RELA KHA++AA AIDSLPES+D +VR SRRAL+DLT+RVITRTK
Sbjct: 360 DYLGKSRGIQRARELAKKHANIAAEAIDSLPESNDEDVRKSRRALLDLTERVITRTK 416
>gb|AAN86061.1| geranylgeranyl diphosphate synthase [Citrus unshiu]
Length = 426
Score = 596 bits (1536), Expect = e-169
Identities = 321/432 (74%), Positives = 365/432 (84%), Gaps = 15/432 (3%)
Query: 1 MLFHRGLSRISRNLRGSLNGTRRLLSIQEQNHQSLQSHHYHTPTGSTAKVMRSL----AF 56
ML +RGLSRISR + + G R L S H L + + ++ L A+
Sbjct: 1 MLIYRGLSRISRISKKTPFGRRWLPS-----HPLLSGASHSAAAAAADSSVKVLGCREAY 55
Query: 57 SKGLPALHSSRYQIHDQSSSI-----SEEELDPFSLVADELSFIANNLRAMVIAEVPKLA 111
S LPALH R+QIH QSSS+ S+E+LDPFSLVADELS +A LR+MV+AEVPKLA
Sbjct: 56 SWSLPALHGIRHQIHHQSSSVIEDTDSQEQLDPFSLVADELSILAKRLRSMVVAEVPKLA 115
Query: 112 SGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKPPPSNDLGGTFTADLRSRQQRIAEIT 171
S AEYFFKMGVEGKRFRPTVLLLM+TALN+ +P+P + + +LR+RQQ IAEIT
Sbjct: 116 SAAEYFFKMGVEGKRFRPTVLLLMATALNVRVPEPL-HDGVEDALATELRTRQQCIAEIT 174
Query: 172 EMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVS 231
EMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVV+
Sbjct: 175 EMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVT 234
Query: 232 LLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVA 291
LLA VVEHLVTGETMQM+T+SDQRCSM+YYMQKTYYKTASLISNSCKAIA+LAGQTA+VA
Sbjct: 235 LLATVVEHLVTGETMQMTTSSDQRCSMDYYMQKTYYKTASLISNSCKAIALLAGQTAEVA 294
Query: 292 MLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRTV 351
+LAFDYGKNLGLA+QLIDDVLDFTGTSASLGKGSLSDI+HGI+TAPILFAMEEFPQLRTV
Sbjct: 295 ILAFDYGKNLGLAYQLIDDVLDFTGTSASLGKGSLSDIQHGIITAPILFAMEEFPQLRTV 354
Query: 352 VDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRAL 411
V++GF++ NVDIALEYLGKSRGIQ+TRELA+KHA+LAAAAIDSLPE++D +VR SRRAL
Sbjct: 355 VEQGFEDSSNVDIALEYLGKSRGIQKTRELAVKHANLAAAAIDSLPENNDEDVRKSRRAL 414
Query: 412 VDLTQRVITRTK 423
+DLT RVITR K
Sbjct: 415 LDLTHRVITRNK 426
>gb|AAM13005.1| putative trans-prenyltransferase [Arabidopsis thaliana]
Length = 423
Score = 533 bits (1374), Expect = e-150
Identities = 289/433 (66%), Positives = 347/433 (79%), Gaps = 20/433 (4%)
Query: 1 MLFHRGLSRIS------RNLRGSLNG--TRRLLSIQEQNHQSLQSHHYHTPTGSTAKVMR 52
MLF R ++RIS R+ GS + R I +Q H S H V R
Sbjct: 1 MLFTRSVARISSKFLRNRSFYGSSQSLASHRFAIIPDQGHSCSDSPH------KLGYVCR 54
Query: 53 SLAFSKGLPALHSSRYQIHDQSSSISEEELDPFSLVADELSFIANNLRAMVIAEVPKLAS 112
+ +S P +Q++ QSSS+ EEELDPFSLVADELS ++N LR MV+AEVPKLAS
Sbjct: 55 T-TYSLKSPVFGGFSHQLYHQSSSLVEEELDPFSLVADELSLLSNKLREMVLAEVPKLAS 113
Query: 113 GAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKP--PPSNDLGGTFTADLRSRQQRIAEI 170
AEYFFK GV+GK+FR T+LLLM+TALN+ +P+ S D+ T++LR RQ+ IAEI
Sbjct: 114 AAEYFFKRGVQGKQFRSTILLLMATALNVRVPEALIGESTDI---VTSELRVRQRGIAEI 170
Query: 171 TEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVV 230
TEMIHVASLLHDDVLDDADTRRG+GSLN VMGNK++VLAGDFLLSRAC ALA+LKNTEVV
Sbjct: 171 TEMIHVASLLHDDVLDDADTRRGVGSLNVVMGNKMSVLAGDFLLSRACGALAALKNTEVV 230
Query: 231 SLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADV 290
+LLA VEHLVTGETM+++++++QR SM+YYMQKTYYKTASLISNSCKA+A+L GQTA+V
Sbjct: 231 ALLATAVEHLVTGETMEITSSTEQRYSMDYYMQKTYYKTASLISNSCKAVAVLTGQTAEV 290
Query: 291 AMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRT 350
A+LAF+YG+NLGLAFQLIDD+LDFTGTSASLGKGSLSDIRHG++TAPILFAMEEFPQLR
Sbjct: 291 AVLAFEYGRNLGLAFQLIDDILDFTGTSASLGKGSLSDIRHGVITAPILFAMEEFPQLRE 350
Query: 351 VVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRA 410
VVD+ +P NVDIALEYLGKS+GIQR RELA++HA+LAAAAI SLPE+D+ +V+ SRRA
Sbjct: 351 VVDQVEKDPRNVDIALEYLGKSKGIQRARELAMEHANLAAAAIGSLPETDNEDVKRSRRA 410
Query: 411 LVDLTQRVITRTK 423
L+DLT RVITR K
Sbjct: 411 LIDLTHRVITRNK 423
>gb|AAW39025.1| At2g34630 [Arabidopsis thaliana]
Length = 422
Score = 533 bits (1373), Expect = e-150
Identities = 288/433 (66%), Positives = 346/433 (79%), Gaps = 21/433 (4%)
Query: 1 MLFHRGLSRIS------RNLRGSLNG--TRRLLSIQEQNHQSLQSHHYHTPTGSTAKVMR 52
MLF R ++RIS R+ GS + R I +Q H S H +T
Sbjct: 1 MLFTRSVARISSKFLRNRSFYGSSQSLASHRFAIIPDQGHSCSDSPHKGYVCRTT----- 55
Query: 53 SLAFSKGLPALHSSRYQIHDQSSSISEEELDPFSLVADELSFIANNLRAMVIAEVPKLAS 112
+S P +Q++ QSSS+ EEELDPFSLVADELS ++N LR MV+AEVPKLAS
Sbjct: 56 ---YSLKSPVFGGFSHQLYHQSSSLVEEELDPFSLVADELSLLSNKLREMVLAEVPKLAS 112
Query: 113 GAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKP--PPSNDLGGTFTADLRSRQQRIAEI 170
AEYFFK GV+GK+FR T+LLLM+TALN+ +P+ S D+ T++LR RQ+ IAEI
Sbjct: 113 AAEYFFKRGVQGKQFRSTILLLMATALNVRVPEALIGESTDI---VTSELRVRQRGIAEI 169
Query: 171 TEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVV 230
TEMIHVASLLHDDVLDDADTRRG+GSLN VMGNK++VLAGDFLLSRAC ALA+LKNTEVV
Sbjct: 170 TEMIHVASLLHDDVLDDADTRRGVGSLNVVMGNKMSVLAGDFLLSRACGALAALKNTEVV 229
Query: 231 SLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADV 290
+LLA VEHLVTGETM+++++++QR SM+YYMQKTYYKTASLISNSCKA+A+L GQTA+V
Sbjct: 230 ALLATAVEHLVTGETMEITSSTEQRYSMDYYMQKTYYKTASLISNSCKAVAVLTGQTAEV 289
Query: 291 AMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRT 350
A+LAF+YG+NLGLAFQLIDD+LDFTGTSASLGKGSLSDIRHG++TAPILFAMEEFPQLR
Sbjct: 290 AVLAFEYGRNLGLAFQLIDDILDFTGTSASLGKGSLSDIRHGVITAPILFAMEEFPQLRE 349
Query: 351 VVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRA 410
VVD+ +P NVDIALEYLGKS+GIQR RELA++HA+LAAAAI SLPE+D+ +V+ SRRA
Sbjct: 350 VVDQVEKDPRNVDIALEYLGKSKGIQRARELAMEHANLAAAAIGSLPETDNEDVKRSRRA 409
Query: 411 LVDLTQRVITRTK 423
L+DLT RVITR K
Sbjct: 410 LIDLTHRVITRNK 422
>emb|CAC16849.1| geranyl diphosphate synthase [Arabidopsis thaliana]
Length = 422
Score = 531 bits (1368), Expect = e-149
Identities = 287/433 (66%), Positives = 346/433 (79%), Gaps = 21/433 (4%)
Query: 1 MLFHRGLSRIS------RNLRGSLNG--TRRLLSIQEQNHQSLQSHHYHTPTGSTAKVMR 52
MLF R ++RIS R+ GS + R I +Q H S H +T
Sbjct: 1 MLFTRSVARISSKFLRNRSFYGSSQSLASHRFAIIPDQGHSCSDSPHKGYVCRTT----- 55
Query: 53 SLAFSKGLPALHSSRYQIHDQSSSISEEELDPFSLVADELSFIANNLRAMVIAEVPKLAS 112
+S P +Q++ QSSS+ EEELDPFSLVADELS ++N LR MV+AEVPKLAS
Sbjct: 56 ---YSLKSPVFGGFSHQLYHQSSSLVEEELDPFSLVADELSLLSNKLREMVLAEVPKLAS 112
Query: 113 GAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKP--PPSNDLGGTFTADLRSRQQRIAEI 170
AEYFFK GV+GK+FR T+LLLM+TAL++ +P+ S D+ T++LR RQ+ IAEI
Sbjct: 113 AAEYFFKRGVQGKQFRSTILLLMATALDVRVPEALIGESTDI---VTSELRVRQRGIAEI 169
Query: 171 TEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVV 230
TEMIHVASLLHDDVLDDADTRRG+GSLN VMGNK++VLAGDFLLSRAC ALA+LKNTEVV
Sbjct: 170 TEMIHVASLLHDDVLDDADTRRGVGSLNVVMGNKMSVLAGDFLLSRACGALAALKNTEVV 229
Query: 231 SLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADV 290
+LLA VEHLVTGETM+++++++QR SM+YYMQKTYYKTASLISNSCKA+A+L GQTA+V
Sbjct: 230 ALLATAVEHLVTGETMEITSSTEQRYSMDYYMQKTYYKTASLISNSCKAVAVLTGQTAEV 289
Query: 291 AMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRT 350
A+LAF+YG+NLGLAFQLIDD+LDFTGTSASLGKGSLSDIRHG++TAPILFAMEEFPQLR
Sbjct: 290 AVLAFEYGRNLGLAFQLIDDILDFTGTSASLGKGSLSDIRHGVITAPILFAMEEFPQLRE 349
Query: 351 VVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRA 410
VVD+ +P NVDIALEYLGKS+GIQR RELA++HA+LAAAAI SLPE+D+ +V+ SRRA
Sbjct: 350 VVDQVEKDPRNVDIALEYLGKSKGIQRARELAMEHANLAAAAIGSLPETDNEDVKRSRRA 409
Query: 411 LVDLTQRVITRTK 423
L+DLT RVITR K
Sbjct: 410 LIDLTHRVITRNK 422
>emb|CAC16851.1| geranyl diphosphate synthase [Citrus sinensis]
Length = 321
Score = 528 bits (1359), Expect = e-148
Identities = 274/322 (85%), Positives = 301/322 (93%), Gaps = 1/322 (0%)
Query: 102 MVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKPPPSNDLGGTFTADLR 161
MVIAEVPKLAS AEYFFKMGVEGKRFRPTVLLLM+TALN+ +P+P + T +LR
Sbjct: 1 MVIAEVPKLASAAEYFFKMGVEGKRFRPTVLLLMATALNVRVPEPLHDGVEDASAT-ELR 59
Query: 162 SRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVAL 221
+RQQ IAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVAL
Sbjct: 60 TRQQCIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVAL 119
Query: 222 ASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIA 281
ASLKNTEVV+LLA VVEHLVTGETMQM+T+SDQRCSM+YYMQKTYYKTASLISNSCKAIA
Sbjct: 120 ASLKNTEVVTLLATVVEHLVTGETMQMTTSSDQRCSMDYYMQKTYYKTASLISNSCKAIA 179
Query: 282 ILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFA 341
+LAGQTA+VA+LAFDYGKNLGLA+QLIDDVLDFTGTSASLGKGSLSDIRHGI+TAPILFA
Sbjct: 180 LLAGQTAEVAILAFDYGKNLGLAYQLIDDVLDFTGTSASLGKGSLSDIRHGIITAPILFA 239
Query: 342 MEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDD 401
MEEFPQLRTVV++GF++ NVDIALEYLGKSRGIQ+TRELA+KHA+LAAAAIDSLPE++D
Sbjct: 240 MEEFPQLRTVVEQGFEDSSNVDIALEYLGKSRGIQKTRELAVKHANLAAAAIDSLPENND 299
Query: 402 GEVRLSRRALVDLTQRVITRTK 423
+V SRRAL+DLT RVITR K
Sbjct: 300 EDVTKSRRALLDLTHRVITRNK 321
>dbj|BAD45931.1| putative geranyl diphosphat synthase [Oryza sativa (japonica
cultivar-group)] gi|52076633|dbj|BAD45534.1| putative
geranyl diphosphate synthase [Oryza sativa (japonica
cultivar-group)]
Length = 430
Score = 518 bits (1333), Expect = e-145
Identities = 267/363 (73%), Positives = 315/363 (86%), Gaps = 5/363 (1%)
Query: 63 LHSSRYQIHDQSSSISEEELDPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGV 122
LH ++YQ+ S SEE+ DPF LVADELS +AN LR+MV AEVPKLAS AEYFFK+G
Sbjct: 71 LHDAQYQVRQDGLSRSEEQQDPFELVADELSLLANRLRSMVAAEVPKLASAAEYFFKVGA 130
Query: 123 EGKRFRPTVLLLMSTALNLPIPKPPPSNDLGG-TFTAD-LRSRQQRIAEITEMIHVASLL 180
EGKRFRPTVLLLM++AL P+ S ++G T A+ LR+RQQ IAEITEMIHVASLL
Sbjct: 131 EGKRFRPTVLLLMASALKFPLSD---STEVGVLTILANKLRTRQQNIAEITEMIHVASLL 187
Query: 181 HDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHL 240
HDDVLDDADTRRG+ SLN +MGNKL+VLAGDFLLSRACVALA+L NTEVVSL+A VEHL
Sbjct: 188 HDDVLDDADTRRGVSSLNCIMGNKLSVLAGDFLLSRACVALAALGNTEVVSLMATAVEHL 247
Query: 241 VTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKN 300
VTGETMQ+ST+ +QR SM+YY+QKTYYKTASLISNSCKA+AILAG TADV+MLA++YG+N
Sbjct: 248 VTGETMQISTSREQRRSMDYYLQKTYYKTASLISNSCKAVAILAGHTADVSMLAYEYGRN 307
Query: 301 LGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPE 360
LGLAFQLIDDVLDFTGTSASLGKGSL+DIRHGI+TAP+L+AMEEFPQL VVD GFDNP
Sbjct: 308 LGLAFQLIDDVLDFTGTSASLGKGSLTDIRHGIITAPMLYAMEEFPQLHEVVDRGFDNPA 367
Query: 361 NVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVIT 420
NV++AL+YL KSRGI++T+ELA +HA+ A AI++LP+SDD +V SRRAL+D+T+RVIT
Sbjct: 368 NVELALDYLQKSRGIEKTKELAREHANRAIKAIEALPDSDDEDVLTSRRALIDITERVIT 427
Query: 421 RTK 423
RTK
Sbjct: 428 RTK 430
>gb|AAR08151.1| geranyl diphosphate synthase [Vitis vinifera]
Length = 321
Score = 511 bits (1316), Expect = e-143
Identities = 263/322 (81%), Positives = 296/322 (91%), Gaps = 1/322 (0%)
Query: 102 MVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKPPPSNDLGGTFTADLR 161
MV+AEVPKLAS AEYFFKMGVEGKR RPTVLLLM+TALN+P+P+P + ++ T + +LR
Sbjct: 1 MVVAEVPKLASAAEYFFKMGVEGKRXRPTVLLLMATALNVPLPRPALA-EVPETLSTELR 59
Query: 162 SRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVAL 221
+RQQ IAEITEMIHVASLLHDDVLDDA+TRRGIGSLN +MGNK+AVLAGDFLLSRACVAL
Sbjct: 60 TRQQCIAEITEMIHVASLLHDDVLDDAETRRGIGSLNIMMGNKVAVLAGDFLLSRACVAL 119
Query: 222 ASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIA 281
ASLKNTEVVSLLA VVEHLVTGETMQM++TS+QR SMEYY+QKTYYKTASLISNSCKAIA
Sbjct: 120 ASLKNTEVVSLLATVVEHLVTGETMQMTSTSEQRVSMEYYLQKTYYKTASLISNSCKAIA 179
Query: 282 ILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFA 341
+LAGQTA+V+MLAF+YGKNLGLAFQLIDD LDFTGTSASLGKGSLSDIRHGI+TAPILFA
Sbjct: 180 LLAGQTAEVSMLAFEYGKNLGLAFQLIDDXLDFTGTSASLGKGSLSDIRHGIITAPILFA 239
Query: 342 MEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDD 401
+EEFPQL VV G DNP ++D+AL+YLG+SRGIQRTRELA+KHA+LAA AIDSLPES D
Sbjct: 240 IEEFPQLDAVVKRGLDNPADIDLALDYLGRSRGIQRTRELAMKHANLAAEAIDSLPESGD 299
Query: 402 GEVRLSRRALVDLTQRVITRTK 423
+V SRRAL+DLT RVITRTK
Sbjct: 300 EDVLRSRRALIDLTHRVITRTK 321
>ref|NP_850234.1| geranyl diphosphate synthase, putative / GPPS, putative /
dimethylallyltransferase, putative / prenyl transferase,
putative [Arabidopsis thaliana]
Length = 321
Score = 479 bits (1234), Expect = e-134
Identities = 246/324 (75%), Positives = 291/324 (88%), Gaps = 5/324 (1%)
Query: 102 MVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKP--PPSNDLGGTFTAD 159
MV+AEVPKLAS AEYFFK GV+GK+FR T+LLLM+TALN+ +P+ S D+ T++
Sbjct: 1 MVLAEVPKLASAAEYFFKRGVQGKQFRSTILLLMATALNVRVPEALIGESTDI---VTSE 57
Query: 160 LRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACV 219
LR RQ+ IAEITEMIHVASLLHDDVLDDADTRRG+GSLN VMGNK++VLAGDFLLSRAC
Sbjct: 58 LRVRQRGIAEITEMIHVASLLHDDVLDDADTRRGVGSLNVVMGNKMSVLAGDFLLSRACG 117
Query: 220 ALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKA 279
ALA+LKNTEVV+LLA VEHLVTGETM+++++++QR SM+YYMQKTYYKTASLISNSCKA
Sbjct: 118 ALAALKNTEVVALLATAVEHLVTGETMEITSSTEQRYSMDYYMQKTYYKTASLISNSCKA 177
Query: 280 IAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPIL 339
+A+L GQTA+VA+LAF+YG+NLGLAFQLIDD+LDFTGTSASLGKGSLSDIRHG++TAPIL
Sbjct: 178 VAVLTGQTAEVAVLAFEYGRNLGLAFQLIDDILDFTGTSASLGKGSLSDIRHGVITAPIL 237
Query: 340 FAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPES 399
FAMEEFPQLR VVD+ +P NVDIALEYLGKS+GIQR RELA++HA+LAAAAI SLPE+
Sbjct: 238 FAMEEFPQLREVVDQVEKDPRNVDIALEYLGKSKGIQRARELAMEHANLAAAAIGSLPET 297
Query: 400 DDGEVRLSRRALVDLTQRVITRTK 423
D+ +V+ SRRAL+DLT RVITR K
Sbjct: 298 DNEDVKRSRRALIDLTHRVITRNK 321
>gb|AAC26705.1| putative trans-prenyltransferase [Arabidopsis thaliana]
gi|25314274|pir||A84759 probable trans-prenyltransferase
[imported] - Arabidopsis thaliana
Length = 297
Score = 428 bits (1101), Expect = e-118
Identities = 226/324 (69%), Positives = 268/324 (81%), Gaps = 29/324 (8%)
Query: 102 MVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKP--PPSNDLGGTFTAD 159
MV+AEVPKLAS AEYFFK GV+GK+FR T+LLLM+TALN+ +P+ S D+ T++
Sbjct: 1 MVLAEVPKLASAAEYFFKRGVQGKQFRSTILLLMATALNVRVPEALIGESTDI---VTSE 57
Query: 160 LRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACV 219
LR RQ+ IAEITEMIHVASLLHDDVLDDADTRRG+GSLN VMGNK
Sbjct: 58 LRVRQRGIAEITEMIHVASLLHDDVLDDADTRRGVGSLNVVMGNK--------------- 102
Query: 220 ALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKA 279
VV+LLA VEHLVTGETM+++++++QR SM+YYMQKTYYKTASLISNSCKA
Sbjct: 103 ---------VVALLATAVEHLVTGETMEITSSTEQRYSMDYYMQKTYYKTASLISNSCKA 153
Query: 280 IAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPIL 339
+A+L GQTA+VA+LAF+YG+NLGLAFQLIDD+LDFTGTSASLGKGSLSDIRHG++TAPIL
Sbjct: 154 VAVLTGQTAEVAVLAFEYGRNLGLAFQLIDDILDFTGTSASLGKGSLSDIRHGVITAPIL 213
Query: 340 FAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPES 399
FAMEEFPQLR VVD+ +P NVDIALEYLGKS+GIQR RELA++HA+LAAAAI SLPE+
Sbjct: 214 FAMEEFPQLREVVDQVEKDPRNVDIALEYLGKSKGIQRARELAMEHANLAAAAIGSLPET 273
Query: 400 DDGEVRLSRRALVDLTQRVITRTK 423
D+ +V+ SRRAL+DLT RVITR K
Sbjct: 274 DNEDVKRSRRALIDLTHRVITRNK 297
>emb|CAD42868.1| solanesyl pyrophosphate synthase [Mucor circinelloides f.
lusitanicus]
Length = 471
Score = 259 bits (663), Expect = 8e-68
Identities = 159/397 (40%), Positives = 235/397 (59%), Gaps = 49/397 (12%)
Query: 62 ALHSSRYQIHDQSSSISEEELDPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMG 121
AL ++ + D I +DP LV D+L I N+ ++ + P L + A ++F
Sbjct: 85 ALSEAQSLVKDSDDRI----IDPAKLVGDDLKEIKANISKLLGSGHPFLNTVARHYFSG- 139
Query: 122 VEGKRFRPTVLLLMSTALNLPIPKP------------PPSNDLGGTFTADLRSR------ 163
EGK RP ++LL++ A ++ K P S+ L T D+ R
Sbjct: 140 -EGKHVRPLLVLLIAQATSIADKKSLASNIDYKSIDTPISHGLKNITTNDVFDRKLHYTP 198
Query: 164 ------------QQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGD 211
Q+R+AEI+EMIH ASLLHDDV+D + TRR + S N GNK+AVL GD
Sbjct: 199 SVTDQGCIILPTQRRLAEISEMIHTASLLHDDVIDASMTRRNLPSANASFGNKMAVLGGD 258
Query: 212 FLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSD-------QRCSMEYYMQK 264
FLL+RA +ALA L+N E + L+A + +LV GE MQ+ T + + + ++YM+K
Sbjct: 259 FLLARASLALARLRNAECIELMATCIANLVEGEFMQLRNTKEGESGKVKKLSTFDHYMEK 318
Query: 265 TYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKG 324
TY KT SLI+ SCKA A+L G T +VA +A+D+GKNLGLAFQL+DD+LDFT T+A LGK
Sbjct: 319 TYMKTGSLIAQSCKASAVLGGSTKEVANIAYDFGKNLGLAFQLVDDMLDFTVTAAELGKP 378
Query: 325 SLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIK 384
+ +D++ G+ TAP+LFA EE+P+L ++ F + + A + + +S G+++T +LA
Sbjct: 379 AGADLKLGLATAPVLFAWEEYPELEPLIKRKFSVKGDEEKARDLVYQSDGLKKTLDLAQI 438
Query: 385 HADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITR 421
H LA A+ LP SD +R ALV +T++++TR
Sbjct: 439 HCKLATDALYKLPASD------ARSALVQITEKLLTR 469
>ref|NP_001017656.1| hypothetical protein LOC550349 [Danio rerio]
gi|62202649|gb|AAH93175.1| Hypothetical protein
LOC550349 [Danio rerio]
Length = 411
Score = 258 bits (660), Expect = 2e-67
Identities = 143/341 (41%), Positives = 220/341 (63%), Gaps = 19/341 (5%)
Query: 83 DPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLP 142
DPFSLV +L I ++++ ++ +L + +Y+F +GK FRP +++LM+ A N+
Sbjct: 90 DPFSLVQKDLQNIYDDIKQQLLVSKAELKALCDYYFDG--KGKAFRPMIVILMARACNVH 147
Query: 143 IPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMG 202
SN G A Q+ IA I+EMIH ASL+HDDV+DD+D RRG ++N V G
Sbjct: 148 ------SNKEGVLLPA-----QRSIAMISEMIHTASLVHDDVIDDSDKRRGKNTINNVWG 196
Query: 203 NKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYM 262
+ A+LAGDF+LS A +ALA + NT VVS+L++V+E LV GE MQ+ + ++ ++Y+
Sbjct: 197 ERKAILAGDFILSAASMALARIGNTTVVSVLSQVIEDLVRGEFMQLGSKENENERFKHYL 256
Query: 263 QKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLG 322
+KT+ KTASLI+NSCKA++IL +V +A+ YG+N+G+AFQL+DD+LDFT + LG
Sbjct: 257 EKTFKKTASLIANSCKAVSILVNSDPEVHEIAYQYGRNVGIAFQLVDDILDFTSNANCLG 316
Query: 323 KGSLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELA 382
K S +D++ G+ T P+LFA ++FP+L +++ F + +VD A +Y+ KS G+++T LA
Sbjct: 317 KPSAADLKLGLATGPVLFACQQFPELHSMIMRRFSSDGDVDRAWQYVLKSDGVEQTNYLA 376
Query: 383 IKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITRTK 423
+ A I L S + R AL+ LT+ V+ R K
Sbjct: 377 QHYCQEAIRQISRLRPSSE------RDALIRLTELVLRRDK 411
>gb|EAL92956.1| solanesyl pyrophosphate synthase [Aspergillus fumigatus Af293]
Length = 450
Score = 258 bits (659), Expect = 2e-67
Identities = 159/382 (41%), Positives = 226/382 (58%), Gaps = 48/382 (12%)
Query: 82 LDPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTA--- 138
+DP +V EL F+ N+R ++ + P L A+Y+ + EGK RP ++LLMS A
Sbjct: 77 VDPLRIVGKELKFLTKNIRQLLGSGHPTLDKVAKYYTRS--EGKHMRPLLVLLMSQATAL 134
Query: 139 --------------LNLPIPKP-------PPSNDLGGT-------FTAD--LRSRQQRIA 168
+N PI P P +N L F D + Q+R+A
Sbjct: 135 TPGKSRPNDVAPISVNDPISSPSILADTNPDTNSLVSESAEAQYDFVGDENILPSQRRLA 194
Query: 169 EITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTE 228
EITE+IH ASLLHDDV+D+A +RR S N GNK+AVLAGDFLL RA VALA L++ E
Sbjct: 195 EITELIHTASLLHDDVIDNAVSRRSSNSANLQFGNKMAVLAGDFLLGRASVALARLRDPE 254
Query: 229 VVSLLAKVVEHLVTGETMQM-STTSDQRC------SMEYYMQKTYYKTASLISNSCKAIA 281
V LLA V+ +LV GE MQ+ +T +D++ ++ YY+QKTY KTASLIS SC+A A
Sbjct: 255 VTELLATVIANLVEGEFMQLKNTVADEKNPVFTDETISYYLQKTYLKTASLISKSCRAAA 314
Query: 282 ILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFA 341
+L G +V A+ YG+NLGLAFQL+DD+LD+T + LGK + +D+ G+ TAP+LFA
Sbjct: 315 LLGGSAPEVVEAAYSYGRNLGLAFQLVDDLLDYTVSGVELGKPAGADLELGLATAPLLFA 374
Query: 342 MEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDD 401
++ P+L +V F +V A E + S GI++TR LA ++AD A AAI P+S+
Sbjct: 375 WKQNPELGPLVGRKFSQEGDVQRAREIVYNSDGIEQTRALAQEYADKAVAAIADFPDSE- 433
Query: 402 GEVRLSRRALVDLTQRVITRTK 423
++ L+ + ++ + R K
Sbjct: 434 -----AKSGLIQMCEKTMNRRK 450
>gb|EAA63201.1| hypothetical protein AN2767.2 [Aspergillus nidulans FGSC A4]
gi|67524619|ref|XP_660371.1| hypothetical protein
AN2767_2 [Aspergillus nidulans FGSC A4]
gi|49090886|ref|XP_406904.1| hypothetical protein
AN2767.2 [Aspergillus nidulans FGSC A4]
Length = 1092
Score = 257 bits (656), Expect = 5e-67
Identities = 158/387 (40%), Positives = 225/387 (57%), Gaps = 53/387 (13%)
Query: 82 LDPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNL 141
+DP +V EL F+ N+R ++ + P L A+Y+ + EGK RP ++LLMS A L
Sbjct: 707 VDPLRIVGKELKFLTKNIRQLLGSGHPTLDKVAKYYTRS--EGKHMRPLLVLLMSQATAL 764
Query: 142 PIPKPPPS-----------ND-------LGGT------------------FTAD--LRSR 163
P+ S ND L T F D +
Sbjct: 765 TTPRSNRSTSDAALSSVTVNDPITSPSVLADTNPDLDRLTSQSPTEGQYDFAGDENILPS 824
Query: 164 QQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALAS 223
Q+R+AEITE+IH ASLLHDDV+D+A TRR S N GNK+AVLAGDFLL RA VALA
Sbjct: 825 QRRLAEITELIHTASLLHDDVIDNALTRRSSNSANIEFGNKMAVLAGDFLLGRASVALAR 884
Query: 224 LKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCS-------MEYYMQKTYYKTASLISNS 276
L++ EV LLA V+ +LV GE MQ+ T++ + + YY+QKTY KTASLIS S
Sbjct: 885 LRDPEVTELLATVIANLVEGEFMQLKNTAEDEKNPVFTDDTISYYLQKTYLKTASLISKS 944
Query: 277 CKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTA 336
C+A A+L T DV A+ YG+NLGLAFQL+DD+LD+T + LGK + +D+ G+ TA
Sbjct: 945 CRAAALLGHSTPDVVEAAYSYGRNLGLAFQLVDDMLDYTVSGVELGKPAGADLELGLATA 1004
Query: 337 PILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSL 396
P+LFA ++ P+L +V F +V+ A E + ++ G+++TR LA ++AD A AAI
Sbjct: 1005 PLLFAWKQHPELGPLVGRKFCREGDVEKARELVYRANGVEQTRALAQEYADKAIAAISPF 1064
Query: 397 PESDDGEVRLSRRALVDLTQRVITRTK 423
P+S+ ++ L+++ ++ + R K
Sbjct: 1065 PDSE------AKAGLIEMCRKTMNRRK 1085
>gb|EAA73657.1| hypothetical protein FG10933.1 [Gibberella zeae PH-1]
gi|46138837|ref|XP_391109.1| hypothetical protein
FG10933.1 [Gibberella zeae PH-1]
Length = 444
Score = 254 bits (650), Expect = 3e-66
Identities = 156/372 (41%), Positives = 218/372 (57%), Gaps = 46/372 (12%)
Query: 82 LDPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNL 141
+DP VA E+ F+ N+R ++ + P L A+Y+ + EGK RP ++LLMS A L
Sbjct: 73 IDPLRSVAKEMKFLTGNIRKLLGSGHPSLDRAAKYYTQ--AEGKHVRPLIVLLMSRATYL 130
Query: 142 PIPKPP---------------------------------PSNDLGGTFTADLRSRQQRIA 168
PK P P ++ +D+ Q+R+A
Sbjct: 131 -CPKTPATVPTVTHRGVDTSLSPAQILADVNPAAHPLSSPEQEIADA-NSDILPSQRRLA 188
Query: 169 EITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTE 228
EI E+IH ASLLHDDV+D + +RRG S N GNK+AVLAGDFLL RA VALA L+N E
Sbjct: 189 EIAELIHTASLLHDDVIDHSVSRRGSPSANLEFGNKMAVLAGDFLLGRASVALARLRNPE 248
Query: 229 VVSLLAKVVEHLVTGETMQMSTTS-DQRC------SMEYYMQKTYYKTASLISNSCKAIA 281
VV LLA V+ +LV GE MQ+ T D+R ++ YY+QKTY KTASLIS SC+A A
Sbjct: 249 VVELLATVIANLVEGEFMQLKNTERDERNPKWSEETVTYYLQKTYLKTASLISKSCRAAA 308
Query: 282 ILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFA 341
+L A A+ YG+NLGLAFQL+DD+LD+T + + LGK + +D+ G+ TAP+LFA
Sbjct: 309 LLGNTDAVTVDAAYSYGRNLGLAFQLVDDLLDYTQSGSDLGKPAGADLELGLATAPLLFA 368
Query: 342 MEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESD- 400
++ P+L +V F +V A E + +S GI++TR LA + D A A+I PES+
Sbjct: 369 WKQMPELGALVGRKFAQEGDVQRARELVLQSDGIEQTRALAQDYVDKAIASIADFPESEA 428
Query: 401 -DGEVRLSRRAL 411
DG + ++ ++L
Sbjct: 429 KDGLIEMAHKSL 440
>ref|ZP_00325129.1| COG0142: Geranylgeranyl pyrophosphate synthase [Trichodesmium
erythraeum IMS101]
Length = 323
Score = 252 bits (643), Expect = 2e-65
Identities = 138/337 (40%), Positives = 218/337 (63%), Gaps = 23/337 (6%)
Query: 85 FSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIP 144
FS V +L + NNL+ +V A P L + AE+ F+ V+GKR RP ++LL+S L
Sbjct: 8 FSPVEADLQLLTNNLKQLVGARHPILYAAAEHLFR--VKGKRIRPAIVLLISRTTML--- 62
Query: 145 KPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNK 204
T ++ + +R+AEITEMIH ASL+HDD++D+++ RRG+ +++ + N+
Sbjct: 63 ------------TQEITPKHRRLAEITEMIHTASLVHDDIVDESEIRRGVPTVHSLFTNR 110
Query: 205 LAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQK 264
+AVLAGDFL +++ LA+L N EVV LL+KV+ L GE Q + S+E Y++K
Sbjct: 111 IAVLAGDFLFAQSSWYLANLDNLEVVKLLSKVIMDLAEGEIQQGLHRFNTDLSIEAYLEK 170
Query: 265 TYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKG 324
+YYKT+SLI+NS KA AI++ ++ + YG+ +GLAFQ++DD+LDFT + SLGK
Sbjct: 171 SYYKTSSLIANSSKAAAIISDVPPEIVQDMYYYGRYIGLAFQIVDDILDFTSATESLGKP 230
Query: 325 SLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIK 384
+ SD++ G +TAP L+A+EE P L +++ +++ A++ + SRGI+R++ELA K
Sbjct: 231 ACSDLKSGNLTAPTLYALEEKPALEKLLERELAQDSDLEEAIQLIKNSRGIKRSQELATK 290
Query: 385 HADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITR 421
+A +A + SLP+ D SR+AL++LT V++R
Sbjct: 291 YAKMAVEHLQSLPDCD------SRQALINLTDYVLSR 321
>ref|XP_418592.1| PREDICTED: similar to trans-prenyltransferase; polyprenyl
pyrophosphate synthetase; 2610203G20Rik; 2700031G06Rik
[Gallus gallus]
Length = 408
Score = 251 bits (642), Expect = 2e-65
Identities = 141/361 (39%), Positives = 226/361 (62%), Gaps = 20/361 (5%)
Query: 64 HSSRYQIHDQSSSISEEEL-DPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGV 122
H+S + S ++S+E+ DPF L +L + +++ ++ +L EY+F
Sbjct: 67 HTSTALMCSCSKTVSDEKYQDPFQLGRKDLKNLYEDIKKELLVSTAELREMCEYYFDG-- 124
Query: 123 EGKRFRPTVLLLMSTALNLPIPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHD 182
+GK FRP +++LM+ A N+ SN + ++++ Q+ +A I EMIH ASL+HD
Sbjct: 125 KGKAFRPMIVVLMARACNIH-----HSN------SREVQASQRSVAIIAEMIHTASLVHD 173
Query: 183 DVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVT 242
DV+DDA++RRG ++N + G + AVLAGDF+LS A +ALA + NT ++S+L +V+E LV
Sbjct: 174 DVIDDANSRRGKMTVNQIWGERKAVLAGDFILSAASLALARIGNTTIISVLTQVIEDLVR 233
Query: 243 GETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLG 302
GE +Q+ + ++ +Y++KT+ KTASLI+NSCKA++IL V +A+ YGKN+G
Sbjct: 234 GEFLQLGSKENENERFAHYLEKTFKKTASLIANSCKAVSILGCPDPKVHEIAYQYGKNVG 293
Query: 303 LAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENV 362
+AFQLIDDVLDFT + LGK + +D++ G+ T P+LFA +FP++ ++ F P +V
Sbjct: 294 IAFQLIDDVLDFTSCADHLGKPAAADLKLGLATGPVLFACRQFPEMNAMIMRRFSKPGDV 353
Query: 363 DIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITRT 422
+ A +Y+ +S G+Q+T LA ++ A I L S + R AL+ LT+ V+ R
Sbjct: 354 ERARKYVLQSDGVQQTTYLAQRYCHEATREISKLRPSPE------REALIQLTEMVLMRD 407
Query: 423 K 423
K
Sbjct: 408 K 408
>emb|CAG05473.1| unnamed protein product [Tetraodon nigroviridis]
Length = 418
Score = 251 bits (640), Expect = 4e-65
Identities = 141/341 (41%), Positives = 215/341 (62%), Gaps = 19/341 (5%)
Query: 83 DPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLP 142
DPF L +L + ++R + +L +Y+F +GK RP +++LM+ ALN+
Sbjct: 97 DPFMLAQKDLKSLYEDIRKELFVSKEELKFLCDYYFDG--KGKAIRPMIVVLMARALNIH 154
Query: 143 IPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMG 202
SN G DL Q+ IA I+EMIH ASL+HDDV+D +D RRG ++N + G
Sbjct: 155 ------SNRSG-----DLLPGQRAIAMISEMIHTASLVHDDVIDGSDQRRGKTTINEIWG 203
Query: 203 NKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYM 262
+ A+LAGDF+LS A +ALA + N VV +L++V+E LV GE MQ+ + +++ ++Y+
Sbjct: 204 ERKAILAGDFILSAASMALARIGNITVVKVLSQVIEDLVRGEFMQLGSKENEKERFKHYL 263
Query: 263 QKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLG 322
+KT+ KTASLI+NSCKA++IL +V +AF YGKN+G+AFQL+DDVLDFT + LG
Sbjct: 264 EKTFKKTASLIANSCKAVSILVNSDPEVHEIAFQYGKNVGIAFQLVDDVLDFTSGANQLG 323
Query: 323 KGSLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELA 382
K S +D++ G+ T P+LFA ++FP+L ++ F + +VD A +Y+ +S G+++T LA
Sbjct: 324 KPSAADLKLGLATGPVLFACQQFPELHAMIMRRFSSKGDVDRAWQYVLQSDGVEQTNYLA 383
Query: 383 IKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITRTK 423
++ A I L S + R AL+ LT+ V+TR K
Sbjct: 384 RRYCQEAIRQISLLRPSAE------RDALIRLTEMVLTRDK 418
>ref|ZP_00158124.1| COG0142: Geranylgeranyl pyrophosphate synthase [Anabaena variabilis
ATCC 29413]
Length = 323
Score = 250 bits (638), Expect = 7e-65
Identities = 137/337 (40%), Positives = 216/337 (63%), Gaps = 23/337 (6%)
Query: 85 FSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIP 144
F+ V +L +A+NL+ +V P L + AE+ F G GKR RP ++LL+S A L
Sbjct: 8 FTPVEADLRILADNLKQLVGNRHPILFAAAEHLF--GAGGKRIRPAIVLLISRATMLE-- 63
Query: 145 KPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNK 204
D+ R +R+AEITEMIH ASL+HDDV+D+++ RRG+ +++ + GN+
Sbjct: 64 -------------QDITPRHRRLAEITEMIHTASLVHDDVVDESEVRRGVPTVHSLFGNR 110
Query: 205 LAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQK 264
+A+LAGDFL +++ LA+L N +VV LL++V+ L TGE Q D S+E Y++K
Sbjct: 111 IAILAGDFLFAQSSWYLANLDNLQVVKLLSEVIMDLATGEIQQGLNRFDASISIENYIEK 170
Query: 265 TYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKG 324
+YYKTASLI+NS KA +L+ + + A + YG++LG+AFQ++DD+LDFT T+ +LGK
Sbjct: 171 SYYKTASLIANSSKAAGLLSEVSPETAEHLYAYGRHLGIAFQIVDDILDFTSTTDTLGKP 230
Query: 325 SLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIK 384
+ SD++ G +TAP+LFA+ E P L +++ F +++ ALE + S+GIQ+ R+LA
Sbjct: 231 AGSDLKSGNLTAPVLFALAEKPYLEVLIEREFAQEGDLEQALELIQNSQGIQKARDLAAH 290
Query: 385 HADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITR 421
H LA + +LP S+ S +AL+++ + ++R
Sbjct: 291 HTKLAIEHLTTLPTSE------SHQALINIAEYTLSR 321
>emb|CAI17280.1| trans-prenyltransferase (TPT) [Homo sapiens]
gi|50659086|ref|NP_055132.2| trans-prenyltransferase
[Homo sapiens]
Length = 415
Score = 248 bits (632), Expect = 3e-64
Identities = 139/352 (39%), Positives = 216/352 (60%), Gaps = 19/352 (5%)
Query: 72 DQSSSISEEELDPFSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTV 131
D + E+ DPF L +L + ++R ++ +L +EY+F +GK FRP +
Sbjct: 83 DSKTHSGEKYTDPFKLGWRDLKGLYEDIRKELLISTSELKEMSEYYFDG--KGKAFRPII 140
Query: 132 LLLMSTALNLPIPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADTR 191
+ LM+ A N+ + +++ Q+ IA I EMIH ASL+HDDV+DDA +R
Sbjct: 141 VALMARACNIHHNN-----------SRHVQASQRAIALIAEMIHTASLVHDDVIDDASSR 189
Query: 192 RGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTT 251
RG ++N + G K AVLAGD +LS A +ALA + NT V+S+L +V+E LV GE +Q+ +
Sbjct: 190 RGKHTVNKIWGEKKAVLAGDLILSAASIALARIGNTTVISILTQVIEDLVRGEFLQLGSK 249
Query: 252 SDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDV 311
++ +Y++KT+ KTASLI+NSCKA+++L V +A+ YGKN+G+AFQLIDDV
Sbjct: 250 ENENERFAHYLEKTFKKTASLIANSCKAVSVLGCPDPVVHEIAYQYGKNVGIAFQLIDDV 309
Query: 312 LDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGK 371
LDFT S +GK + +D++ G+ T P+LFA ++FP++ ++ F P +VD A +Y+ +
Sbjct: 310 LDFTSCSDQMGKPTSADLKLGLATGPVLFACQQFPEMNAMIMRRFSLPGDVDRARQYVLQ 369
Query: 372 SRGIQRTRELAIKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITRTK 423
S G+Q+T LA ++ A I L S + R AL+ L++ V+TR K
Sbjct: 370 SDGVQQTTYLAQQYCHEAIREISKLRPSPE------RDALIQLSEIVLTRDK 415
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.134 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 646,733,080
Number of Sequences: 2540612
Number of extensions: 26079393
Number of successful extensions: 73924
Number of sequences better than 10.0: 979
Number of HSP's better than 10.0 without gapping: 803
Number of HSP's successfully gapped in prelim test: 176
Number of HSP's that attempted gapping in prelim test: 71614
Number of HSP's gapped (non-prelim): 1072
length of query: 423
length of database: 863,360,394
effective HSP length: 131
effective length of query: 292
effective length of database: 530,540,222
effective search space: 154917744824
effective search space used: 154917744824
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0250.7


