

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
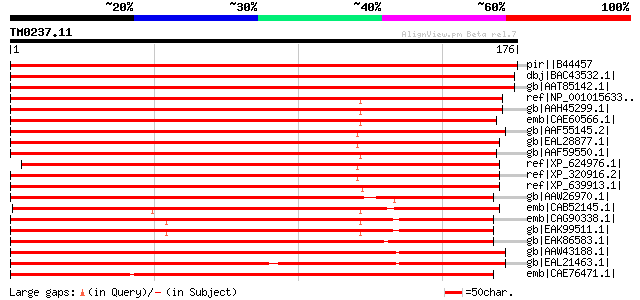
Query= TM0237.11
(176 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||B44457 RNA polymerase II fifth largest subunit - soybean gi... 352 2e-96
dbj|BAC43532.1| putative RNA polymerase II [Arabidopsis thaliana... 334 7e-91
gb|AAT85142.1| putative RNA polymerase II subunit 5 [Oryza sativ... 333 2e-90
ref|NP_001015633.1| polymerase (RNA) II (DNA directed) polypepti... 209 3e-53
gb|AAH45299.1| Polymerase (RNA) II (DNA directed) polypeptide G ... 209 3e-53
emb|CAE60566.1| Hypothetical protein CBG04195 [Caenorhabditis br... 206 2e-52
gb|AAF55145.2| CG31155-PA [Drosophila melanogaster] gi|24647020|... 205 4e-52
gb|EAL28877.1| GA16051-PA [Drosophila pseudoobscura] 204 9e-52
gb|AAF59550.1| Hypothetical protein Y54E10BR.6 [Caenorhabditis e... 203 1e-51
ref|XP_624976.1| PREDICTED: similar to GA16051-PA, partial [Apis... 203 2e-51
ref|XP_320916.2| ENSANGP00000017695 [Anopheles gambiae str. PEST... 198 5e-50
ref|XP_639913.1| RNA polymerase II core subunit [Dictyostelium d... 197 1e-49
gb|AAW26970.1| unknown [Schistosoma japonicum] 185 4e-46
emb|CAB52145.1| SPAPYUK71.02 [Schizosaccharomyces pombe] gi|1911... 181 1e-44
emb|CAG90338.1| unnamed protein product [Debaryomyces hansenii C... 177 1e-43
gb|EAK99511.1| likely RNA Pol II core complex subunit [Candida a... 175 4e-43
gb|EAK86583.1| hypothetical protein UM05334.1 [Ustilago maydis 5... 174 1e-42
gb|AAW43188.1| conserved hypothetical protein [Cryptococcus neof... 171 1e-41
gb|EAL21463.1| hypothetical protein CNBD1580 [Cryptococcus neofo... 160 1e-38
emb|CAE76471.1| probable DNA-directed RNA polymerase II chain Rp... 158 5e-38
>pir||B44457 RNA polymerase II fifth largest subunit - soybean
gi|1173137|sp|P46279|RPB7_SOYBN DNA-directed RNA
polymerase II 19 kDa polypeptide (RNA polymerase II
subunit 5) gi|170052|gb|AAA34005.1| RNA polymerase II
Length = 176
Score = 352 bits (904), Expect = 2e-96
Identities = 171/176 (97%), Positives = 174/176 (98%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFHIVLERNMQLHPRYFGRNLRDNLV+KLMKDVEGTCSGRHGFVVAVTGIEN+GKGLIR
Sbjct: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVSKLMKDVEGTCSGRHGFVVAVTGIENIGKGLIR 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ
Sbjct: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPVAI 176
SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDP +
Sbjct: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPATV 176
>dbj|BAC43532.1| putative RNA polymerase II [Arabidopsis thaliana]
gi|9759239|dbj|BAB09763.1| DNA-directed RNA polymerase
II 19 kD polypeptide (RNA polymerase II subunit 5)
[Arabidopsis thaliana] gi|28973525|gb|AAO64087.1|
putative RNA polymerase II [Arabidopsis thaliana]
gi|15237831|ref|NP_200726.1| DNA-directed RNA polymerase
II [Arabidopsis thaliana] gi|585917|sp|P38421|RPB7_ARATH
DNA-directed RNA polymerase II 19 kDa polypeptide (RNA
polymerase II subunit 5) gi|166854|gb|AAA32861.1| RNA
polymerase II
Length = 176
Score = 334 bits (856), Expect = 7e-91
Identities = 159/175 (90%), Positives = 170/175 (96%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFHIVLERNMQLHPR+FGRNL++NLV+KLMKDVEGTCSGRHGFVVA+TGI+ +GKGLIR
Sbjct: 1 MFFHIVLERNMQLHPRFFGRNLKENLVSKLMKDVEGTCSGRHGFVVAITGIDTIGKGLIR 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
DGTGFVTFPVKYQCVVFRPFKGEILEAVVT+VNKMGFFAEAGPVQIFVS HLIPDDMEFQ
Sbjct: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTLVNKMGFFAEAGPVQIFVSKHLIPDDMEFQ 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPVA 175
+GDMPNYTTSDGSVKIQK+ EVRLKIIGTRVDAT IFC+GTIKDDFLGVINDP A
Sbjct: 121 AGDMPNYTTSDGSVKIQKECEVRLKIIGTRVDATAIFCVGTIKDDFLGVINDPAA 175
>gb|AAT85142.1| putative RNA polymerase II subunit 5 [Oryza sativa (japonica
cultivar-group)]
Length = 177
Score = 333 bits (853), Expect = 2e-90
Identities = 161/175 (92%), Positives = 172/175 (98%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFHIVLERNMQLHPR+FG +LRD LV+KL+KDVEGTCSGRHGFVVA+TG+E+VGKGLIR
Sbjct: 1 MFFHIVLERNMQLHPRHFGPHLRDKLVSKLIKDVEGTCSGRHGFVVAITGVEDVGKGLIR 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
+GTG+VTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ
Sbjct: 61 EGTGYVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPVA 175
SGD+PNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI+DP A
Sbjct: 121 SGDVPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVISDPGA 175
>ref|NP_001015633.1| polymerase (RNA) II (DNA directed) polypeptide G [Bos taurus]
gi|16758826|ref|NP_446400.1| polymerase (RNA) II (DNA
directed) polypeptide G [Rattus norvegicus]
gi|13385826|ref|NP_080605.1| polymerase (RNA) II (DNA
directed) polypeptide G [Mus musculus]
gi|57099729|ref|XP_533260.1| PREDICTED: similar to
polymerase (RNA) II (DNA directed) polypeptide G [Canis
familiaris] gi|38051973|gb|AAH60550.1| Polymerase (RNA)
II (DNA directed) polypeptide G [Rattus norvegicus]
gi|38014576|gb|AAH60595.1| Polymerase (RNA) II (DNA
directed) polypeptide G [Rattus norvegicus]
gi|33243991|gb|AAH55278.1| Polymerase (RNA) II (DNA
directed) polypeptide G [Mus musculus]
gi|37514866|gb|AAH05580.3| Polymerase (RNA) II (DNA
directed) polypeptide G [Mus musculus]
gi|59858369|gb|AAX09019.1| DNA directed RNA polymerase
II polypeptide G [Bos taurus] gi|1360132|emb|CAA96468.1|
RNA polymerase II RPB7 subunit-like protein [Rattus
norvegicus] gi|50403602|sp|P62488|RPB7_MOUSE
DNA-directed RNA polymerase II 19 kDa polypeptide (RPB7)
gi|50403601|sp|P62487|RPB7_HUMAN DNA-directed RNA
polymerase II 19 kDa polypeptide (RPB7)
gi|50403597|sp|P62489|RPB7_RAT DNA-directed RNA
polymerase II 19 kDa polypeptide (RPB7)
gi|1924974|gb|AAB96827.1| RNA polymerase II seventh
subunit [Homo sapiens] gi|929921|gb|AAA86500.1| RNA
polymerase II subunit hsRPB7 gi|12856725|dbj|BAB30760.1|
unnamed protein product [Mus musculus]
gi|12850308|dbj|BAB28670.1| unnamed protein product [Mus
musculus] gi|12847875|dbj|BAB27743.1| unnamed protein
product [Mus musculus] gi|12846312|dbj|BAB27119.1|
unnamed protein product [Mus musculus]
gi|1098305|prf||2115374A RNA polymerase II
gi|4505947|ref|NP_002687.1| DNA directed RNA polymerase
II polypeptide G [Homo sapiens]
Length = 172
Score = 209 bits (532), Expect = 3e-53
Identities = 95/172 (55%), Positives = 131/172 (75%), Gaps = 1/172 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG NL + + KL +VEGTC+G++GFV+AVT I+N+G G+I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPNLLNTVKQKLFTEVEGTCTGKYGFVIAVTTIDNIGAGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PVKY+ +VFRPFKGE+++AVVT VNK+G F E GP+ F+S H IP +MEF
Sbjct: 61 PGRGFVLYPVKYKAIVFRPFKGEVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFD 120
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIN 171
+ + P Y T D + IQ+D E+RLKI+GTRVD +IF IG++ DD+LG+++
Sbjct: 121 PNSNPPCYKTMDEDIVIQQDDEIRLKIVGTRVDKNDIFAIGSLMDDYLGLVS 172
>gb|AAH45299.1| Polymerase (RNA) II (DNA directed) polypeptide G [Danio rerio]
gi|50401534|sp|Q7ZW41|RPB7_BRARE DNA-directed RNA
polymerase II 19 kDa polypeptide (RPB7)
gi|62858849|ref|NP_001017065.1| hypothetical protein
LOC549819 [Xenopus tropicalis]
gi|41054447|ref|NP_955963.1| polymerase (RNA) II (DNA
directed) polypeptide G [Danio rerio]
Length = 172
Score = 209 bits (532), Expect = 3e-53
Identities = 96/172 (55%), Positives = 131/172 (75%), Gaps = 1/172 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG NL + + KL +VEGTC+G++GFV+AVT I+N+G G+I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPNLLNTVKQKLFTEVEGTCTGKYGFVIAVTTIDNIGAGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PVKY+ +VFRPFKGE+++AVVT VNK+G F E GP+ F+S H IP +MEF
Sbjct: 61 PGRGFVLYPVKYKAIVFRPFKGEVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFD 120
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIN 171
+ + P Y T D V IQ+D E+RLKI+GTRVD +IF IG++ DD+LG+++
Sbjct: 121 PNSNPPCYKTVDEDVVIQQDDEIRLKIVGTRVDKNDIFAIGSLMDDYLGLVS 172
>emb|CAE60566.1| Hypothetical protein CBG04195 [Caenorhabditis briggsae]
Length = 197
Score = 206 bits (525), Expect = 2e-52
Identities = 92/170 (54%), Positives = 129/170 (75%), Gaps = 1/170 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFH+ L+ + LHP+YFG NL + + AKL +VEGTC+G++GFV+AVT I+ +G GLI+
Sbjct: 24 MFFHLSLDHEVCLHPKYFGPNLNETIKAKLFNEVEGTCTGKYGFVIAVTTIDTIGHGLIQ 83
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PV+Y+ +VFRPFKG++++ VVT VNK+G F + GP+ F+S H IP DMEF
Sbjct: 84 PGRGFVIYPVRYKAIVFRPFKGQVVDGVVTQVNKVGIFCDIGPLSCFISRHCIPPDMEFD 143
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGV 169
+ + P Y T+D + I+ D E+R+K+IGTRVDA +IF IGT+ DDFLG+
Sbjct: 144 PNSEKPCYKTNDEATVIRNDDEIRVKLIGTRVDANDIFAIGTLMDDFLGL 193
>gb|AAF55145.2| CG31155-PA [Drosophila melanogaster] gi|24647020|ref|NP_731983.1|
CG31155-PA [Drosophila melanogaster]
gi|66772833|gb|AAY55728.1| IP02321p [Drosophila
melanogaster]
Length = 173
Score = 205 bits (522), Expect = 4e-52
Identities = 94/173 (54%), Positives = 132/173 (75%), Gaps = 1/173 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE+ + LHPRYFG L + + KL +VEGTC+G++GFV+AVT I+ +G G+I+
Sbjct: 1 MFYHISLEQEILLHPRYFGPQLLETVKQKLYSEVEGTCTGKYGFVIAVTTIDQIGSGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEF- 119
G GFV +PVKY+ +VFRPFKGE+L+AVV +NK+G FAE GP+ F+S+H IP DM+F
Sbjct: 61 PGQGFVVYPVKYKAIVFRPFKGEVLDAVVKQINKVGMFAEIGPLSCFISHHSIPADMQFC 120
Query: 120 QSGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIND 172
+G+ P Y + D V I + ++RLKI+GTRVDAT IF IGT+ DD+LG++++
Sbjct: 121 PNGNPPCYKSKDEDVVISGEDKIRLKIVGTRVDATGIFAIGTLMDDYLGLVSN 173
>gb|EAL28877.1| GA16051-PA [Drosophila pseudoobscura]
Length = 173
Score = 204 bits (519), Expect = 9e-52
Identities = 94/171 (54%), Positives = 129/171 (74%), Gaps = 1/171 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG L + + KL +VEGTC+G++GFV+AVT I+ +G G+I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPQLLETVKQKLYSEVEGTCTGKYGFVIAVTTIDQIGSGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEF- 119
G GFV +PVKY+ +VFRPFKGE+L+AVV +NK+G FAE GP+ F+S+H IP DM+F
Sbjct: 61 PGQGFVVYPVKYKAIVFRPFKGEVLDAVVKQINKVGMFAEIGPLSCFISHHSIPADMQFC 120
Query: 120 QSGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI 170
+G+ P Y + D V I + ++RLKI+GTRVDAT IF IGT+ DD+LG++
Sbjct: 121 PNGNPPCYKSKDEDVVISGEDKIRLKIVGTRVDATGIFAIGTLMDDYLGLV 171
>gb|AAF59550.1| Hypothetical protein Y54E10BR.6 [Caenorhabditis elegans]
gi|17510349|ref|NP_491093.1| polymerase II (1D600)
[Caenorhabditis elegans]
Length = 197
Score = 203 bits (517), Expect = 1e-51
Identities = 91/170 (53%), Positives = 128/170 (74%), Gaps = 1/170 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFH+ L+ + LHP+YFG NL + + KL +VEGTC+G++GFV+AVT I+ +G GLI+
Sbjct: 24 MFFHLSLDHEVCLHPKYFGPNLNETIKMKLFNEVEGTCTGKYGFVIAVTTIDTIGHGLIQ 83
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PV+Y+ +VFRPFKG++++ VVT VNK+G F + GP+ F+S H IP DMEF
Sbjct: 84 PGRGFVIYPVRYKAIVFRPFKGQVVDGVVTQVNKVGIFCDIGPLSCFISRHCIPPDMEFD 143
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGV 169
+ + P Y T+D + I+ D E+R+K+IGTRVDA +IF IGT+ DDFLG+
Sbjct: 144 PNSEKPCYKTNDEANVIRNDDEIRVKLIGTRVDANDIFAIGTLMDDFLGL 193
>ref|XP_624976.1| PREDICTED: similar to GA16051-PA, partial [Apis mellifera]
Length = 169
Score = 203 bits (516), Expect = 2e-51
Identities = 97/167 (58%), Positives = 127/167 (75%), Gaps = 1/167 (0%)
Query: 5 IVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIRDGTG 64
I LE + LHPRYFG L D + KL +VEGTC+G++GFVVAVT I+N+G G+I+ G G
Sbjct: 1 ISLEHEILLHPRYFGPQLLDTVKQKLYTEVEGTCTGKYGFVVAVTTIDNIGAGIIQPGQG 60
Query: 65 FVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEF-QSGD 123
FV +PVKY+ +VFRPFKGE+L+A+VT VNK+G FAE GP+ F+S+H IP DM+F + +
Sbjct: 61 FVVYPVKYKAIVFRPFKGEVLDAIVTQVNKVGMFAEIGPLSCFISHHSIPADMQFCPNIN 120
Query: 124 MPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI 170
P Y T + V IQ D E+RLKI+GTRVDAT IF IGT+ DD+LG++
Sbjct: 121 PPCYKTKEEDVIIQPDDEIRLKIVGTRVDATGIFAIGTLMDDYLGLV 167
>ref|XP_320916.2| ENSANGP00000017695 [Anopheles gambiae str. PEST]
gi|55233251|gb|EAA00949.2| ENSANGP00000017695 [Anopheles
gambiae str. PEST]
Length = 173
Score = 198 bits (504), Expect = 5e-50
Identities = 93/171 (54%), Positives = 126/171 (73%), Gaps = 1/171 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG L + + KL +VEGTC+G++GFV+AVT I+++G G I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPQLIETVKQKLYTEVEGTCTGKYGFVIAVTTIDDIGSGTIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEF- 119
G GFV +PVKY+ +VFRPFKGE+L+A V VNK+G FAE GP+ F+S+H IP DM+F
Sbjct: 61 PGQGFVVYPVKYKAIVFRPFKGEVLDATVKQVNKVGMFAEIGPLSCFISHHSIPADMQFC 120
Query: 120 QSGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI 170
+G P Y +G I + ++RLKI+GTRVDAT IF IGT+ DD+LG++
Sbjct: 121 PNGAPPCYRAINGESVIAAEDKIRLKIVGTRVDATGIFAIGTLMDDYLGLV 171
>ref|XP_639913.1| RNA polymerase II core subunit [Dictyostelium discoideum]
gi|60466865|gb|EAL64909.1| RNA polymerase II core
subunit [Dictyostelium discoideum]
Length = 172
Score = 197 bits (501), Expect = 1e-49
Identities = 90/171 (52%), Positives = 129/171 (74%), Gaps = 1/171 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFH+ LE+++ +HP++ G NL +L +VEGTC+GR+GF++ +T ++ + KG +
Sbjct: 1 MFFHLTLEKDLHMHPKHCGPNLFTIATQQLYSEVEGTCTGRYGFIITITSVDFLSKGKVL 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
+ +G+V F VKY+ ++F+PFKGE+L+A+VT V +GFFAEAGP+ IFVS LIP DM F
Sbjct: 61 ESSGYVVFNVKYKAIIFKPFKGEVLDAIVTKVTNLGFFAEAGPLSIFVSTQLIPSDMIFD 120
Query: 121 S-GDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI 170
+ +P + + DGS KI KD EVRL+I GTRVDATEIF IG+I++D+LGVI
Sbjct: 121 AQSAVPCFVSEDGSSKISKDDEVRLQIKGTRVDATEIFAIGSIREDYLGVI 171
>gb|AAW26970.1| unknown [Schistosoma japonicum]
Length = 175
Score = 185 bits (470), Expect = 4e-46
Identities = 84/176 (47%), Positives = 127/176 (71%), Gaps = 12/176 (6%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHP+YFG NL + + KL DVEGTCSG++GF++AVT IE++G G+I+
Sbjct: 1 MFYHIKLEHEILLHPKYFGPNLTETVKTKLFSDVEGTCSGKYGFIIAVTNIEHIGAGIIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
GFV + + Y+ +V+RPFKGE+++A+VT V K+G FAEAGP+ IF+S + +P +++F+
Sbjct: 61 PNRGFVQYRIVYRAIVYRPFKGEVVDAIVTQVTKVGVFAEAGPLTIFISKYSVPSNIKFE 120
Query: 121 SGDMPNYTTSDG--------SVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLG 168
+ TT+DG S +Q + +R++IIG RVDA++IF +GT+ DD+LG
Sbjct: 121 GAE----TTTDGLNDDDEEESQTVQIEDRIRIRIIGLRVDASDIFAVGTLMDDYLG 172
>emb|CAB52145.1| SPAPYUK71.02 [Schizosaccharomyces pombe]
gi|19114885|ref|NP_593972.1| hypotDNA-directed RNA
polymerase II subunit [Schizosaccharomyces pombe 972h-]
gi|3334317|sp|O14459|RPB7_SCHPO DNA-directed RNA
polymerase II 19 kDa polypeptide
gi|4091821|gb|AAC99317.1| RNA polymerase II subunit Rpb7
[Schizosaccharomyces pombe] gi|2708528|gb|AAB92516.1|
Rpb7 [Schizosaccharomyces pombe]
gi|2529241|dbj|BAA22802.1| RNA polymerase II subunit
Rpb7 [Schizosaccharomyces pombe]
gi|2529245|dbj|BAA22804.1| RNA polymerase II subunit
Rpb7 [Schizosaccharomyces pombe]
Length = 172
Score = 181 bits (458), Expect = 1e-44
Identities = 89/172 (51%), Positives = 124/172 (71%), Gaps = 5/172 (2%)
Query: 2 FFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAV--TGIENVGKGLI 59
FF L + LHP YFG ++D L AKL+ DVEGTCSG++G+++ V + ++ KG +
Sbjct: 3 FFLKELSLTISLHPSYFGPRMQDYLKAKLLADVEGTCSGQYGYIICVLDSNTIDIDKGRV 62
Query: 60 RDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEF 119
G GF F VKY+ V++RPF+GE+++A+VT VNKMGFFA GP+ +FVS+HL+P DM+F
Sbjct: 63 VPGQGFAEFEVKYRAVLWRPFRGEVVDAIVTTVNKMGFFANIGPLNVFVSSHLVPPDMKF 122
Query: 120 Q-SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI 170
+ + PNY+ D I+K S VRLKI+GTR DATEIF I T+K+D+LGV+
Sbjct: 123 DPTANPPNYSGED--QVIEKGSNVRLKIVGTRTDATEIFAIATMKEDYLGVL 172
>emb|CAG90338.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50426557|ref|XP_461875.1| unnamed protein product
[Debaryomyces hansenii]
Length = 173
Score = 177 bits (448), Expect = 1e-43
Identities = 91/171 (53%), Positives = 119/171 (69%), Gaps = 5/171 (2%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIEN--VGKGL 58
MFF L N+ LHP YFG + L KL+ DVEGTC+G+ G++V V N VGKG
Sbjct: 1 MFFLKDLSLNLTLHPSYFGPQMDQYLRDKLLSDVEGTCTGQFGYIVCVLDCMNIDVGKGR 60
Query: 59 IRDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDME 118
+ G+G F VKY+ VV++PFKGE+++AVVT VNKMGFFA+ GP+ +FVS HLIP DM+
Sbjct: 61 VIPGSGVAEFEVKYRAVVWKPFKGEVVDAVVTTVNKMGFFADVGPLSVFVSTHLIPSDMK 120
Query: 119 FQ-SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLG 168
F S + P Y + D + I+K S VRLKI+GTR D EI+ IG+IK+D+LG
Sbjct: 121 FNPSANPPAYVSPDEN--IEKGSRVRLKIVGTRTDVNEIYAIGSIKEDYLG 169
>gb|EAK99511.1| likely RNA Pol II core complex subunit [Candida albicans SC5314]
gi|46439926|gb|EAK99238.1| likely RNA Pol II core
complex subunit [Candida albicans SC5314]
gi|12655858|gb|AAK00627.1| RNA polymerase II subunit 7
[Candida albicans]
Length = 173
Score = 175 bits (444), Expect = 4e-43
Identities = 91/171 (53%), Positives = 118/171 (68%), Gaps = 5/171 (2%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIEN--VGKGL 58
MFF L N+ LHP +FG + L KL+ DVEGTC+G+ G++V V N VGKG
Sbjct: 1 MFFLKDLSLNLTLHPSFFGPQMDQYLREKLLSDVEGTCTGQFGYIVCVLDSMNIDVGKGR 60
Query: 59 IRDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDME 118
I TG F VKY+ VV++PFKGE+++AVVT VNKMGFFA+ GP+ +FVS HLIP DM+
Sbjct: 61 IIPSTGMAEFEVKYRAVVWKPFKGEVVDAVVTTVNKMGFFADVGPLSVFVSTHLIPSDMK 120
Query: 119 FQ-SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLG 168
F S + P Y + D + I+K S VRLKI+GTR D EI+ IG+IK+D+LG
Sbjct: 121 FNPSANPPAYVSPDEN--IEKGSRVRLKIVGTRTDVNEIYAIGSIKEDYLG 169
>gb|EAK86583.1| hypothetical protein UM05334.1 [Ustilago maydis 521]
gi|49078380|ref|XP_402949.1| hypothetical protein
UM05334.1 [Ustilago maydis 521]
Length = 170
Score = 174 bits (440), Expect = 1e-42
Identities = 86/168 (51%), Positives = 117/168 (69%), Gaps = 1/168 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFF L +LHP YFG ++ + L KL DVEGTC+G+HG+++ V G+ +VG+G +
Sbjct: 1 MFFLKHLTHKTELHPSYFGPSMVEFLNQKLRDDVEGTCTGKHGYIIKVIGLVDVGQGKVI 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G+G F YQ VV +PFKGE+++A +T VNKMGFFAE GP+ IFVS+HLIP + +FQ
Sbjct: 61 PGSGLAEFNSTYQAVVLKPFKGEVMDAKITNVNKMGFFAEVGPLNIFVSSHLIPIEYKFQ 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLG 168
P S S ++ K +VR+KI+GTRVDA EIF IGT+K+D+LG
Sbjct: 121 PESNPPEFVS-ASDRLVKGRKVRIKIVGTRVDANEIFAIGTMKEDYLG 167
>gb|AAW43188.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58266678|ref|XP_570495.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 172
Score = 171 bits (432), Expect = 1e-41
Identities = 85/172 (49%), Positives = 115/172 (66%), Gaps = 1/172 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFF L + LHP YFG L D L KL +DVEGTCSG+HG++++V I ++G+G I
Sbjct: 1 MFFLRELTHTILLHPSYFGAQLEDYLRQKLYEDVEGTCSGKHGYIISVITITDIGEGKII 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
TG F +Y +V +PFKGE+++A V VNKMGFFA GP+Q+FVS HL DM+F
Sbjct: 61 PSTGQAKFKTRYTAIVMKPFKGEVVDAKVVNVNKMGFFAMVGPLQVFVSCHLTHSDMKFD 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIND 172
P S+ + IQKD++VR++I+G RV+A ++F IGTIK D+LG I D
Sbjct: 121 PSVSPPCYRSNDEI-IQKDTKVRIQIVGCRVEANDMFAIGTIKKDYLGQIRD 171
>gb|EAL21463.1| hypothetical protein CNBD1580 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 169
Score = 160 bits (405), Expect = 1e-38
Identities = 82/172 (47%), Positives = 112/172 (64%), Gaps = 4/172 (2%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFF L + LHP YFG L D L KL +DVEGTCSG+HG++++V I ++G+G I
Sbjct: 1 MFFLRELTHTILLHPSYFGAQLEDYLRQKLYEDVEGTCSGKHGYIISVITITDIGEGKII 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
TG F +Y +V +PFKGE+++A V MGFFA GP+Q+FVS HL DM+F
Sbjct: 61 PSTGQAKFKTRYTAIVMKPFKGEVVDAKVV---NMGFFAMVGPLQVFVSCHLTHSDMKFD 117
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIND 172
P S+ + IQKD++VR++I+G RV+A ++F IGTIK D+LG I D
Sbjct: 118 PSVSPPCYRSNDEI-IQKDTKVRIQIVGCRVEANDMFAIGTIKKDYLGQIRD 168
>emb|CAE76471.1| probable DNA-directed RNA polymerase II chain Rpb7 [Neurospora
crassa]
Length = 167
Score = 158 bits (400), Expect = 5e-38
Identities = 76/168 (45%), Positives = 113/168 (67%), Gaps = 1/168 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFF LER + LHP YFGRN+ + + KL+KDVEGTC+G + +++A+ +V +G I
Sbjct: 1 MFFLHNLERRVTLHPSYFGRNMHELVTTKLVKDVEGTCAGDY-YIIAIMDAFDVSEGRIL 59
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G G F VKY+ VV+RPFKGE+++A+V +N GFF +AGP+ IFVS+HL+P+++ F
Sbjct: 60 PGNGLAEFTVKYRAVVWRPFKGEVVDAIVFSINPHGFFCQAGPLSIFVSSHLMPEEIHFD 119
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLG 168
P T++ + I+ + VR+KI G R + E++ IG+I DFLG
Sbjct: 120 PNATPPQFTNNADMVIEPGTHVRVKIGGLRTELGEMYAIGSINGDFLG 167
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 297,556,242
Number of Sequences: 2540612
Number of extensions: 11615128
Number of successful extensions: 27304
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 27149
Number of HSP's gapped (non-prelim): 126
length of query: 176
length of database: 863,360,394
effective HSP length: 119
effective length of query: 57
effective length of database: 561,027,566
effective search space: 31978571262
effective search space used: 31978571262
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0237.11


